Abstract
Carbapenem resistance in Bacteroides fragilis is associated with cfiA-encoded class B metallo-beta-lactamase. cfiA-negative and cfiA-positive isolates belong to genotypically distinct groups. Of a total of 248 B. fragilis isolates included in this study, 214 were susceptible, 10 were intermediate, and 24 were resistant to meropenem. We show that matrix-assisted laser desorption ionization–time of flight mass spectrometry is able to differentiate between cfiA-negative and cfiA-positive isolates and predict carbapenem resistance in a routine laboratory setting.
TEXT
Bacteroides fragilis is a strictly anaerobic Gram-negative bacillus present in the human gut. It is recovered from various infection sites and frequently associated with abscess formation and sepsis. Carbapenem resistance in B. fragilis is emerging and associated with cfiA-encoded class B metallo-beta-lactamase, which is activated by insertion sequence (IS) elements. However, elevated MICs or resistance was also observed in strains that did not have activating IS elements in the upstream region of cfiA (17). In the 2003-2005 Belgian multicenter survey (18), the prevalence of the cfiA resistance gene was 7.4% (10/135) (19), which is high compared with the prevalence of cfiA positivity (2 to 7%) in other countries (5, 6, 13, 15, 20). In the survey, 4% (6/135) of B. fragilis isolates were intermediate or resistant to meropenem according to CLSI breakpoints (18).
By using various molecular typing methods, such as arbitrarily primed PCR, ribotyping, multilocus enzyme electrophoresis, and sequencing of recA and glnA genes, cfiA-negative and cfiA-positive strains were shown to belong to two genotypically distinct groups (9, 10, 13). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has the capacity to discriminate closely related species based on the spectrum of constantly expressed highly abundant proteins, such as ribosomal proteins. This technique was recently introduced as a rapid and accurate method for the identification of bacteria (2, 7, 14) and can also be applied in the identification of Bacteroides isolates (12). The aim of the present study was to examine the discriminatory power of MALDI-TOF MS to differentiate cfiA-positive from cfiA-negative B. fragilis strains in order to predict carbapenem susceptibility.
The 135 B. fragilis clinical isolates from the survey mentioned above (18, 19), originating from nine Belgian hospitals, as well as 113 B. fragilis isolates from routine cultures collected at our laboratory since 2005 were analyzed. Identification was performed by appropriate biochemical and enzymatic tests (11) and confirmed by MALDI-TOF MS. Meropenem susceptibility was determined by Etest methodology (bioMérieux, Marcy l'Etoile, France). The CLSI breakpoints for susceptible and resistant strains are ≤4 mg/liter and ≥16 mg/liter, respectively (4). PCR analysis was performed to detect the presence of the cfiA gene. The annealing temperature of the cfiA gene detection method described by Sóki et al. (16) was increased to 62°C to avoid aspecific reactions. In cfiA-positive strains, the cfiA promoter region was amplified. A PCR product of more than 0.4 kb was considered indicative of the presence of an IS element and was identified by sequencing (3, 20).
Susceptibility to meropenem and genotypic characteristics of the studied isolates are represented in Table 1. Out of 248 B. fragilis isolates included in the study, 214 were susceptible, 10 were intermediate, and 24 were resistant to meropenem. Although previous studies reported that the cfiA gene is not always activated, our 41 cfiA-positive isolates had high MICs of meropenem, ranging from 2 to >32 mg/liter (MIC90, ≥32 mg/liter), while the MICs for cfiA-negative isolates ranged from 0.064 to 4 mg/liter (MIC90, 0.5 mg/liter). Using CLSI breakpoints, only 7 of 41 cfiA-positive isolates were classified as susceptible, while all cfiA-negative isolates were susceptible. IS elements were detected only in 5 isolates, all with a MIC of ≥32 mg/liter, while 28 of 34 meropenem-intermediate or -resistant isolates lacked IS elements in the cfiA promoter region. In another study, this was the case for 19 of 25 isolates, and alternative mechanisms of activation, such as cfiA gene activation by its own promoter, were suggested (17).
Table 1.
