Abstract
HIV-infected persons are at heightened risk for recurrent community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) infections, but there are limited data regarding the molecular characterization of these events. We describe an HIV-infected patient with 24 soft tissue infections and multiple colonization events. Molecular genotyping from 33 nonduplicate isolates showed all strains were USA300, Panton-Valentine leukocidin (PVL) and arginine catabolic mobile element (ACME) positive, and genetically related.
CASE REPORT
A 27-year-old African-American male was diagnosed with HIV/AIDS in September 1997 after an episode of cryptococcal meningitis and a CD4 cell count of 16 cells/mm3. His clinical course was complicated by antiretroviral medication nonadherence; the development of dermatologic conditions, including eosinophilic folliculitis and xerosis; esophageal candidiasis; HIV-associated wasting; and chronic renal dysfunction. The patient denied intravenous drug use or recent sexual activity, lived alone, and had no pets.
He was admitted in July 2005 for left lower extremity and neck abscesses which were culture positive for methicillin-resistant Staphylococcus aureus (MRSA); there was no history of MRSA, and there had been no hospital admissions in the prior 90 days. He was successfully treated with oral clindamycin and linezolid and received a 7-day course of nasal mupirocin for decolonization.
During the next 5 years (July 2005 to June 2010), the patient was diagnosed and treated for a total of 24 culture-proven MRSA skin and soft tissue infection (SSTI) events (Table 1). Seven (22%) of the SSTI events involved >1 body site; the total number of distinct culture-proven MRSA infections was 32. Regarding the site of infection, 38% occurred on the lower extremities, 22% on the upper extremities, 18% on the head/face, 16% on the trunk, and 6% on the buttocks/scrotum. All SSTI events were treated with antibiotics selected by the patient's provider (Table 1); of note, the patient was allergic to vancomycin and trimethoprim-sulfamethoxazole. In addition to antibiotics, incision and drainage procedures were performed for fluctuant abscesses. Seven MRSA SSTIs required hospital admission, totaling 50 days. These included a life-threatening MRSA necrotizing myositis of the lower extremity with septic shock, which required a 25-day hospital admission and the performance of multiple surgical debridements followed by three skin graft procedures.
Table 1.
Recurrent MRSA events in an HIV-infected person
| Event no. | MRSA eventsa |
Molecular analysisc |
Susceptibilityd |
HIV-associated factors |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date (mo/day/yr) | Presentation | Site | Antibiotic treatmentb | USA strain type | SCCmec type | PVL | ACME | msrA | Clindamycin | Tetracyclinee | CD4 count (cells/mm3 [±2 wk]) | HIV RNA level (copies/ml [± 2 wk]) | Antiretroviral therapyf | |
| 1g | 7/14/05 | SSTI | Lower extremity | Clindamycin, linezolid, mupirocin (2%) | Sensitive | Regimen 1 | ||||||||
| SSTI | Head/face | Clindamycin, linezolid, mupirocin (2%) | Sensitive | |||||||||||
| 2 | 8/16/05 | Colonization | Nares | 138 | 1,823 | Regimen 1 | ||||||||
| Colonization | Groin | |||||||||||||
| 3 | 8/25/05 | Colonization | Nares | Mupirocin (2%) | Regimen 1 | |||||||||
| 4 | 9/13/05 | SSTI | Lower extremity | Minocycline → linezolid | Intermediate | Regimen 1 | ||||||||
| 5g | 2/14/06 | SSTI | Head/face | Daptomycin, minocycline → linezolid | Sensitive | |||||||||
| SSTI | Trunk | Daptomycin, minocycline → linezolid | Intermediate | |||||||||||
| SSTI | Buttocks/scrotum | Daptomycin, minocycline → linezolid | Sensitive | |||||||||||
| 6 | 8/29/06 | SSTI | Head/face | Linezolid | Sensitive | |||||||||
| SSTI | Upper extremity | Mupirocin (2%) | Sensitive | |||||||||||
| Colonization | Nares | Mupirocin (2%) | ||||||||||||
| 7g | 1/23/07 | SSTI | Upper extremity | Daptomycin → linezolid, rifampin | Resistant | 0 | >100,000 | |||||||
| SSTI | Buttocks/scrotum | Daptomycin → linezolid, rifampin | Resistant | |||||||||||
| 8 | 4/10/07 | SSTI | Lower extremity | Daptomycin → linezolid | Resistant | |||||||||
| Colonization | Nares | Daptomycin → linezolid | ||||||||||||
| 9 | 6/19/07 | SSTI | Upper extremity | Linezolid → daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | 2 | >100,000 | |
