Abstract
Anaplasma ovis is an intraerythrocytic rickettsial pathogen of small ruminants. Loop-mediated isothermal amplification (LAMP) is a nucleic acid detection method in which the target DNA can be efficiently amplified with high specificity and sensitivity under isothermal conditions. In this study, a LAMP method was developed for the specific detection of A. ovis, using LAMP primers designed on the basis of the major surface protein 4 gene. LAMP was performed at 65°C for 30 min. Its specificity was confirmed by successful amplification of several A. ovis isolates and through EcoRI restriction analysis of LAMP products. No cross-reactivity with the A. marginale Lushi isolate, Mycoplasma mycoides subsp. capri, Chlamydophila psittaci, Theileria ovis, T. luwenshuni, T. uilenbergi, or the Babesia sp. Xinjiang isolate was observed. Detection using the LAMP method was compared with that using conventional PCR in 227 field samples; LAMP demonstrated a sensitivity of 95.45%. In summary, LAMP is a specific, sensitive, and rapid test for the diagnosis of A. ovis infection, with the potential to be standardized as a detection method for A. ovis in areas of endemicity.
INTRODUCTION
Anaplasma ovis is an intraerythrocytic rickettsial pathogen of sheep, goats, and some wild ruminants (2, 11, 24). It is classified in the genus Anaplasma (Rickettsiales: Anaplasmataceae), along with A. marginale, A. phagocytophilum, and A. bovis, which also infect ruminants, and A. platys, which infects dogs (2, 5, 6). In China, A. ovis was first found in sheep in the Xinjiang Uygur Autonomous Region (17) and in goats in Liaoning Province (4). A later investigation revealed that A. ovis was widely distributed in sheep and goats in northwest China, including Inner Mongolia, Gansu, Shaanxi, Ningxia, and Qinghai Provinces (16). Experimental transmission studies showed that Dermacentor nuttalli, Hyalomma asiaticum kozlovi, and Rhipicephalus pumilio ticks act as biological vectors of A. ovis in China, transmitting A. ovis by transient infestation when they move from one host to another (16, 22). A recent study demonstrated that ruminant-infective Anaplasma species may also be widely present in the southwest of China (25).
The most frequently used method for diagnosing A. ovis infection involves microscopic detection of the A. ovis pathogen in blood following Giemsa staining. Serologically, the complement fixation test has also been used for field sample investigations in China (16); however, this test is difficult to perform and has fallen into disuse. A competitive enzyme-linked immunosorbent assay has also been used for diagnosis of A. ovis infections (21). Nucleic acid-based molecular tools, such as PCR using the 16S rRNA gene (15) and major surface protein 4 (MSP4) gene (2) and the reverse line blotting method (1), have been shown to be of great diagnostic value in the identification of A. ovis infections. However, these methods are time-consuming and require expensive experimental devices and a high degree of laboratory experience in molecular biology. The present study aimed to develop a loop-mediated isothermal amplification (LAMP) method (20) for the rapid diagnosis of A. ovis infection and to evaluate its applicability by testing field samples and comparison with PCR.
MATERIALS AND METHODS
Preparation of template DNA.
Genomic DNA (g-DNA) of Anaplasma species was prepared using a commercial kit, as described previously (15), and stored at −70°C until use. A. ovis DNA samples from the Yuzhong, Zhangjiachuan, and Yongjing isolates were used as positive samples for the establishment of the LAMP assay, while DNA samples from the A. marginale Lushi isolate, Mycoplasma mycoides subsp. capri, Chlamydophila psittaci, Theileria ovis, T. luwenshuni, T. uilenbergi, and the Babesia sp. Xinjiang isolate and g-DNA of sheep were used as controls. Among the control DNA samples, Mycoplasma and Chlamydia DNAs were gifts from J. Zhou of the zoonotic disease group at our institute, and the others were reference samples collected by us (as members of the Vector and Vector-Borne Diseases Group). The concentrations of the DNA samples were measured using a Nanodrop 2000 spectrophotometer (Thermo Scientific, China) and then diluted to 50 mg/ml.
Design of primer set for LAMP assay.