Meropenem susceptibility and genotypic characteristics of the 248 isolates studieda
| MIC (mg/liter) | No. of isolates that were cfiA PCR |
Length of cfiA promoter (kb) | Insertion sequence identification | |
|---|---|---|---|---|
| Negative (n = 207) | Positive (n = 41) | |||
| ≤0.125 | 146 | 0 | ND | |
| 0.25 | 30 | 0 | ND | |
| 0.5 | 21 | 0 | ND | |
| 1 | 3 | 0 | ND | |
| 2 | 4 | 2 | For cfiA-negative isolates, ND; for cfiA-positive isolates, 0.4 kb | |
| 4 | 3 | 5 | For cfiA-negative isolates, ND; for cfiA-positive isolates, 0.4 kb | |
| 8 | 0 | 10 | 0.4 kb in all isolates | |
| 16 | 0 | 9 | 0.4 kb in all isolates | |
| ≥32 | 0 | 15 | 0.4 kb in 9 isolates | |
| 1.5-2 kb in 5 isolates | IS612B like (1 isolate) | |||
| IS1187 like (2 isolates) | ||||
| IS1169 like (2 isolates) | ||||
| ND (we failed to amplify a cfiA promoter of 1 isolate) | ID | |||
MICs of cfiA-negative isolates ranged from 0.064 to 4 mg/liter (MIC90, 0.5 mg/liter), and MICs of cfiA-positive isolates ranged from 2 to ≥32 mg/liter (MIC90, ≥32 mg/liter). The expected length of a cfiA promoter amplification product is 0.4 kb; longer fragments indicate the presence of an insertion element. ND, not determined; ID, indeterminate.
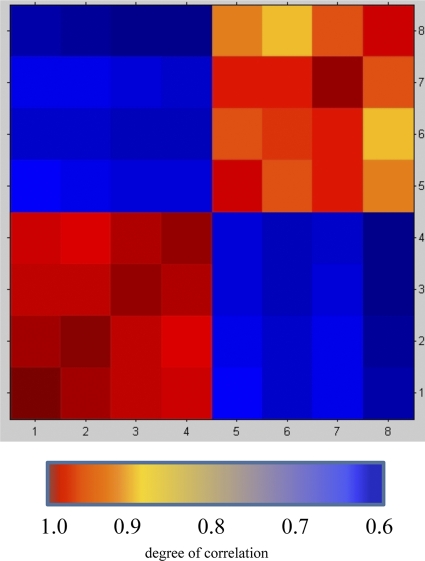
For MALDI-TOF MS analysis, all strains were cultured on fastidious anaerobic agar (Lab M, Bury, United Kingdom) at 35°C in an anaerobic chamber for 24 to 48 h. As a first step, four cfiA-positive (isolates 1 to 4) and four cfiA-negative (isolates 5 to 8) isolates were spotted on the target plate after ethanol-formic acid extraction (7) to obtain high-quality spectra. All spots were overlaid with 1 μl alfa-cyano-4-hydroxycinnamic acid matrix in 50% acetonitrile and 2.5% trifluoroacetic acid. Spectra were obtained with a Microflex LT mass spectrometer in the linear positive mode within a mass range of 2,000 to 20,000 Da and analyzed with MALDI Biotyper 2.0 software and reference library 3.1.1.0 (Bruker Daltonik GmbH, Bremen, Germany). Spectra were internally calibrated each week and controlled every day using Escherichia coli ribosomal proteins (bacterial test standard; Bruker Daltonik GmbH). On each plate, a blank spot overlaid with 1 μl matrix was used as a negative control. Visual inspection of the mass spectra revealed about 10 peak differences between the spectra of the two groups. Since determining qualitative differences between spectra is subject to personal bias, the relatedness between spectra was determined using the composite correlation index (CCI) tool of MALDI Biotyper. It is a modification of the mathematical algorithm to compare and distinguish MALDI mass spectra of whole bacterial cells described by Arnold and Reilly (1). CCI values around 1 represent a high relationship between spectra. A CCI matrix was created using 48 raw spectra of isolates 1 to 8 (Fig. 1). Means of CCI values between cfiA-positive (1 to 4) and cfiA-negative (5 to 8) isolates were 0.98 and 0.93, respectively, and were higher than the mean (0.63) of CCI values if cfiA-positive isolates were compared with cfiA-negative isolates (unpaired t test; P < 0.001) (MedCalc software, version 11.4.4.0; MedCalc Software bvba, Mariakerke, Belgium).
