Abstract
Previously developed methods for noninvasive PCR diagnosis of visceral leishmaniasis (VL) have significant limitations. Diagnosis of VL using PCR and buccal swabs was evaluated in 307 subjects, including 148 patients confirmed to have VL. This method is simple and well tolerated and has good potential for development, showing 83% sensitivity with 90.56% specificity in control groups.
TEXT
Visceral leishmaniasis (VL) is a fatal disease in the absence of treatment. The definitive diagnosis of VL is the demonstration of amastigotes (Giemsa-stained slides) in splenic smears (sensitivity of 95% to 98%) or bone marrow aspirates (sensitivity of 53% to 95%) (2, 3, 29, 30). Serology-based tests, such as direct agglutination test, rk39 enzyme-linked immunosorbent assay (ELISA), and dipstick, have high sensitivities and specificities (24, 25) but cannot discriminate between past and current infections. The molecular diagnosis of VL by PCR on blood samples and PCR is accurate, but it is invasive and requires phlebotomists, which may not be feasible in large epidemiologic studies. Also, subjects are often reluctant to provide blood samples, which reduces participation rates. Therefore, efforts have been made to develop noninvasive, simpler, and sensitive techniques by using conjunctival swabs or urine samples (13, 20, 23). In canine visceral leishmaniasis, conjunctival swabs have shown 90 to 92% sensitivity (23), but with humans, this method is not well tolerated and is associated with the possibility of corneal damage. Exfoliated buccal epithelial cells are a promising alternative source of genomic DNA (4–7, 9, 10, 14, 16–18, 27, 28), as they have a high yield (6, 14).
We evaluated buccal swabs from four groups of subjects: (i) 148 patients parasitologically confirmed to have VL were recruited from the Kala-azar Medical Research Center, Muzaffarpur, Bihar, India; (ii) 159 controls, including 92 healthy subjects from a region where leishmaniasis is endemic; (iii) 39 healthy subjects; and (iv) 28 patients suffering from other infectious diseases from regions of Varanasi, India, where leishmaniasis is not endemic. All subjects were subjected to thorough clinical examinations for the presence of signs, symptoms, and laboratory indicators (pancytopenia and hypergammaglobulinemia) of VL before inclusion in the study. Ethical clearance for the study was obtained from the Institute of Medical Sciences, Banaras Hindu University, and written informed consent was obtained from the subjects. Buccal swabs were collected by Hi-Media sterile transport viscose swab sticks and kept in 2 ml transport buffer (10 mM Tris base, 10 mM EDTA, 0.5% sodium Sarkosyl; Merck) and stored at 4°C. DNA was isolated from these samples within 48 h of collection.
The parasitic DNA was used as a positive control using promastigotes, cultured as given in reference 15. A buccal swab from a healthy individual was seeded with decreasing concentrations of parasites, quantified using a hemocytometer (Rohem, India), from 1 × 105 parasites to a single parasite per ml in 10-fold dilutions. DNA isolation from parasites, spiked samples, and clinical buccal swab samples was done using the QIAamp DNA minikit (Qiagen) per the manufacturer's protocol.
Primers were designed using Vector NTI version 9 (Invitrogen). The 18S rRNA gene sequence from Leishmania donovani (GenBank accession no. X07773) was retrieved from PubMed. The specificity of the primers was examined using the BLAST program at the NCBI site. In silico PCR was performed using the UCSC genome browser (http://genome.ucsc.edu/) to ensure that the primers did not show any cross-reactivity with human sequences. The forward primer BHUleiF (5′GTTTGTTCCTGGTCGTCCCGT3′) and reverse primer BHUleiR (5′AAGACGAACTACAGCGAAGGC3′) amplify a region of 490 bp and were synthesized by Metabion International AG (Germany).