The primer set was designed on the basis of the MSP4 gene of A. ovis (Table 1). The MSP4 genes of A. ovis from the Yongjing, Yuzhong, Zhangjiachuan, and Chengde isolates were amplified by PCR, as described previously (2). PCR products were cloned into a pGEM-T Easy vector (Promega, Beijing, China) and subjected to sequencing (Sangon Biotech, Shanghai, China). The forward outer primer (F3), backward outer primer (B3), forward inner primer (FIP), and backward inner primer (BIP) were designed on the basis of the sequences of these MSP4 genes using the Primer Explorer (version 4) program (http://primerexplorer.jp/elamp4.0.0/index.html), while the loop primers (LF and LB) were manually designed. To confirm the specificity of the LAMP reaction, two EcoRI restriction enzyme cleavage sites were created in the F1 complementary and F2 primers and the B1 complementary and B2 (Table 1) primers for subsequent restriction analysis of LAMP products. The primers were synthesized by TaKaRa (Dalian, China).
Table 1.
Nucleotide sequences of LAMP primers for detection of A. ovis
| Primer | Type | Sequence (5′–3′)a |
|---|---|---|
| MSP4F3 | Forward outer primer | GTGTTGCACACAGATTTGCC |
| MSP4B3 | Backward outer primer | AGGCTTTTGCTTCTCCGG |
| MSP4FIP | Forward inner primer (F1c + F2) | GCCCCTGTAGGCTAGCTTTGTGgaattcCCCATATGTGTGTGCCGG |
| MSP4BIP | Backward inner primer (B1c + B2) | TGGTGGTAGGTGGGTTCTACCAgaattcATGTGCGGGTATGTCCTTG |
| MSP4LF | Loop primer F | TGTCGACAAAGCTAGCACC |
| MSP4LB | Loop primer B | CGGACTCTTTGACGAGTCTT |
Lowercase italics in FIP and BIP primer sequences indicate EcoRI restriction sites.
Establishment, specificity, and sensitivity of LAMP.
LAMP was performed in a final volume of 25 μl, containing 12.5 μl 2× LAMP reaction buffer [20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, and 0.2% Tween 20], 125 μM each deoxynucleoside triphosphate (dNTP), 0.8 M betaine (Sigma-Aldrich), 8 U of Bst DNA polymerase large fragment (New England BioLabs, United Kingdom), 40 pmol each FIP and BIP primers, 20 pmol LF and LB primers, 5 pmol each F3 and B3 primers, and 2 μl of target DNA. The reaction mixture was incubated at 65°C for 30 min using a conventional heating block (Eppendorf, Germany) and then heated at 80°C for 5 min to terminate the reaction. The specificity of LAMP was examined by testing 2 μl of g-DNA of A. ovis Yongjing, A. ovis Yuzhong, A. ovis Zhangjiachuan, the A. marginale Lushi isolate, M. mycoides subsp. capri, C. psittaci, T. ovis, T. luwenshuni, T. uilenbergi, and the Babesia sp. Xinjiang isolate, as well as sheep g-DNA and a water control. The sensitivity of the reaction was evaluated by measuring the concentration of plasmid DNA containing the MSP4 gene of A. ovis Yuzhong using a Nanodrop 2000 spectrophotometer (Thermo Scientific), and the corresponding copy number was calculated using the method described by Lee et al. (12). The plasmid was diluted to contain 500 copies/μl and then serially diluted 10-fold. Two microliters was used in each reaction mixture when the sensitivity of the assay was evaluated. The LAMP products were subjected to electrophoresis on a 2% agarose gel containing 0.5 μg/ml ethidium bromide, followed by visualization under UV light.
EcoRI restriction digestion of LAMP products.
The LAMP products were purified using a PCR purification kit (TaKaRa, China) to remove the oligonucleotide residues. The purified LAMP products were digested with EcoRI (New England BioLabs, United Kingdom) at 37°C for 1 h. Digested products were analyzed by a 2.0% agarose gel electrophoresis, as described above.
Analysis of field samples using LAMP and PCR methods.