| SSTI | Lower extremity | Linezolid → daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Nares | Linezolid → daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 10 | 7/13/07 | SSTI | Upper extremity | Linezolid → daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | 2 | >100,000 | |
| SSTI | Upper extremity | Linezolid → daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 11 | 8/1/07 | SSTI | Upper extremity | Daptomycin | USA300 | IV | + | + | − | Resistant | Resistant | |||
| Colonization | Nares | Daptomycin | ||||||||||||
| 12 | 8/15/07 | SSTI | Upper extremity | Linezolid | USA300 | IV | + | + | − | Resistant | Resistant | Regimen 2 | ||
| Colonization | Nares | Linezolid | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Axilla | Linezolid | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Perirectal | Linezolid | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 13 | 8/17/07 | SSTI | Trunk | Daptomycin → linezolid | Resistant | Regimen 2 | ||||||||
| 14 | 8/27/07 | Colonization | Axilla | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | 28 | 62,166 | Regimen 3 |
| Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 15 | 9/11/07 | Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | Regimen 4 | ||
| Colonization | Groin | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Perirectal | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 9/18/07 | Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | ||||
| 16 | 9/26/07 | SSTI | Head/face | Mupirocin (2%; to wound site and nares) | USA300 | IV | + | + | − | Resistant | Resistant | 20 | 62,166 | |
| Colonization | Nares | Mupirocin (2%; to wound site and nares) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Axilla | Mupirocin (2%; to wound site and nares) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| Colonization | Groin | Mupirocin (2%; to wound site and nares) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 17 | 10/10/07 | Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | 6 | 34,483 | |
| Colonization | Groin | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 18 | 11/30/07 | SSTI | Lower extremity | Linezolid | USA300 | IV | + | + | − | Resistant | Resistant | 8 | >100,000 | |
| Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | − | Resistant | Resistant | |||||
| 19 | 1/8/08 | Colonization | Nares | |||||||||||
| 20g | 3/11/08 | SSTI (including necrotizing myositis) | Lower extremity | Clindamycin, daptomycin | Resistant | |||||||||
| Colonization | Nares | Clindamycin, daptomycin | ||||||||||||
| 21g | 6/19/09 | SSTI | Head/face | Linezolid | USA300 | IV | + | + | + | Sensitive | Sensitive | 214,568 | Regimen 5 | |
| SSTI | Lower extremity | Mupirocin (2%) | USA300 | IV | + | + | + | Sensitive | Sensitive | |||||
| Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | + | Sensitive | Sensitive | |||||
| 22 | 11/2/09 | SSTI | Lower extremity | Linezolid | Sensitive | 14 | 135,382 | |||||||
| 23 | 11/17/09 | SSTI | Lower extremity | Daptomycin → minocycline | Sensitive | 7 | 1,328,761 | |||||||
| 24g | 1/7/10 | SSTI | Trunk | Daptomycin → minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | |||
| Colonization | Nares | Daptomycin → minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | |||||
| 25 | 2/10/10 | SSTI | Trunk | Minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | 7 | 203,175 | |
| 26g | 3/8/10 | SSTI | Lower extremity | Daptomycin → linezolid → minocycline | Sensitive | 4 | 515,260 | |||||||
| 27 | 3/16/10 | Colonization | Nares | |||||||||||
| 28 | 3/23/10 | SSTI | Trunk | Linezolid → minocycline | Sensitive | |||||||||
| Colonization | Nares | Mupirocin (2%) | ||||||||||||
| 29 | 4/14/10 | SSTI | Lower extremity | Linezolid → minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | |||
| 30 | 5/4/10 | SSTI | Lower extremity | Linezolid → minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | 14 | 798,625 | |
| Colonization | Nares | Mupirocin (2%) | USA300 | IV | + | + | + | Sensitive | Sensitive | |||||
| 31 | 5/18/10 | SSTI | Head/face | Daptomycin → minocycline | USA300 | IV | + | + | + | Sensitive | Sensitive | |||
MRSA events were differentiated based on site and presentation date: SSTI and colonization of the same site occurring within 1 week were grouped as a single event.