Fig. 1.
The composite correlation index (CCI) values between cfiA-positive (1 to 4) and cfiA-negative (5 to 8) isolates were higher than CCI values if cfiA-positive isolates were compared with cfiA-negative isolates.
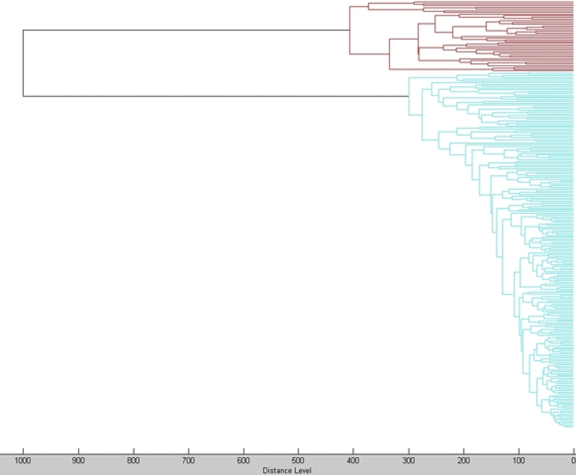
After these preliminary tests, main spectra (MSP) containing information on average peak masses, average peak intensities, and peak frequencies were created for one cfiA-negative (A1) and one cfiA-positive (05/0113) isolate by processing at least 25 mass spectra after ethanol-formic acid extraction with MALDI Biotyper according to the manufacturer's instructions. All isolates were analyzed by the direct-transfer method routinely used in our laboratory. A small amount of a single colony was smeared directly on the target plate in a thin film using an inoculation needle, and spots were overlaid with 1 μl matrix (7). They were identified as B. fragilis by MALDI-TOF MS using the Bruker reference spectra database, with a log(score) of ≥2. The spectra of all isolates were matched with the CCI matrix. These results classified unequivocally all strains in the expected group. In the dendrogram (Fig. 2), the spectra of these isolates clustered in cfiA-positive and cfiA-negative groups without overlap. The mean log(score) for cfiA-positive isolates against cfiA-positive MSP (05/0113) was 2.33 and against cfiA-negative MSP (A1) was 1.76, with a mean difference of 0.56. Inversely, the mean log(score) for cfiA-negative isolates against cfiA-positive MSP (05/0113) was 1.81 and against cfiA-negative MSP (A1) was 2.45, with a mean difference of 0.63.
Fig. 2.
Dendrogram of all tested isolates. cfiA-positive B. fragilis isolates are represented in red, and cfiA-negative isolates are in blue.
Our data suggest that it is possible to differentiate cfiA-positive from cfiA-negative isolates in a routine laboratory setting and so pinpoint B. fragilis strains potentially resistant to carbapenems. This discrimination is not based on the absence or occurrence of one specific peak. The protein profiles of these two genotypically distinct groups differ at approximately 10 peaks. Since the two separate genetic divisions of B. fragilis are not clustered geographically (13), the rapid detection of carbapenem resistance can probably be applied universally.