A total of 227 field blood samples were collected from sheep in Yongjing County of Gansu Province. DNA extraction and preparation were performed using a DNeasy blood and tissue kit (Qiagen, China), following the manufacturer's instructions. These DNA samples were analyzed using both LAMP and a PCR method (2). The PCR primers were forward primer MSP4-5 (5′-GGGAGCTCCTATGAATTACAGAGAATTGTTTAC-3′) and reverse primer MSP4-3 (5′-CCGGATCCTTAGCTGAACAGGAATCTTGC-3′). The reactions took place in a final volume of 50 μl, which contained 1.0 mM each primer, 5 μl PCR buffer, 4 μl dNTPs, 0.25 μl Taq (5 U/ml; TaKaRa, China), and 1 μl of DNA sample. Reactions were performed in an automated DNA C1000 thermal cycler (Bio-Rad, Beijing, China) for 35 cycles. After an initial denaturation step of 30 s at 94°C, each cycle consisted of a denaturing step of 30 s at 94°C, an annealing step of 30 s at 60°C, and an extension step of 1 min at 68°C. Reaction mixtures included g-DNA of A. ovis Yongjing as a positive control, and sheep g-DNA and distilled water were used as negative controls. The LAMP and PCR products were subjected to electrophoresis on agarose gels containing 0.5 μg/ml ethidium bromide, followed by visualization under UV light.
Nucleotide sequence accession numbers.
The sequences obtained were submitted to GenBank under accession numbers HQ456347 to HQ456350.
RESULTS
Optimum reaction conditions, specificity, and sensitivity of LAMP.
The optimum incubation temperature for LAMP with the A. ovis primer set was established using a range of temperatures from 61 to 65°C. All temperatures gave positive results (data not shown), but 65°C was chosen as the reaction temperature for all applications. Time periods for the reactions ranging from 15 to 45 min were then tested at this reaction temperature. Analysis of the results indicated that incubation for 30 min was sufficient to allow the reaction to occur (Fig. 1). Under these conditions, six LAMP primers produced LAMP amplicons from g-DNA of A. ovis isolates (Yongjing, Yuzhong, and Zhangjiachuan) in a ladder pattern, while there were no products from DNA of other related pathogens, uninfected sheep DNA, or the water control (Fig. 2). To confirm that LAMP products correspond to the correct target sequence, the amplified products were digested with EcoRI enzyme (there are no EcoRI restriction sites in the target MSP4 gene sequence) and were introduced by use of FIP and BIP primers. LAMP products before and after digestion were analyzed by agarose gel electrophoresis (Fig. 3). The sensitivity of LAMP was evaluated by testing 10-fold serial dilutions of plasmid (starting from 1,000 copies) containing the MSP4 gene of A. ovis. The LAMP assay had a DNA detection limit of 1 copy of plasmid DNA (Fig. 4).
Fig. 1.
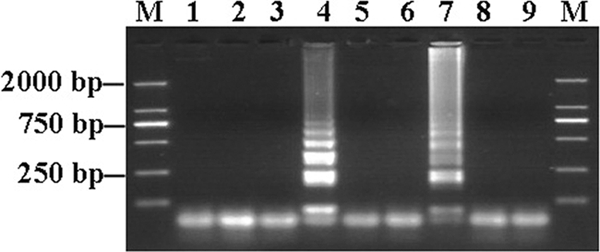
Establishment of optimal incubation time for LAMP at 65°C. Lanes: M, DNA marker; 1, 4, and 7, DNA of A. ovis Yongjing; 2, 5, and 8, sheep g-DNA; 3, 6, and 9, water control; 1 to 3, reactions for 15 min; 4 to 6, reactions for 30 min; and 7 to 9, reactions for 45 min.
Fig. 2.
