Abstract
Expense inherent to molecular diagnostics may prevent laboratories from utilizing real-time PCR for Clostridium difficile infection. Frozen master mix and overnight aliquot modifications of the BD GeneOhm Cdiff assay failed to impact performance indices compared to the package insert protocol (P ≥ 0.31), provided accurate results, and decreased reagent expenditure.
TEXT
Traditional laboratory methods detecting toxigenic Clostridium difficile to confirm Clostridium difficile infection (CDI) suffer from deficiencies in analytical sensitivity and/or time to final result (14). Recent consensus clinical practice guidelines for CDI developed by the Society for Healthcare Epidemiology of America (SHEA) and Infectious Diseases Society of America (IDSA) suggest that molecular testing for toxigenic C. difficile may ultimately address laboratory diagnostic concerns (3). In spite of this, some clinical microbiology laboratories may not adopt rapid, molecular biology-based methods due to financial considerations (17, 21) or reservations about intricate processing protocols (particularly with specimens possessing nucleic acid amplification inhibitors [9]). The real-time BD GeneOhm Cdiff assay (BD Diagnostics, Sainte-Foy, Quebec, Canada) has demonstrated high levels of clinical sensitivity for detection of tcdB from primary clinical specimens (1, 4, 8, 15, 19, 20). In this report, we demonstrate the economical and logistical impact of two protocol modifications on this assay.
(Part of this work was presented at the 110th General Meeting of the American Society for Microbiology, San Diego, CA, 23 to 27 May 2010 [2, 11].)
In an institutional review board (IRB)-approved protocol, 251 consecutive unpreserved liquid fecal specimens from patients symptomatic for CDI were processed to a lysate form for the BD GeneOhm Cdiff assay per the manufacturer's specifications (same-day aliquot). Specimen aliquots were also transferred into kit-provided buffer tubes 1 day prior to lysate generation and stored at 2 to 8°C (overnight aliquot). Freshly reconstituted and frozen master mix were prepared and analyzed as previously described (10, 12). tcdB (positive)- and buffer (negative)-based control material was prepared and analyzed analogous to previously described methods (12). A prospective clinical study analyzed 502 lysates in tandem using freshly reconstituted master mix and master mix that had been frozen for 1 week.
Primary specimen aliquots were transferred to kit-provided diluent for the ProSpecT C. difficile toxin A/B microplate assay (Remel, Lenexa, KS). Spectrophotometric optical density (OD) values ≥ 0.080 were interpreted as positive results. All components of enzyme immunoassay (EIA) and PCR testing, including specimen aliquoting, were timed for workflow analysis. Consensus positive and negative results generated by both EIA and PCR constituted reference results. Discrepancies were resolved upon utilization of the toxigenic culture reference standard (P. Riska; Montefiore Medical Center, Bronx, NY [8]). Toxigenic culture, following a 30-min 100% alcohol treatment, comprised two concurrent methods: (i) culture of stool specimens in cooked meat broth with cycloserine, cefoxitin, and taurocholate additives, followed by toxin A/B EIA if there was visible growth in the broth; and (ii) anaerobic culture on cycloserine-cefoxitin-fructose agar, followed by confirmation of the identity and toxigenicity of isolates resembling C. difficile with an internally validated multiplex PCR for the C. difficile toxin repressor gene tcdC, glutamate dehydrogenase gene gdh, and binary toxin genes (8).
tcdB-positive/negative lysates generated expected results in freshly reconstituted master mix and master mix frozen 1 to 5 weeks. The cycle threshold (CT) for the tcdB-positive lysates (Table 1) did not exhibit changes in frozen master mix potency (P = 0.18 by one-way analysis of variance). In contrast, the temporal potency of frozen master mix increased with internal control nucleic acid (P = 0.02; Table 1). Preparation of frozen control material resulted in expected amplification of tcdB and internal control sequences. The data suggest a temporal decrease in CT for frozen control material compared to freshly reconstituted control material (P ≤ 0.02 by t test for independent samples; Table 1).
Table 1.
