Abstract
Approximately 45% of untreated United States patients with early Lyme disease associated with erythema migrans have a positive blood culture based on microscopic detection of Borrelia burgdorferi in Barbour-Stoenner-Kelly medium after 2 to 12 weeks of incubation. In this study we demonstrate that the yield of blood cultures can be significantly increased to 70.8% by the use of a combined culture-quantitative PCR technique and that among those patients found to have a positive blood culture, positivity was detected in over 90% within just 7 days of incubation. Patients with multiple erythema migrans were almost uniformly culture positive by this technique.
INTRODUCTION
Early studies of the yield of blood cultures in United States patients with early Lyme disease associated with erythema migrans found disappointing results of approximately 5% (2, 5). Subsequent studies, however, have demonstrated that the yield can be markedly improved, to approximately 45%, simply by culturing the plasma fraction of blood specifically and increasing the volume of material cultured to 9 ml (1, 6). The findings of these investigations suggested that if the yield can still be enhanced further, this would have to be accomplished by developing innovative approaches to enhance the sensitivity of the culture method per se, rather than by further increases in the volume of material cultured (6). In the current study we demonstrate that the yield can be significantly increased by the use of a combined culture-quantitative PCR (qPCR) technique.
MATERIALS AND METHODS
Patients and blood cultures.
In EDTA-blood collection tubes, three 3-ml plasma samples were obtained from adult patients with a clinical diagnosis of erythema migrans that had been established at the Lyme Disease Diagnostic Center (Valhalla, NY) in the years 2005, 2006, and 2007. All eligible patients with erythema migrans were invited to participate in this study. Patient samples were excluded from this analysis if the patients had recently been treated with a beta-lactam or tetracycline antibiotic before the plasma sample was obtained. The protocol was approved by the Institutional Review Board of New York Medical College, and written informed consent was obtained from all participants.
Plasma was separated by centrifugation at 260 × g for 15 min. Within 3 h of the time of collection, each 3-ml aliquot of plasma was inoculated into a 70-ml screw-cap plastic flask containing 60 ml of antibiotic-free BSK medium, which was prepared as described elsewhere (5, 6). Cultures were incubated at 32°C to 33°C for up to 12 weeks in 2005 and 2006 and for up to 8 weeks in 2007. The cultures were examined by fluorescence microscopy at 2 weeks and thereafter at 2- to 4-week intervals. Sampling for each culture was done as follows. A 10-μl aliquot of culture material was mixed with 10 μl of an acridine orange staining solution (100 μl in PBS [pH 7.41]). Ten microliters of this mixture was placed on a slide overlaid with a coverslip and was examined with a microscope (magnification, ×400). A minimum of 20 high-power fields were viewed for the presence of spirochetes.
In addition, after gently shaking the culture flasks, at multiple time points, one 1-ml aliquot was removed from each of the three blood culture flasks (i.e., three 1-ml aliquots in total per sampling day) to be analyzed by qPCR. Although it was intended for these aliquots to be removed daily on weekdays for the first week and weekly for the subsequent 2 weeks, a number of logistical factors sometimes reduced the number of aliquots that were submitted or underwent qPCR testing. Furthermore, in years 2 and 3 of the study, aliquots were tested only until a positive qPCR result was obtained. Thus, qPCR was not performed on culture aliquots removed on all sampling days; the median number of sampling days per patient on which aliquots were tested was 4.
qPCR of blood culture aliquots.
One-milliliter aliquots were centrifuged at 12,000 × g, the pellet was resuspended in 100 μl of IsoQuick sample buffer, and DNA was extracted by means of a commercial nucleic acid extraction kit (IsoQuick; Orca Research) as described previously (4) and suspended in 50 μl of water according to the manufacturer's instructions. Two microliters of DNA suspension was used for PCR amplification. Real-time qPCR was performed in an ABI Prism 7900HT sequence detection system (TaqMan) using two flaB-specific primers designated FL-571F (GCAGCTAATGTTGCAAATCTTTTC) and FL-662R (TGAGCTCCTTCCTGTTGA) based on those described earlier (3). Reaction mixtures contained 2 μl of DNA solution and 20 pmol of each primer in FastStart universal probe master buffer (Roche Diagnostics, Indianapolis IN). Genomic copy numbers were quantitated with a 6-carboxy-fluorescein (FAM)/6-carboxy-tetramethyl-rhodamine-labeled probe, FL-611P (FAM-AAACTGCTCAGGCTGCACCGGTTC-minor groove binder [MGB]).
Standard curves relating flaB copy number and spirochete number were generated using a dilution series (containing 10 to 105) of a culture, for which the titer had been determined, of B. burgdorferi B31 MI assayed by dark-field microscopy. Real-time qPCRs were run in 96-well plates; all qPCRs (with both standards and blood culture-derived samples) were performed in triplicate, and reaction mixtures containing no DNA were also included as controls. Samples lacking DNA were uniformly qPCR negative. Standard curves and patient spirochete loads were calculated using SDS 2.1 software (Applied Biosystems).