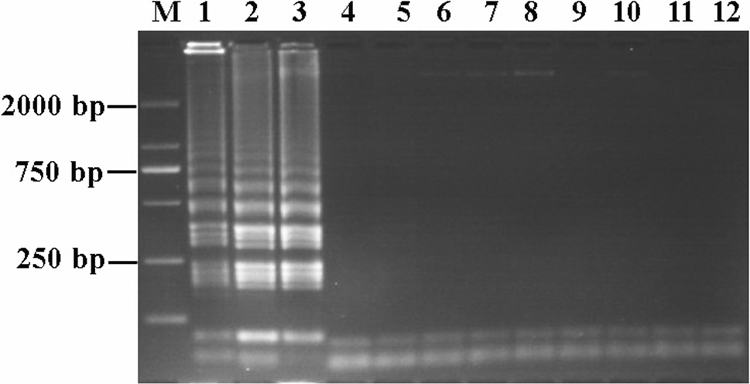
Specificity of LAMP primers for A. ovis. g-DNA from the indicated species was incubated for 30 min at 65°C. Lanes: M, DNA marker (2 kb); 1, A. ovis Yuzhong isolate; 2, A. ovis Yongjing isolate; 3, plasmid containing MSP4 gene; 4, M. mycoides, 5; C. psittaci, 6; A. marginale Lushi isolate; 7, T. ovis; 8, T. luwenshuni; 9, T. uilenbergi; 10, Babesia sp. Xinjiang isolate; 11, sheep g-DNA; 12, water control.
Fig. 3.
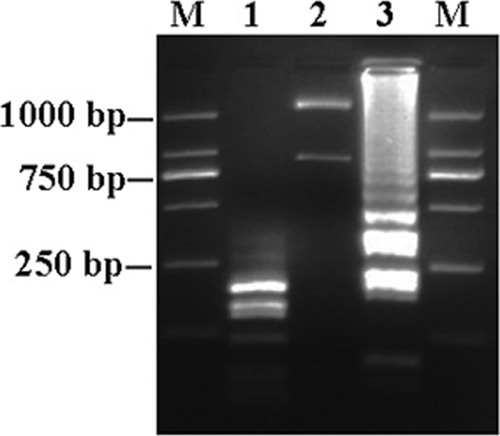
Restriction digestion of LAMP products from A. ovis. EcoRI restriction sites were introduced in FIP and BIP primers, allowing restriction fragment analysis of the specificity of the LAMP reaction. Lanes: M, DNA marker; 1, EcoRI digest of A. ovis LAMP product; 2, EcoRI enzyme activity control by digesting reconstructed plasmid DNA in pGEM-T Easy vector; 3, LAMP products from g-DNA of A. ovis.
Fig. 4.

Sensitivity of LAMP primers for A. ovis using 10-fold serial dilutions of plasmid DNA containing the MSP4 gene of A. ovis. Lanes: M, DNA marker; 1, 1,000 copies; 2, 100 copies; 3, 10 copies; 4, 1 copy; 5, 0.1 copies; 6, 0.01 copies; 7, 0.001 copies.
Detection of field samples using LAMP assay and PCR.
Detection of field samples collected from 227 sheep in Yongjing County of Gansu Province was conducted using both the LAMP and the PCR methods. A comparison of the results is presented in Table 2, where LAMP detected A. ovis DNA from 157 samples (69.16%) and PCR detected DNA from 66 samples (29.07%). However, 94 samples found to be positive by LAMP were negative when tested by PCR. Likewise, 3 samples found to be negative by LAMP were positive when tested by PCR. Moreover, results for 63 positive samples and 67 negative samples were in agreement with both techniques. On the basis of the data in Table 2, using PCR as the reference method, the sensitivity and specificity of LAMP are 95.45% and 41.61%, respectively.
Table 2.
Comparative evaluation of LAMP and PCR methods for detection of field samples
| LAMP result | No. (%) of specimens with the following PCR result: |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 63 | 94 | 157 (69.16) |
| Negative | 3 | 67 | 70 |
| Total | 66 (29.07) | 161 | 227 |
DISCUSSION
LAMP is a nucleic acid amplification method that has demonstrated the advantages of being rapid, easy to use, and less expensive than other nucleic acid-based tests for the diagnosis of infectious diseases (18, 20). It therefore has the potential for use in resource-limited veterinary laboratories in developing countries, such as China, where many endemic diseases exist. A. ovis has been found to be distributed over 44 counties in Gansu Province, with infection rates of 30.1 to 60.8% (16). Ovine Theileria and Babesia infections have also been found in regions of Gansu Province in recent years (13, 23). It is difficult to differentiate between Piroplasma and Anaplasma pathogens on the basis of clinical signs and microscopic examination, and a rapid diagnostic method, LAMP, was therefore developed in the present study. LAMP primers were designed on the basis of the MSP4 gene sequence of A. ovis, which is reportedly highly conserved among A. ovis isolates (2). Alignment of the MSP4 sequences from four A. ovis isolates from Yongjing, Jintai, Zhangjiachuan, and Chengde confirmed 99.8% sequence identity. Using such a conserved gene as a target gene for LAMP is important, because for the LAMP reaction to occur, all eight primers must specifically bind to the target sequences (19). The primers used in this study allowed the LAMP reaction to be completed within half an hour. The high level of efficiency was probably attributable to the introduction of the loop primers (LP and LB), which may have reduced the time taken for amplification by half (19).