Arrows indicate changes in antibiotic treatment. In some instances, antibiotic courses may have overlapped. All listed antibiotics were systemic, except for mupirocin, which was topical.
All available MRSA isolates were the same with regard to other molecular and susceptibility patterns.
“Resistant” was defined as having a clindamycin MIC of ≥2 μg/ml and a tetracycline MIC of ≥8 μg/ml. “Sensitive” was defined as having a MIC of ≤0.5 μg/ml for both antibiotics.
For SSTIs without isolates, tetracycline susceptibility was tested by disk diffusion as part of standard clinical practice.
Regimen 1, atazanavir, didanosine, emtricitabine, ritonavir, and tenofovir; regimen 2, atazanavir, didanosine, fosamprenavir, lamivudine, and ritonavir; regimen 3, atazanavir, didanosine, lamivudine, and ritonavir; regimen 4, atazanavir, didanosine, and lamivudine; regimen 5, darunavir, etravirine, lamuvidine, raltegravir, and ritonavir.
MRSA event requiring hospital admission.
Throughout the study period, screening for MRSA colonization was performed on 31 occasions, of which 19 (61%) were positive for MRSA at one or more body sites, with recovery of a total of 29 individual MRSA isolates. The frequency of MRSA colonization at each body site was examined: the nares were colonized on 67% (20/30) of swabs, groin on 21% (4/19), axilla on 16% (3/19), perirectal area on 14% (2/14), and throat on 0% (0/14). Seven of the colonization events occurred without an associated SSTI, and topical nasal mupirocin was prescribed in four instances. Twelve SSTI events had concurrent colonization; mupirocin was prescribed for seven of these events. In addition, the patient received 10 prescriptions of 3% hexachlorophene–4% chlorhexidine body washes and also had multiple courses of 10% bleach solutions, during this period in an attempt at decolonization.
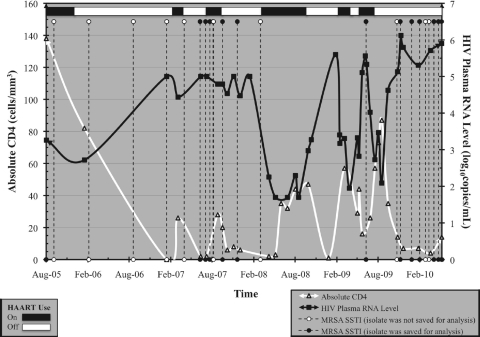
Data regarding the HIV status of the patient were abstracted from the medical records; CD4 cell counts and plasma HIV RNA levels were performed as part of routine clinical practice. A review of laboratory results demonstrated an overall mean CD4 cell count of 30 cells/mm3 (standard deviation [SD], 31) and a mean plasma HIV RNA level of 137,520 copies/ml (SD, 267,090) during this time period. Twenty-six (81%) of the MRSA SSTIs presented at a severely immunosuppressed state (CD4, <50 cells/mm3), and all 32 SSTIs (100%) occurred at an HIV plasma RNA level of >1,000 copies/ml. Poor adherence to antiretroviral therapy was noted, with only brief periods of adherence, during this period (Table 1). Increasing numbers of MRSA events appeared temporarily associated with low CD4 cell counts and elevated HIV RNA levels (Fig. 1).
Fig. 1.
MRSA infections in relation to HIV-specific factors, including CD4 count, HIV RNA level, and antiretroviral use. HAART, highly active antiretroviral therapy.
MRSA isolates were preserved in a random fashion at our hospital among all patients beginning in 2007. Thirty-three nonduplicate isolates (15 SSTIs and 18 colonization) from 15 different time points were available from this patient. MRSA isolates were preserved at −70°C and tested at a single time point for molecular characterization and susceptibility to antimicrobial agents. All MRSA isolates underwent molecular analyses, which included pulsed-field gel electrophoresis (PFGE) following SmaI digestion. PFGE findings were resolved and analyzed with BioNumerics software (Applied Maths, Inc., Austin, TX) and grouped into pulsed-field types using established criteria (15, 26). A USA300 reference strain from the Centers for Disease Control and Prevention (CDC) was included for comparison.