Statistics.
Categorical variables were compared by the Fisher exact test (two tailed), and continuous variables by the Wilcoxon rank-sum test or the Brown-Mood test, since the data were not normally distributed. A P value of <0.05 was considered to be significant.
RESULTS
The 65 patients in this study were adults between the ages of 19 and 84 years, 36 (55.4%) of whom were male. Thirty (46.2%) had a positive plasma blood culture for B. burgdorferi based on microscopic detection of the spirochete. For each of these 30 patients, the fla gene was also detected by qPCR of a culture aliquot. An additional 16 patients had a positive qPCR result in the absence of detectable spirochetes by microscopic examination, raising the yield of the combined culture-qPCR technique to 70.8%. Overall, a significantly greater proportion of patients had a positive blood culture based on qPCR than on microscopic examination (46/65 [70.8%] versus 30/65 [46.2%], P = 0.007).
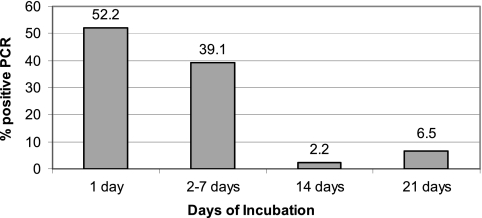
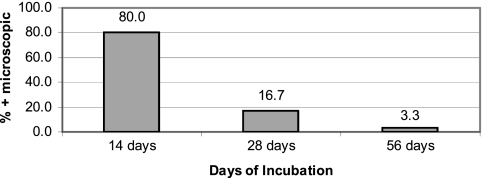
Of the 46 patients with a positive qPCR assay of culture medium, 42 (91.3%) had a positive qPCR result within 7 days after the cultures were incubated. Indeed, of the 38 culture-positive patients for whom qPCR testing was performed on a culture aliquot at 1 day after incubation, 24 (63.2%) were already positive (Fig. 1). For 4 (8.7%) patients, the qPCR of culture aliquots was negative at 7 or fewer days, 1 of the patients had a positive blood culture based on qPCR of culture aliquots at 14 days, and the other 3 patients had a positive qPCR of one or more culture aliquots at day 21. Culture aliquots from the latter 3 patients, however, were not tested at day 14. There was no significant difference in the median time to qPCR positivity between cases with visual detection of spirochetes (2 days) and those in which spirochetes were not seen (1 day) (P = 0.182). For purposes of comparison, the time to culture positivity based on microscopic detection is shown in Fig. 2.
Fig. 1.
A total of 46 patients had a positive qPCR of an aliquot of culture supernatant. For these 46 patients the percentage with a positive qPCR test (y axis) is shown in relation to the duration of incubation of the culture (x axis) at the time of first detection of qPCR positivity.
Fig. 2.
A total of 30 patients had a positive blood culture based on microscopic detection of spirochetes in culture medium. The percentage of patients with a positive blood culture based on microscopic detection (y axis) is shown in relation to the duration of incubation of the culture (x axis) at the time of detection.
A positive qPCR result was not always found with all three of the 1-ml aliquots of culture medium that were evaluated at a given time point; at the time point of first qPCR positivity, all three aliquots were positive in 25/45 (55.6%) cases, and 10 cases each had either 2 positive aliquots (22.2%) and 1 positive aliquot (22.2%). The remaining case was positive for 2 of 2 aliquots.
At the time of first detection, the median number of fla gene copies was 4,100/ml, with a range of 460 to 731,000/ml. For the cases with qPCR results reported at day 14, the first time point that the blood cultures were examined microscopically, the median number of fla gene copies per milliliter in the 10 microscopically positive samples was 559,500 versus 7,240 in the 8 samples for which the spirochete could not be detected visually (P = 0.018). Over 10,000 fla gene copies were found in all 10 of the microscopically proven cases compared with just 3 of the 8 cultures that were positive by PCR alone (P = 0.007).
In the evaluation of serial samples obtained at different time points from the same culture, it does appear in some instances that growth of the borrelial strain was aborted. Of the 18 qPCR-positive cases in which culture medium was tested specifically at 14 days of incubation, all 18 were positive, whereas of the 13 qPCR-positive cases in which the culture medium was tested specifically at day 21 of incubation, only 9 (69.2%) were positive. It is possible that some cultures that contained spirochetes at least transiently were misclassified as qPCR negative because testing was not done sufficiently frequently. It seems unlikely that misclassification was very common, however, since 15 (78.9%) of the 19 qPCR-negative cultures were tested at at least 3 different time points and all but 2 were tested at least once between days 7 and 21 of culture incubation.