Mean cycle threshold values generated by analysis of reconstituted BD GeneOhm Cdiff assay master mix and control material that was frozen at −70°C for up to 5 weeksa
| Specimen | Reconstituted reagent stored at −70°C | Mean CT ± SEM following storage interval of: |
|||
|---|---|---|---|---|---|
| 0 wkb | 1 wk | 3 wk | 5 wk | ||
| Positive lysatec | Master mix | 30.51 ± 1.09 | 30.30 ± 1.00 | 30.29 ± 0.95 | 30.84 ± 1.14 |
| Negative lysatec | Master mix | 36.29 ± 0.21 | 35.99 ± 0.21 | 35.94 ± 0.23 | 35.99 ± 0.13 |
| None | Positive controld | 34.72 ± 0.08 | 34.55 ± 0.09 | 34.55 ± 0.09 | 34.43 ± 0.08e |
| None | Negative controld | 36.04 ± 0.04 | 35.90 ± 0.05 | 35.84 ± 0.05f | 35.81 ± 0.07f |
Mean cycle threshold values generated by analysis of reconstituted BD GeneOhm Cdiff assay master mix and control material frozen for up to 5 weeks at −70°C. The cycle threshold (CT) values derived from negative lysates and negative-control material indicate internal control amplification.
Freshly reconstituted master mix.
Ten lysates were incubated with frozen master mix from each storage interval.
Twenty replicates each of control material frozen for 1 week, 3 weeks, and 5 weeks.
Significantly different (P = 0.02) from the value for freshly reconstituted positive control.
Significantly different (P ≤ 0.009) from the value for freshly reconstituted negative control.
A total of 502 lysates from 251 clinical specimens were prospectively tested in tandem utilizing the master mix paradigms. Paired results from two lysates were excluded from further analysis because of at least one final unresolved result within the tandem pair. Of the remaining 500 evaluable results, 491 (98.2%) demonstrated concordant PCR results between freshly reconstituted and frozen master mix (4 and 5 lysates tested positive via only freshly reconstituted and frozen master mix, respectively). Initial unresolved rates for both master mix systems were 2.99%; these rates decreased to ≤0.4% following a single freeze-thaw cycle.
After EIA and PCR (package insert protocol) were performed on 251 primary clinical specimens, 250 data pairs could be analyzed (1 final unresolved PCR result); 80.8% of the results by the two methods were concordant (Table 2). Following adjudication by toxigenic culture, Cdiff assay performance characteristics (package insert protocol) exceeded those of EIA in this population of 15.9% C. difficile toxin prevalence (40 true-positive specimens). Deficiencies in both EIA sensitivity and specificity translated into an EIA positive predictive value of 45.3% (Table 2). The mean OD reading from specimens yielding false-positive EIA results was 1.395 (median, 1.018; range, 0.081 to 3.249).
Table 2.
Comparison of Clostridium difficile toxin A/B enzyme immunoassay performance characteristics with those of the BD GeneOhm Cdiff assay processed via the package insert protocol or via modified PCR methodsa
| Performance characteristic | Toxin A/B EIAb | BD GeneOhm Cdiff assay |
P valued | |||
|---|---|---|---|---|---|---|
| Same-day aliquot |
Overnight aliquot |
|||||
| Fresh master mixc | Frozen master mix | Fresh master mix | Frozen master mix | |||
| Sensitivity (%) | 58.5 | 90.0 | 90.0 | 85.0 | 87.5 | ≥0.50 |
| Specificity (%) | 86.2 | 99.0 | 98.6 | 99.1 | 99.5 | ≥0.31 |
| Positive predictive value (%) | 45.3 | 94.7 | 92.3 | 94.4 | 97.2 | ≥0.34 |
| Negative predictive value (%) | 91.4 | 98.1 | 98.1 | 97.2 | 97.7 | ≥0.54 |
| Initial unresolved results (%) | NA | 3.19 | 3.98 | 2.79 | 2.00 | ≥0.19 |
| Final unresolved results (%) | NA | 0.40 | 0.80 | 0.00 | 0.00 | ≥0.16 |
| % agreemente | 80.8f | NA | 97.2f | 99.2f | 98.4f | ≥0.09 |
The final performance characteristic data were generated following adjudication of discordant results by toxigenic culture.
ProSpecT C. difficile toxin A/B microplate assay. NA, not applicable.
Package insert protocol (PCR reference method).
P values assess differences in PCR data generated from modified protocols compared to the BD GeneOhm Cdiff assay package insert protocol (PCR reference method).
Following repeat testing of initial unresolved specimens.
Compared to package insert PCR reference method.
PCR analysis of lysates from 251 same-day fecal aliquots with frozen master mix yielded 2 final unresolved results, allowing for analysis of tandem results from 249 lysates. A concordance of 97.2% (242/249) was observed between results derived from freshly reconstituted and frozen master mix. No significant differences in sensitivity, specificity, and predictive value (Table 2) were noted between the master mix systems (P ≥ 0.65 by significance test of proportions). Procurement of 251 aliquots that had been refrigerated overnight yielded 99.2% concordance for BD GeneOhm Cdiff assay analysis compared to the package insert protocol, translating into largely unchanged sensitivity, specificity, and predictive value indices (P ≥ 0.50; Table 2). Processed lysates from overnight aliquots (compared to EIA) demonstrated analogous ∼30% and ∼40% increases in sensitivity and positive predictive value, respectively, upon incubation in master mix frozen for 1 week (Table 2).