PCR was performed to determine the staphylococcal cassette chromosome mec (SCCmec) type and to detect the presence of genes for Panton-Valentine leukocidin (PVL) and the arginine catabolic mobile element (ACME), as well as the accessory gene regulator (agr) locus. Antimicrobial susceptibility testing was performed with the BD Phoenix automated microbiology system (Becton, Dickinson and Co., Franklin Lakes, NJ). In cases of erythromycin resistance and clindamycin susceptibility, D-tests were performed to confirm clindamycin sensitivity. In addition, PCR was conducted to detect antimicrobial resistance genes, including msrA, qacA/B, smr, aac6, and blaZ.
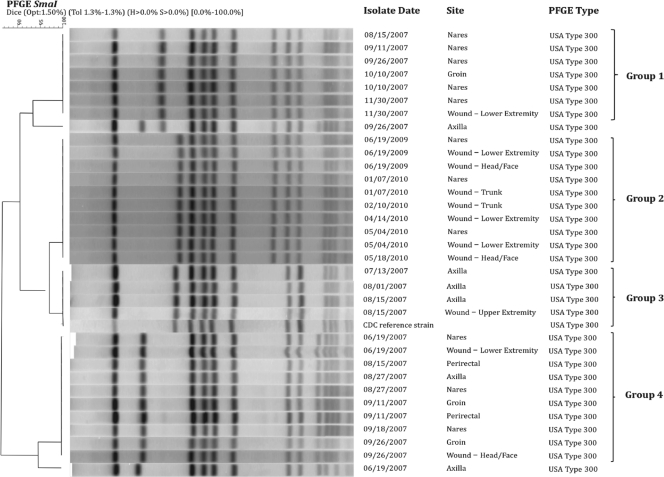
On molecular testing, all isolates (n = 33) were confirmed to be MRSA, carrying the SCCmec type IV allele, and were USA300 strains by PFGE (Table 1). In addition, the presence of ACME and agr (group I), and the genes (lukS-PV and lukF-PV), which encode PVL, were detected in all available isolates. Figure 2 depicts the genetic relatedness of the isolates. Analyses show that all collected isolates were >85% genetically similar by PFGE. Within each of the four groups (Fig. 2), isolates were 100% similar; for example, all isolates from 2009 and 2010 were genetically identical by PFGE.
Fig. 2.
Genetic relatedness of MRSA isolates from pulsed-field gel electrophoresis (PFGE). All isolates within a group were 100% genetically similar.
All available isolates were sensitive to vancomycin (MIC, ≤1 μg/ml), daptomycin (MIC, ≤1 μg/ml), quinupristin-dalfopristin (MIC, ≤1 μg/ml), linezolid (MIC, ≤2 μg/ml), rifampin (MIC, ≤1 μg/ml), trimethoprim-sulfamethoxazole (MIC, ≤0.5 μg/ml), and gentamicin (MIC, ≤1 μg/ml). Resistance was detected among all isolates to erythromycin (MIC, ≥4 μg/ml), and all were intermediate resistant to levofloxacin (MIC, ≥4 μg/ml).
The only variation in antimicrobial susceptibility noted among isolates over time was that all available MRSA isolates from June 2007 to June 2009 (n = 23) were resistant to clindamycin (MIC, ≥2 μg/ml) and tetracycline (MIC, ≥8 μg/ml), whereas all isolates after this date (n = 10) were sensitive for both antimicrobials, with MICs of ≤0.5 μg/ml. We also examined data regarding antibiotic susceptibilities obtained as part of clinical practice prior to the first preserved isolates (July 2005 to April 2007) and noted that the MRSA isolates were initially sensitive to tetracyclines, but that resistance developed over time, as shown in Table 1. Hence the patient initially had a tetracycline-sensitive strain, followed by a resistant strain and then subsequently was infected with a sensitive strain.
Genetic testing for antimicrobial resistance indicated that MRSA isolates from June 2007 to June 2009 were negative for the msrA gene. After this point, the MRSA isolates expressed the msrA gene and thus conferred susceptibility to the lincosamides, including clindamycin, as evidenced by the antimicrobial susceptibility data. Additional PCR results demonstrated a lack of resistance genes for qacA/B (cationic aseptic agents, including chlorhexidine), smr (quaternary ammonium compounds), and aac6 (aminoglycosides), but the presence of blaZ (β-lactamase) among all isolates tested.