Of the 65 patients evaluated, 23 (35.4%) had multiple erythema migrans skin lesions, a clinical manifestation of hematogenous dissemination, and 42 (64.6%) had a single skin lesion. Blood culture positivity was documented for 22 (95.7%) of the 23 patients with multiple erythema migrans skin lesions compared with 24 (57.1%) of the 42 patients with a single skin lesion (P = 0.001). Among the patients with multiple skin lesions, blood culture positivity based on qPCR detection of the fla gene was found for 22 (95.7%) patients and based on microscopic detection for 18 (78.3%) patients (nonsignificant difference [P = 0.19]). For the 42 patients with a single skin lesion, however, detection of blood culture positivity was significantly more likely by qPCR than by microscopy (57.1% versus 28.6%, P = 0.01). Examination of these data from a slightly different perspective shows that negative microscopic findings were significantly more frequent among culture-positive patients with a single erythema migrans skin lesion compared with those who had multiple erythema migrans lesions. Of the 22 culture-positive patients with multiple erythema migrans skin lesions, only 4 (18.2%) were negative by microscopy, whereas among the 24 culture-positive patients with a single erythema migrans lesion, 12 (50.0%) were negative by microscopy (P = 0.03).
DISCUSSION
This study suggests that the yield of plasma blood cultures from adult patients with erythema migrans may be as high as 70.8% and that performing qPCR of incubating cultures will increase the detection rate by more than 50%. Moreover, in the vast majority of cases there was a positive PCR result by 7 days of incubation, often within 24 h.
In a prior in vitro study in which a limited number of strains of B. burgdorferi were inoculated into BSK medium, we estimated that growth to a level of 106 spirochetes per milliliter was necessary for reliable microscopic detection of spirochetes, given the assumption that the doubling time during exponential growth was 12 h (4). Consistent with these results, the median number of fla gene copies when spirochetes were successfully visualized by microscopic examination in this study at day 14 of culture incubation (the time they were first examined) was 559,500/ml, compared with 4,100/ml at time of first detection by qPCR (P = 0.002). Although more rapid detection would be expected by qPCR, what was surprising is that microscopic detection proved entirely unsuccessful through at least 8 weeks of incubation in 34.8% of patients for whom the spirochete was detectable by qPCR of a culture aliquot. An apparent explanation for this is that some borrelia strains grow poorly in BSK and never reach the quantity required for visual identification. Furthermore, our findings further suggest that some strains die out and disappear over the course of time in BSK medium before they reach the threshold for microscopic detection and that there may be a narrow window of opportunity to detect such strains even by qPCR. An alternative explanation is that in some instances the qPCR method has detected only the presence of B. burgdorferi DNA originally present in plasma, rather than even transiently viable intact spirochetes. Since detection of blood culture positivity was significantly more likely by qPCR than by microscopy for patients with a single erythema migrans skin lesion, and not for those with multiple skin lesions, it is possible that strains which grow poorly in BSK medium are also less pathogenic, being less likely to disseminate to additional skin sites.
A previous study employed a PCR assay of culture aliquots and also showed that detection of B. burgdorferi could be increased by a combination of PCR and culture (4). There are a number of differences between the current and previous studies. First, the earlier experiment involved culture of skin biopsy specimens from erythema migrans lesions. In addition, standard PCR was employed rather than qPCR, as in the current study. Use of qPCR provides the advantages of increased sensitivity, quantitation of B. burgdorferi copy number, and decreased possibility of contamination. Finally, in the present study aliquots were removed from culture as early as 1 day (as opposed to 14 days in the previous study), which facilitated more rapid detection of positive cultures. Thus, the current study provides several enhancements over the previously reported methodology.
Further studies should be done to determine if the time from blood collection to inoculation in culture medium could be extended beyond 3 h, as this time constraint limits applicability. In addition, to maximize sensitivity and cost-effectiveness, the optimal quantity of culture aliquots and durations of incubation for assessing cultures by qPCR should be defined.
ACKNOWLEDGMENTS
This work was supported by NIH grant AR41511.
We thank Lisa Giarratano and Joseph DeMarco for their assistance.
There are no conflicts of interest.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Coulter P., et al. 2005. Two-year evaluation of Borrelia burgdorferi culture and supplemental tests for definitive diagnosis of Lyme disease. J. Clin. Microbiol. 43:5080–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goodman J. L., et al. 1995. Bloodstream invasion in early Lyme disease: results from a prospective, controlled, blinded study using polymerase chain reaction. Am. J. Med. 99:6–12 [DOI] [PubMed] [Google Scholar]
- 3. Hodzic E., Feng S., Freet K. J., Borjesson D. L., Barthold S. W. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70:3382–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartz I., et al. 1993. Polymerase chain reaction amplification of culture supernatants for rapid detection of Borrelia burgdorferi. Eur. J. Clin. Microbiol. Infect. Dis. 12:879–882 [DOI] [PubMed] [Google Scholar]
- 5. Wormser G. P., et al. 2000. Comparison of the yields of blood cultures using serum or plasma from patients with early Lyme disease. J. Clin. Microbiol. 38:1648–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wormser G. P., et al. 2001. Yield of large-volume blood cultures in patients with early Lyme disease. J. Infect. Dis. 184:1070–1072 [DOI] [PubMed] [Google Scholar]