As the cfiA gene is not always activated, the positive predictive value for the presence of this gene in the isolates of the Belgian 2003-2005 multicenter survey for the detection of meropenem-intermediate or -resistant isolates was 60% and the negative predictive value was 100% according to CLSI breakpoints. EUCAST breakpoints differed only in that the lower breakpoint was 1 dilution lower (≤2 mg/liter) than the CLSI breakpoint (8). Using EUCAST breakpoints, positive and negative predictive values for the presence of the cfiA gene are 90 and 99.2%, respectively, for this collection of isolates. Although the possibility of obtaining false positives exists because of the silent presence of the cfiA gene, it would be interesting to add cfiA-positive isolates to the MALDI-TOF MS databases used for bacterial identification and use them as surrogate markers for the detection of carbapenem resistance. Because MALDI-TOF MS analysis can be performed from the primary plate and is available in less than 10 min, this information can be used to guide empirical treatment of B. fragilis infections, as susceptibility testing takes another 48 h.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Arnold R. J., Reilly J. P. 1998. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12:630–636 [DOI] [PubMed] [Google Scholar]
- 2. Bizzini A., Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 3. Bogaerts P., et al. 2008. Evaluation of a new meropenem-EDTA double-ended Etest strip for the detection of the cfiA metallo-beta-lactamase gene in clinical isolates of Bacteroides fragilis. Clin. Microbiol. Infect. 14:973–977 [DOI] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard, 7th ed. CLSI document M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. das Graças Silva e Souza W., et al. 2000. Resistance profile of Bacteroides fragilis isolated in Brazil. Do they shelter the cfiA gene? J. Antimicrob. Chemother. 45:475–481 [DOI] [PubMed] [Google Scholar]
- 6. Edwards R., Read P. N. 2000. Expression of the carbapenemase gene (cfiA) in Bacteroides fragilis. J. Antimicrob. Chemother. 46:1009–1012 [DOI] [PubMed] [Google Scholar]
- 7. Eigner U., et al. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 8. European Committee on Antimicrobial Susceptibility Testing 2010. Breakpoints tables for interpretation of MICs and zone diameters. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/clinical_breakpoints/ Accessed 20 January 2011 [Google Scholar]
- 9. Gutacker M., Valsangiacomo C., Piffaretti J.-C. 2000. Identification of two genetic groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology 146:1241–1254 [DOI] [PubMed] [Google Scholar]
- 10. Gutacker M., Valsangiacomo C., Bernasconi M. V., Piffaretti J.-C. 2002. recA and glnA sequences separate the Bacteroides fragilis population into two genetic divisions associated with the antibiotic resistance genotypes cepA and cfiA. J. Med. Microbiol. 51:123–130 [DOI] [PubMed] [Google Scholar]
- 11. Jousimies-Somer H. R., et al. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed. Star Publishing Company, Belmont, CA [Google Scholar]
- 12. Nagy E., Maier T., Urban E., Terhes G., Kostrzewa M. on behalf of the ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria 2009. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 15:796–802 [DOI] [PubMed] [Google Scholar]
- 13. Podglajen I., Breuil J., Casin I., Collatz E. 1995. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J. Bacteriol. 177:5270–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 15. Sóki J., Urbán E., Szöke I., Fodor E., Nagy E. 2000. Prevalence of the carbapenemase gene (cfiA) among clinical and normal flora isolates in Bacteroides species in Hungary. J. Med. Microbiol. 49:427–430 [DOI] [PubMed] [Google Scholar]
- 16. Sóki J., et al. 2004. Molecular characterization of imipenem-resistant, cfiA-positive Bacteroides fragilis isolates from the U.S.A., Hungary and Kuwait. J. Med. Microbiol. 53:413–419 [DOI] [PubMed] [Google Scholar]
- 17. Sóki J., et al. 2006. Examination of cfiA-mediated carbapenem resistance in Bacteroides fragilis strains from a European antibiotic susceptibility survey. Int. J. Antimicrob. Agents 28:497–502 [DOI] [PubMed] [Google Scholar]
- 18. Wybo I., et al. 2007. Third Belgian multicentre survey of antibiotic susceptibility of anaerobic bacteria. J. Antimicrob. Chemother. 59:132–139 [DOI] [PubMed] [Google Scholar]
- 19. Wybo I., Soetens O., Vandoorslaer K., Piérard D., Lauwers S. 2008. Carbapenem (cfiA) resistance gene in Bacteroides fragilis group strains in Belgium, abstr. P912. Abstr. 18th Eur. Congr. Clin. Microbiol. Infect. Dis., Barcelona, Spain, 19 to 22 April 2008 http://www.blackwellpublishing.com/eccmid18/abstract.asp?id=68812 [Google Scholar]
- 20. Yamazoe K., et al. 1999. Distribution of the cfiA gene among Bacteroides fragilis strains in Japan and relatedness of cfiA to imipenem resistance. Antimicrob. Agents Chemother. 43:2808–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]