The PCR was performed in a final reaction mixture volume of 25 μl containing 1 U Taq DNA polymerase (Promega), 1.5 mM MgCl2 (Promega), 10 mM deoxynucleoside triphosphates (dNTPs) (NEB), 10 pmol forward and reverse primers (each), 5× buffer (Promega), and 3 μl (50 to 150 ng) of DNA template. Multiple negative controls with no template were used to detect any plausible contamination. The temperature conditions for PCR were as follows: 35 cycles, with 1 cycle consisting of denaturation at 95°C for 5 min, annealing at 60°C for 45 s, extension at 72°C for 1 min, and final extension for 5 min at 72°C. The PCR product was digested with restriction enzymes, gel purified, and sequenced (22).
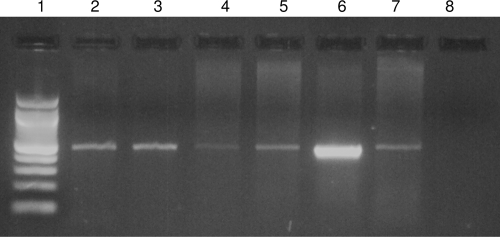
The sensitivity of this assay was 83.11% (Fig. 1), and the specificity varied according to the subject control group (Table 1). The lower detection limit of the developed assay determined by serially diluting parasitic DNA was 0.1 femtogram. The sensitivity of spiked samples was up to 1 parasite. Restriction digestion profiles and sequencing results (data not shown) confirm that the region amplified by BHUleiF/R primers in buccal swab samples is 100% homologous with the small-subunit (SSU) rRNA gene of L. donovani (GenBank accession no. X07773). Microscopy of buccal smears showed no evidence of whole amastigotes, and this was further confirmed by no growth of promastigotes in in vitro culture from buccal cells.
Fig. 1.
PCR assay of buccal swabs from patients confirmed to have VL. Amplified products of 490 bp in buccal swabs from patients confirmed to have VL (lanes 2, 3, 4, 5, and 7). Lane 6, positive control; lane 8, negative control; lane 1, 100-bp DNA ladder.
Table 1.
Comparative sensitivity and specificity of buccal swab samples from patients confirmed to have VL and from different control groups
| Subjects | No. of patients with the following result by PCR: |
Sensitivity (%) | Specificity (%) | 95% CIa (%) | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Healthy controls from areas where leishmaniasis is not endemic (n = 39) | 39 | 0 | 100 | 95.00–100 | |
| Healthy controls from areas where leishmaniasis is endemic (n = 92)b | 79 | 13 | 86.00 | 77.31–91.55 | |
| Subjects with a different disease (n = 28) | 26 | 2 | 92.85 | 77.00–98.00 | |
| Patients confirmed to have VL (n = 148) | 25 | 123 | 83.11 | 76.25–88.29 | |
95% CI, 95% confidence interval.
For 42 healthy controls from areas where leishmaniasis is endemic, PCR was also performed on blood samples, and there was complete agreement in results obtained from buccal swabs.
In this study, we introduce a noninvasive method for molecular diagnosis of VL using buccal swabs. PCR using more-intrusive sampling methods, such as the use of whole-blood or buffy coat samples or blood spots on filter paper, have been described innumerable times with variable reported sensitivities and specificities (11, 12, 19, 21, 26). Noninvasive methods of obtaining biospecimens are very useful for diagnostics and clinical trials, as well as epidemiological screening studies. Among different noninvasive sampling techniques, buccal cells have the advantages of ease of collection and storage. It is possible to obtain sensitivities of >90% using noninvasive molecular diagnosis as shown by a study of canine VL in Israel using conjunctival swabs (23).
In this initial study of PCR and buccal swabs, we obtained encouraging results. The specificity for controls from areas where leishmaniasis is endemic (86%) is comparable to that of PCR on whole-blood samples in a large sample size over a period of 2 years (unpublished data). The lower specificity for controls from areas where leishmaniasis is endemic compared to the specificity for controls from areas where leishmaniasis is not endemic may represent subclinical infections, as it is a well-established fact that only 5 to 10% of infected people develop VL, while the rest remain asymptomatic (1). This is an ongoing challenge in VL diagnosis.