To our knowledge, this case represents the most MRSA SSTIs reported in a single patient and exemplifies the potential risk of recurrent community-acquired (CA)-MRSA infections among HIV-infected persons, especially among those with severe immunosuppression. Despite multiple antibiotic courses, repeated incision and drainage procedures, and decolonization attempts, our patient experienced recurrent infections, suggesting that poor immune status may contribute to recurrent MRSA infections. In addition, our study adds novel data regarding the molecular characterization of recurrent MRSA events demonstrating that repeat infections are often due to genetically similar strains.
HIV-infected persons are known to have a higher rate of MRSA colonization and infection (3, 4, 5, 7, 14, 20, 23). In addition, this group may be at higher risk for recurrent disease (1, 6, 11, 24). Reports have shown that 41 to 71% of HIV patients with an initial MRSA SSTI develop a recurrent infection (1, 6, 11). Our HIV-infected patient developed 24 distinct CA-MRSA events in discrete locations and after resolution of the previous infection. In addition to SSTIs, he developed a life-threatening MRSA infection with necrotizing myositis and septic shock. As such, our case adds to the existing literature of recurrent MRSA infections and exemplifies the challenge regarding the prevention and management of these cases among HIV-infected persons.
The reason for recurrent MRSA infections in our patient is unknown, but may be related to severe immunosuppression, poor antiretroviral adherence, absence of trimethoprim-sulfamethoxazole prophylaxis (due to allergy), and dermatologic conditions (1, 14, 21). Prior studies have implicated illicit drug use and high-risk sexual behaviors as potential risk factors for MRSA infections (5, 7, 13, 14), but these were not noted in our patient. Poor virologic control may also be associated with recurrent CA-MRSA infections (6); it has been suggested that the direct antiproliferative effect high levels of HIV have on CD4 cells may influence immunity to S. aureus (14, 16). We noted a correlation in our patient between low CD4 counts and high plasma HIV RNA levels with increasing MRSA SSTI events. Future evaluations of the exact immune disturbances, including the role of T-cell subsets such as Th17 cells, and the risk of MRSA SSTIs are needed.
Colonization may play an important role in repeated MRSA infections. Prior studies have shown that carriage of MRSA increases the risk for subsequent SSTIs (9, 12, 17, 19, 22, 27). Our patient was repeatedly colonized with similar strains that caused the infections. Despite multiple attempts at decolonization with topical antibacterial preparations, infections continued to occur. It is possible that the patient's existing skin conditions and poor immunologic status negatively impacted our decolonization efforts.
The role of decolonization strategies in preventing CA-MRSA infections among HIV-infected persons remains unknown. A recent study among HIV-positive drug users demonstrated no benefit from nares decolonization in reducing the number of subsequent infections (10). However, studies of the role of colonization as reservoirs for infection (including novel sites such as the perirectal area) (25) and the efficacy of their accompanying decolonization procedures are needed, as well as data on the natural displacement of MRSA strains over time.
We performed robust molecular analyses of the available MRSA isolates in our case. All isolates were consistent with community-acquired (CA) strains—USA300 carrying SCCmec IV, PVL, and ACME genes. The propensity for recurrent colonization and pyogenic SSTIs in our case were likely associated with these bacterial factors (8, 9). For example, the faster doubling time of SCCmec IV strains may outcompete other colonizing and infecting strains, and PVL and ACME may be markers for pyogenic/necrotizing skin infections (8, 18).
The MRSA strains in our case were highly related over time, with few differences detected by PFGE, suggesting continued carriage of similar strains or repeated acquisition from the same source. Though the molecular characteristics of the isolates were relatively conserved throughout study duration, there was an alteration in the strain occurring in June 2009. After this juncture, isolates expressed the msrA gene and thus conferred susceptibility to lincosamides, including clindamycin. The appearance of this novel strain is remarkable in that despite extensive exposure to antistaphylococcal antibiotics, this patient was ultimately colonized and infected by a less resistant strain. We speculate that our patient may have subsequently acquired a different, less-resistant USA300 strain which potentially filled the niche when antibiotic pressure was absent. Alternatively, since genes conferring greater antibiotic resistance are often metabolically costly (2), with the absence of pressure from the specific class of antibiotics, resistance may have been reversible.