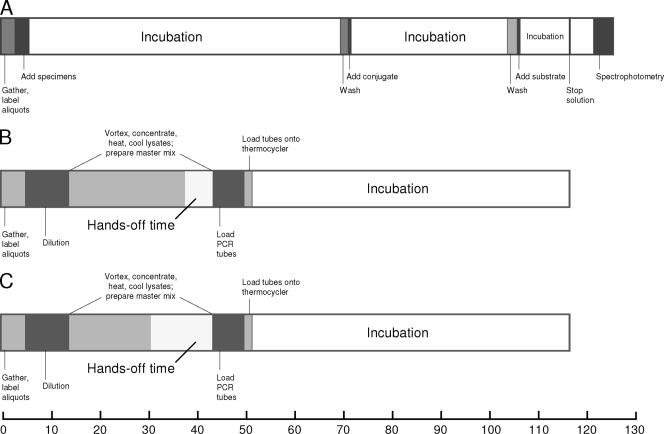
Thirty-eight batches of BD GeneOhm Cdiff assay reaction mixtures were processed throughout the investigation. The mean time to result (Fig. 1) per batch of PCR mixtures (116.92 min) was less than that of EIA (125.80 min; P = 0.003, t test for independent samples). Despite actual hands-on processing time per specimen being longer for PCR than EIA (P < 0.0001; data not illustrated), the frozen master mix modification decreased hands-on processing time by 15.4% (Fig. 1).
Fig. 1.
Illustration of timing and procedural aspects of the Clostridium difficile toxin A/B enzyme immunoassay (A) and BD GeneOhm Cdiff assay (B). (C) Frozen master mix modification impact on hands-on processing time. The x axis shows elapsed time (in minutes).
The advantages of toxigenic C. difficile PCR may be mitigated by financial considerations, particularly for laboratories with a limited testing menu. Because C. difficile testing recipients may largely fall into the paradigm of diagnosis-related group reimbursement, alternative economical C. difficile testing strategies, including two-step testing algorithms incorporating toxin and/or glutamate dehydrogenase (GDH) antigen detection, have been espoused. However, such algorithms incorporating GDH detection are confounded by potentially increased turnaround time and variable assay sensitivities (4, 5, 8, 13, 15–17) that have been attributed to geographical variation in toxigenic C. difficile ribotypes (5, 17) and differences in cytotoxicity reference standard (4).
The adoption of C. difficile PCR by smaller laboratories may be further limited by the composition of commercial kits. Previously reported hypothetical matrices for utilization of BD GeneOhm assays have demonstrated that as much as 23 to 45% of prepared master mix could go unused upon adherence to package insert guidelines (10–12). Our current data extend previous findings from other commercial PCR assays (10, 12) indicating that frozen master mix preparation does not compromise the performance of the BD GeneOhm Cdiff assay (Table 2). Frozen master mix has been used in other microbial and eukaryotic amplification assays (6, 7, 22). Temporal CT data associated with archival lysates and control material (Table 1) suggest that PCR amplification efficiency may be slightly enhanced with frozen master mix. While perhaps less clinically relevant in this venue, similar trends have been reported elsewhere (7, 12).
The logistics of molecular diagnostics may also be a limiting factor to clinical microbiology laboratories. At the same time, process simplicity associated with the highly sensitive random access Xpert C. difficile PCR assay (Cepheid, Sunnyvale, CA) may be offset by the >$30.00 reagent cost per test that may be quoted to some laboratories (17). Kvach et al. (8) cited a reagent cost per test for the manually processed BD GeneOhm Cdiff assay that was ∼20% less than the Xpert C. difficile quotation. This manual process demonstrates acceptable accuracy, as our data extend findings related to PCR (1, 4, 8, 15, 19, 20) and EIA (4, 8, 15, 18, 19). Sloan et al. (18) reported that one toxin EIA product had a proclivity for false-positive results, resulting in 46% positive predictive value. Our data, noteworthy because utilization of spectrophotometry was unlikely to contribute significantly to generation of false-positive results (median OD value of 1.018 for false-positive toxin EIA specimens), further describe shortcomings associated with C. difficile toxin EIA.