This assay is thought to be amplifying nucleotide fragments rather than whole amastigotes. Nucleotide fragments have been previously reported in different biological fluids and tissues (8). The presence of L. donovani nucleotides in buccal epithelia has been confirmed by restriction digestion and sequencing. The primers used are exclusively specific for Leishmania and do not amplify other species of microorganisms like Crithidia, Streptococcus, and Porphyromonas gingivalis which are commonly harbored in the mouth cavity.
To our knowledge, this is the first report of demonstration of Leishmania DNA in buccal swabs from VL patients. The present data emphasize the potential of this noninvasive sampling method for diagnosis. Further work is required to improve the sensitivity to an acceptable >90% level, which may be achieved by applying nested PCR and more-sensitive primers.
Acknowledgments
This study was funded by Tropical Medicine Research Center grant P50AI074321 from the National Institute of Allergy and Infectious Diseases (NIAID), DMID funding mechanism. S.M. received financial support from the Council of Scientific and Industrial Research (CSIR), New Delhi, India.
We thank Rajeev Raman, Cytogenetics Laboratory, Department of Zoology, Banaras Hindu University, Varanasi, India, for allowing us to use the Genetic Analyzer facility for sequencing.
Footnotes
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Boelaert M., Dujardin J. C. 1999. Diagnostic PCR with Leishmania donovani specificity. Trop. Med. Int. Health 4:789. [DOI] [PubMed] [Google Scholar]
- 2. Boelaert M., et al. 2008. Diagnostic tests for kala-azar: a multi-centre study of the freeze-dried DAT, rK39 strip test and KAtex in East Africa and the Indian subcontinent. Trans. R. Soc. Trop. Med. Hyg. 102:32–40 [DOI] [PubMed] [Google Scholar]
- 3. Boelaert M., et al. 1999. Operational validation of the direct agglutination test for diagnosis of visceral leishmaniasis. Am. J. Trop. Med. Hyg. 60:129–134 [DOI] [PubMed] [Google Scholar]
- 4. Cozier Y. C., Palmer J. R., Rosenberg L. 2004. Comparison of methods for collection of DNA samples by mail in the Black Women's Health Study. Ann. Epidemiol. 14:117–122 [DOI] [PubMed] [Google Scholar]
- 5. Etter J. F., Neidhart E., Bertrand S., Malafosse A., Bertrand D. 2005. Collecting saliva by mail for genetic and cotinine analyses in participants recruited through the Internet. Eur. J. Epidemiol. 20:833–838 [DOI] [PubMed] [Google Scholar]
- 6. Feigelson H. S., et al. 2001. Determinants of DNA yield and quality from buccal cell samples collected with mouthwash. Cancer Epidemiol. Biomarkers Prev. 10:1005–1008 [PubMed] [Google Scholar]
- 7. Garcia-Closas M., et al. 2001. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol. Biomarkers Prev. 10:687–696 [PubMed] [Google Scholar]
- 8. Hansen T. V., Simonsen M. K., Nielsen F. C., Hundrup Y. A. 2007. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol. Biomarkers Prev. 16:2072–2076 [DOI] [PubMed] [Google Scholar]
- 9. Harty L. C., et al. 2000. Collection of buccal cell DNA using treated cards. Cancer Epidemiol. Biomarkers Prev. 9:501–506 [PubMed] [Google Scholar]
- 10. King I. B., et al. 2002. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol. Biomarkers Prev. 11:1130–1133 [PubMed] [Google Scholar]
- 11. Lachaud L., et al. 2001. Comparison of various sample preparation methods for PCR diagnosis of visceral leishmaniasis using peripheral blood. J. Clin. Microbiol. 39:613–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lachaud L., et al. 2000. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral leishmaniasis, with special reference to AIDS patients. J. Clin. Microbiol. 38:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leite R. S., Ferreira S. D., Ituassu L. T., de Melo M. N., de Andrade A. S. 2010. PCR diagnosis of visceral leishmaniasis in asymptomatic dogs using conjunctival swab samples. Vet. Parasitol. 170:201–206 [DOI] [PubMed] [Google Scholar]