Our investigation has limitations. We present a single case study, and therefore our findings require confirmation among larger numbers of patients. Second, we did not have isolates available for all MRSA events especially during the latter part of the study period; nonetheless, our study does provide insightful information on the molecular characteristics of recurrent MRSA infections. Third, since the patient was repeatedly infected with the most common type of CA-MRSA (the USA300 strain with SCCmec IV and PVL), it is possible that he reacquired similar strains over time (including after successful decolonization attempts) rather than continued to be infected with the same strain. Finally, although our observational study suggests a relationship between uncontrolled HIV and increased propensity of MRSA infections, causality could not be determined. As such, whether the unfavorable HIV parameters played a causative role in the development of the MRSA infections, or whether the MRSA infections led to reductions in CD4 counts and elevations in HIV RNA levels, requires further investigation.
In summary, we report the largest number of recurrent CA-MRSA infections in a single patient in the literature. The convergence of host (e.g., low CD4 counts) and bacterial factors (USA300 SCCmec IV strain) may have resulted in repeated MRSA infections. Practitioners should be aware of the risks for MRSA recurrences among HIV-infected persons and recognize that despite appropriate treatment and decolonization attempts, infections may recur. Further studies are needed regarding the ideal approaches to reduce recurrent MRSA infections. In the meantime, optimization of HIV control and reduction of potential risk factors are advocated.
Acknowledgments
Support for this work was provided by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed through the Uniformed Services University of the Health Sciences. This project has been funded in whole, or in part, with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under Inter-Agency Agreement Y1-AI-5072.
We certify that all individuals who qualify as authors have been listed; each has participated in the conception and design of this work, the writing of the document, and the approval of the submission of this version; that the document represents valid work; that if we used information derived from another source, we obtained all necessary approvals to use it and made appropriate acknowledgements in the document; and that each takes public responsibility for it. Nothing in the presentation implies any Federal/DOD/DON endorsement.
The authors have no financial interest in this work.
Footnotes
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Anderson E. J., Hawkins C., Bolon M. K., Palella F. J., Jr 2006. A series of skin and soft tissue infections due to methicillin-resistant Staphylococcus aureus in HIV-infected patients. Acquir. Immune Defic. Syndr. 41:125–127 [DOI] [PubMed] [Google Scholar]
- 2. Andersson D. I. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions. Curr. Opin. Microbiol. 9:461–465 [DOI] [PubMed] [Google Scholar]
- 3. Burkey M. D., et al. 2008. The incidence of and risk factors for MRSA bacteraemia in an HIV-infected cohort in the HAART era. HIV Med. 9:858–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cenizal M. J., Hardy R. D., Anderson M., Katz K., Skiest D. J. 2008. Prevalence of and risk factors for methicillin-resistant Staphyloccocus aureus (MRSA) nasal colonization in HIV-infected ambulatory patients. J. Acquir. Immune Syndr. 48:567–571 [DOI] [PubMed] [Google Scholar]
- 5. Crum-Cianflone N. F., Burgi A. A., Hale B. R. 2007. Increasing rates of community-acquired methicillin-resistant Staphyloccocus aureus infections among HIV-infected persons. Int. J. STD AIDS 18:521–526 [DOI] [PubMed] [Google Scholar]
- 6. Crum-Cianflone N. F., Weekes J., Bavaro M. 2009. Recurrent community-acquired methicillin-resistant Staphyloccocus aureus infections among HIV-infected persons: incidence and risk factors. AIDS Patient Care STDs 23:499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diep B. A., et al. 2008. Emergence of multidrug-resistant community-associated methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann. Intern. Med. 148:249–257 [DOI] [PubMed] [Google Scholar]