We demonstrated that the LAMP primers specifically amplified g-DNA of A. ovis isolates but not A. marginale, confirming the high specificity of LAMP for the diagnosis of A. ovis infection. The ability of LAMP to amplify different isolates of A. ovis is critical to its usefulness for detecting the organism in a wide range of areas of endemicity. LAMP did not react with g-DNA of the A. marginale Lushi isolate, probably because of the low level of similarity (91.5 to 91.6%) of the MSP4 gene between A. ovis isolates and A. marginale Lushi (MSP4 gene GenBank accession number HM640938). A. phagocytophilum was not included in the current study, but alignment of the MSP4 gene sequences from A. ovis isolates and A. phagocytophilum strains (3) showed a maximum similarity of 64.4%, suggesting that there is unlikely to be any cross-reaction between these species; however, this should be confirmed experimentally. Digestion of the LAMP products by EcoRI further confirmed the specificity of the assay.
The sensitivity of LAMP using 10-fold serial dilutions of plasmid DNA containing the MSP4 gene indicated that it was able to detect levels of plasmid DNA as low as 1 copy. However, further studies are required to confirm the sensitivity of LAMP for the detection of A. ovis parasitemia. In a comparative evaluation of 227 field samples, the LAMP assay detected more positive samples (69.2%) than PCR (29.1%), implying a higher sensitivity. The higher sensitivity of LAMP than conventional PCR has been demonstrated in several studies (8, 14, 20), and the sensitivity of LAMP has been shown to be less affected by components of the clinical samples than PCR (7, 9). Nevertheless, three samples identified as negative by LAMP produced weak bands with conventional PCR, possibly reflecting the fact that PCR primers amplify g-DNA from A. marginale, as well as A. ovis. A. marginale is known to infect other ruminants (10). Attempts to sequence these three positive PCR products have not yet been successful. However, the MSP4 gene sequence obtained from an Anaplasma sp.-infected sheep in Jingtai County of Gansu Province (GenBank accession number HM195103) showed 98% similarity with the MSP gene sequence of A. marginale, suggesting possible A. marginale infection in sheep in Gansu Province.
In summary, the LAMP method developed in this study shows great potential as a test for the easy diagnosis of ovine anaplasmosis in regions of endemicity in China. It could provide an accurate, sensitive, affordable, and easy-to-use method and a practical alternative to PCR for the routine diagnosis of anaplasmosis caused by A. ovis.