- 14. Le Marchand L., et al. 2001. Feasibility of collecting buccal cell DNA by mail in a cohort study. Cancer Epidemiol. Biomarkers Prev. 10:701–703 [PubMed] [Google Scholar]
- 15. Maurya R., et al. 2010. Evaluation of blood agar microtiter plates for culturing Leishmania parasites to titrate parasite burden in spleen and peripheral blood of patients with visceral leishmaniasis. J. Clin. Microbiol. 48:1932–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Milne E., et al. 2006. Buccal DNA collection: comparison of buccal swabs with FTA cards. Cancer Epidemiol. Biomarkers Prev. 15:816–819 [DOI] [PubMed] [Google Scholar]
- 17. Mulot C., Stucker I., Clavel J., Beaune P., Loriot M. A. 2005. Collection of human genomic DNA from buccal cells for genetics studies: comparison between cytobrush, mouthwash, and treated card. J. Biomed. Biotechnol. 2005:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ng D. P., Koh D., Choo S. G., Ng V., Fu Q. 2004. Effect of storage conditions on the extraction of PCR-quality genomic DNA from saliva. Clin. Chim. Acta 343:191–194 [DOI] [PubMed] [Google Scholar]
- 19. Pereira Ede F., et al. 2008. Molecular diagnosis of leishmaniasis in the Parana state of southern Brazil. Exp. Dermatol. 17:1024–1030 [DOI] [PubMed] [Google Scholar]
- 20. Pilatti M. M., de Almeida Ferreira S., de Melo M. N., Ribeiro de Andrade A. S. 2009. Comparison of PCR methods for diagnosis of canine visceral leishmaniasis in conjunctival swab samples. Res. Vet. Sci. 87:255–257 [DOI] [PubMed] [Google Scholar]
- 21. Reithinger R., Dujardin J. C. 2007. Molecular diagnosis of leishmaniasis: current status and future applications. J. Clin. Microbiol. 45:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Srivastava P., et al. 2010. Detection of Leptomonas sp. parasites in clinical isolates of kala-azar patients from India. Infect. Genet. Evol. 10:1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strauss-Ayali D., Jaffe C. L., Burshtain O., Gonen L., Baneth G. 2004. Polymerase chain reaction using noninvasively obtained samples, for the detection of Leishmania infantum DNA in dogs. J. Infect. Dis. 189:1729–1733 [DOI] [PubMed] [Google Scholar]
- 24. Sundar S., Rai M. 2002. Advances in the treatment of leishmaniasis. Curr. Opin. Infect. Dis. 15:593–598 [DOI] [PubMed] [Google Scholar]
- 25. Sundar S., Rai M. 2002. Laboratory diagnosis of visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 9:951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tavares C. A., Fernandes A. P., Melo M. N. 2003. Molecular diagnosis of leishmaniasis. Expert Rev. Mol. Diagn. 3:657–667 [DOI] [PubMed] [Google Scholar]
- 27. Thoni G. J., et al. 2006. Quality of life in HIV-infected children and adolescents under highly active antiretroviral therapy: change over time, effects of age and familial context. Arch. Pediatr. 13:130–139 (In French.) [DOI] [PubMed] [Google Scholar]
- 28. Vidal-Taboada J. M., Cucala M., Mas Herrero S., Lafuente A., Cobos A. 2006. Satisfaction survey with DNA cards method to collect genetic samples for pharmacogenetics studies. BMC Med. Genet. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zijlstra E. E. 1992. The treatment of kala-azar: old and new options. Trop. Geogr. Med. 44:288. [PubMed] [Google Scholar]
- 30. Zijlstra E. E., et al. 1992. Kala-azar: a comparative study of parasitological methods and the direct agglutination test in diagnosis. Trans. R. Soc. Trop. Med. Hyg. 86:505–507 [DOI] [PubMed] [Google Scholar]