Moreover, we demonstrate that the BD GeneOhm Cdiff assay offers significant workflow benefits. From a laboratory perspective, final results (on a per-batch basis) are turned around faster than a commercial EIA. In addition, hands-on processing time can be reduced by utilization of frozen master mix (Fig. 1). Furthermore, preparation of refrigerated stool aliquots up to 1 day prior to PCR processing can be delegated to nonmolecular microbiologists. These enhancements not only assist in the workflow management of molecular diagnostic laboratories (including those that are entry level) but also augment the aforementioned economic considerations previously described. Taken together, the two modifications described in this report may allow laboratories to more advantageously utilize PCR technology for a real-time contribution to clinical management of Clostridium difficile infection.
Acknowledgments
We express our sincere gratitude to Janice Basile, Bette Berger, Jason Burtch, Janet Egan, Jeanne Klawa, Caitlyn Knapp, Timothy Kramme, Suzanne La Crosse, Cheryl Miller, Kimber Munson, Robin Olson, Maurice Perry, and Rachel Vogel for excellent technical assistance. Additional acknowledgment is given to Walt Earhart for assistance with the illustration.
Footnotes
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Barbut F., Braun M., Burghoffer B., Lalande V., Eckert C. 2009. Rapid detection of toxigenic strains of Clostridium difficile in diarrheal stools by real-time PCR. J. Clin. Microbiol. 47:1276–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bilbo D., et al. 2010. Modifications of a commercial Clostridium difficile PCR assay resulting in enhanced laboratory workflow efficiency, abstr. C-1128. Abstr. 110th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 3. Cohen S. H., et al. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455 [DOI] [PubMed] [Google Scholar]
- 4. Eastwood K., Else P., Charlett A., Wilcox M. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin and cytotoxigenic culture methods. J. Clin. Microbiol. 47:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gilligan P. H. 2008. Is a two-step glutamate dehydrogenase antigen-cytotoxicity neutralization assay algorithm superior to the Premier toxin A and B enzyme immunoassay for laboratory detection of Clostridium difficile? J. Clin. Microbiol. 46:1523–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoorfar J., Ahrens P., Rådström P. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kofler B., Klausegger A. 1999. Simplified PCR set-up using a frozen preformulated mix for the detection of cytomegalovirus. Diagn. Microbiol. Infect. Dis. 34:33–35 [DOI] [PubMed] [Google Scholar]
- 8. Kvach E. J., Ferguson D., Riska P. F., Landry M. L. 2010. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J. Clin. Microbiol. 48:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monteiro L., et al. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munson E., Block T., Voegeli J. T., Hryciuk J. E., Schell R. F. 2009. Cost-effective frozen master mix modification of a commercial methicillin-resistant Staphylococcus aureus PCR assay. J. Clin. Microbiol. 47:1888–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munson E., Culver A., Napierala M. 2010. Cost-effective frozen master mix modification of BD GeneOhm™ Cdiff assay, abstr. C-1127. Abstr. 110th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 12. Munson E., Kramme T., Culver A., Hryciuk J. E., Schell R. F. 2010. Cost-effective modification of a commercial PCR assay for detection of methicillin-resistant or -susceptible Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 48:1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Novak-Weekley S. M., et al. 2010. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J. Clin. Microbiol. 48:889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peterson L. R., Robicsek A. 2009. Does my patient have Clostridium difficile infection? Ann. Intern. Med. 151:176–179 [DOI] [PubMed] [Google Scholar]
- 15. Quinn C. D., et al. 2010. C. Diff Quik Chek complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J. Clin. Microbiol. 48:603–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reller M. E., et al. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 45:3601–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharp S. E., et al. 2010. Evaluation of the C.Diff Quik Chek Complete assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of Clostridium difficile disease. J. Clin. Microbiol. 48:2082–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sloan L. M., Duresko B. J., Gustafson D. R., Rosenblatt J. E. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stamper P. D., et al. 2009. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J. Clin. Microbiol. 47:373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Terhes G., Urbán E., Sóki J., Nacsa E., Nagy E. 2009. Comparison of a rapid molecular method, the BD GeneOhm Cdiff assay, to the most frequently used laboratory tests for detection of toxin-producing Clostridium difficile in diarrheal feces. J. Clin. Microbiol. 47:3478–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wernitz M. H., Keck S., Swidsinski S., Schulz S., Veit S. K. 2005. Cost analysis of a hospital-wide selective screening programme for methicillin-resistant Staphylococcus aureus (MRSA) carriers in the context of diagnosis related groups (DRG) payment. Clin. Microbiol. Infect. 11:466–471 [DOI] [PubMed] [Google Scholar]
- 22. West D. M., Sawyer J. 2006. Freezing complete polymerase chain reaction master mix reagents for routine molecular diagnostics. J. Vet. Diagn. Invest. 18:580–582 [DOI] [PubMed] [Google Scholar]