ACKNOWLEDGMENTS
This study was financially supported by the 973 Program (2010CB530206), the Key Project of Gansu Province (1002NKDA035 and 0801NKDA033), NSFC (30800820, 30972182, 31072130, and 31001061), 948 (2010-S04), Beef and Yak Production System Programme, Specific Fund for Sino-Europe Cooperation, MOST, and State Key Laboratory of Veterinary Etiological Biology Project (SKLVEB2008ZZKT019). The research was also facilitated by EPIZONE (FOOD-CT-2006-016236), ASFRISK (211691), ARBOZOONET (211757), and PIROVAC (KBBE-3-245145) of the European Commission, Brussels, Belgium.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Bekker C. P., de Vos S., Taoufik A., Sparagano O. A., Jongejan F. 2002. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet. Microbiol. 89:223–238 [DOI] [PubMed] [Google Scholar]
- 2. de la Fuente J., et al. 2007. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet. Microbiol. 119:375–381 [DOI] [PubMed] [Google Scholar]
- 3. de la Fuente J., et al. 2005. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43:1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ding Z., Lu F., Han L., Huang J. 1985. The first report of goats anaplasmosis in China. Chi. J. Vet. Sci. Tech. 6:61 (In Chinese.) [Google Scholar]
- 5. Dumler J. S., et al. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145–2165 [DOI] [PubMed] [Google Scholar]
- 6. Gaunt S., et al. 2010. Experimental infection and co-infection of dogs with Anaplasma platys and Ehrlichia canis: hematologic, serologic and molecular findings. Parasit. Vectors 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grab D. J., Lonsdale-Eccles J., Inoue N. 2005. Lamp for tadpoles. Nat. Methods 2:635–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iseki H., et al. 2007. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J. Microbiol. Methods 71:281–287 [DOI] [PubMed] [Google Scholar]
- 9. Kaneko H., Kawana T., Fukushima E., Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70:499–501 [DOI] [PubMed] [Google Scholar]
- 10. Kocan K. M., de la Fuente J., Blouin E. F., Coetzee J. F., Ewing S. A. 2010. The natural history of Anaplasma marginale. Vet. Parasitol. 167:95–107 [DOI] [PubMed] [Google Scholar]
- 11. Krier J. P., Ristic M. 1963. Anaplasmosis. VII. Experimental Anaplasma ovis infection in white-tailed deer (Dama virginiana). Am. J. Vet. Res. 24:567–572 [PubMed] [Google Scholar]
- 12. Lee C., Kim J., Shin S. G., Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273–280 [DOI] [PubMed] [Google Scholar]
- 13. Liu A., et al. 2007. At least two genetically distinct large Babesia species infective to sheep and goats in China. Vet. Parasitol. 147:246–251 [DOI] [PubMed] [Google Scholar]
- 14. Liu Z., et al. 2008. Development of loop-mediated isothermal amplification (LAMP) assay for rapid diagnosis of ovine theileriosis in China. Parasitol. Res. 103:1407–1412 [DOI] [PubMed] [Google Scholar]
- 15. Liu Z., et al. 2005. Amplification of 16S rRNA genes of Anaplasma species in China for phylogenetic analysis. Vet. Microbiol. 107:145–148 [DOI] [PubMed] [Google Scholar]
- 16. Lu W., Lu W., Zhang Q., Yu F., Dou H. 1996. Ovine Anaplasma in Northwest China. Trop. Anim. Health Prod. 29:16S–18S [Google Scholar]
- 17. Ma L., Hua N., Chen W. 1982. Investigation of ovine anaplasmosis. J. Xinjiang Anim. Hus. Sci. Tech. 2:2–13 (In Chinese.) [Google Scholar]
- 18. Mori Y., Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagamine K., Hase T., Notomi T. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223–229 [DOI] [PubMed] [Google Scholar]
- 20. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scoles G. A., Goff W. L., Lysyk T. J., Lewis G. S., Knowles D. P. 2008. Validation of an Anaplasma marginale cELISA for use in the diagnosis of A. ovis infections in domestic sheep and Anaplasma spp. in wild ungulates. Vet. Microbiol. 130:184–190 [DOI] [PubMed] [Google Scholar]
- 22. Yin H., Luo J. 2007. Ticks of small ruminants in China. Parasitol. Res. 101(Suppl. 2):S187–S189 [DOI] [PubMed] [Google Scholar]
- 23. Yin H., et al. 2004. Phylogenetic analysis of Theileria species transmitted by Haemaphysalis qinghaiensis. Parasitol. Res. 92:36–42 [DOI] [PubMed] [Google Scholar]
- 24. Zaugg J. L., Goff W. L., Foreyt W., Hunter D. L. 1996. Susceptibility of elk (Cervus elaphus) to experimental infection with Anaplasma marginale and A. ovis. J. Wildl. Dis. 32:62–66 [DOI] [PubMed] [Google Scholar]
- 25. Zhou Z., et al. 2010. Phylogenetic analysis of the genus Anaplasma in southwestern China based on 16S rRNA sequence. Res. Vet. Sci. 89:262–265 [DOI] [PubMed] [Google Scholar]