- 8. Diep B. A., et al. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530 [DOI] [PubMed] [Google Scholar]
- 9. Ellis M. W., Hospenthal D. R., Dooley D. P., Gray P. J., Murray C. K. 2004. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin. Infect. Dis. 39:971–979 [DOI] [PubMed] [Google Scholar]
- 10. Gordon R. J., et al. 2010. The NOSE study (nasal ointment for Staphylococcus aureus eradication): a randomized controlled trial of monthly mupirocin in HIV-infected individuals. J. Acquir. Immune Defic. Syndr. 55:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graber C. J., Jacobson M. A., Perdreau-Remington F., Chambers H. F., Diep B. A. 2008. Recurrence of skin and soft tissue infection caused by methicillin-resistant Staphyloccocus aureus in a HIV primary clinic. J. Acquir. Immune Defic. Syndr. 49:231–233 [DOI] [PubMed] [Google Scholar]
- 12. Hidron A. I., et al. 2005. Risk factors for colonization with methicillin-resistant Staphyloccocus aureus (MRSA) in patients admitted to an urban hospital: emergence of community-acquired MRSA nasal carriage. Clin. Infect. Dis. 41:159–166 [DOI] [PubMed] [Google Scholar]
- 13. Lee N. E., et al. 2005. Risk factors for community-associated methicillin-resistant Staphyloccocus aureus skin infections among HIV-positive men who have sex with men. Clin. Infect. Dis. 40:1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mathews W. C., et al. 2005. Incidence of and risk factors for clinically significant methicillin-resistant Staphyloccocus aureus infection in a cohort of HIV-infected adults. J. Acquir. Immune Defic. Syndr. 40:155–160 [DOI] [PubMed] [Google Scholar]
- 15. McDougal L. K., et al. 2003. Pulse-field gel electrophoresis typing of oxacillin-resistant Staphylococcous aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McNeil A. C., et al. 2001. High-level HIV-1 viremia suppresses viral antigen-specific CD4 T cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 98:13878–13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen M. H., et al. 1999. Nasal carriage of and infection with Staphylococcus aureus in HIV-infected patients. Ann. Intern. Med. 130:221–225 [DOI] [PubMed] [Google Scholar]
- 18. Okuma K., et al. 2002. Dissemination of new methicillin-resistant Staphyloccocus aureus clones in the community. J. Clin. Microbiol. 40:4289–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan E. S., et al. 2005. Population dynamics of nasal strains of methicillin-resistant Staphyloccocus aureus and their relation to community-acquired disease activity. J. Infect. Dis. 192:811–818 [DOI] [PubMed] [Google Scholar]
- 20. Popovich K. J., Weinstein R. A., Aroutcheva A., Rice T., Hota B. 2010. Community-associated methicillin-resistant Staphylococcus aureus and HIV: intersecting epidemics. Clin. Infect. Dis. 50:979–987 [DOI] [PubMed] [Google Scholar]
- 21. Ramsetty S. K., Stuart L. L., Blake R. T., Parsons C. H., Salgado C. D. 2010. Risks for methicillin-resistant Staphylococcus aureus colonization or infection among patients with HIV infection. HIV Med. 11:389–394 [DOI] [PubMed] [Google Scholar]
- 22. Shet A., et al. 2009. Colonization and subsequent skin and soft tissue infection due to methicillin-resistant Staphylococcus aureus in a cohort of otherwise healthy adults infected with HIV type 1. J. Infect. Dis. 200:88–93 [DOI] [PubMed] [Google Scholar]
- 23. Sissolak D., Geusau A., Heinze G., Witte W., Rotter M. L. 2002. Risk factors for nasal carriage of Staphylococcus aureus in infectious disease patients, including patients infected with HIV, and molecular typing of colonizing strains. Eur. J. Clin. Microbiol. Infect. Dis. 21:88–96 [DOI] [PubMed] [Google Scholar]
- 24. Skiest D., et al. 2006. Community-onset methicillin-resistant Staphylococcus aureus in an urban HIV clinic. HIV Med. 7:361–368 [DOI] [PubMed] [Google Scholar]
- 25. Szumowski J. D., et al. 2009. Methicillin-resistant Staphylococcus aureus colonization, behavioral risk factors, and skin and soft-tissue infection at an ambulatory clinic serving a large population of HIV-infected men who have sex with men. Clin. Infect. Dis. 49:118–121 [DOI] [PubMed] [Google Scholar]
- 26. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulse-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wertheim H. F., et al. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762 [DOI] [PubMed] [Google Scholar]