Abstract
Clostridium difficile is a common cause of infectious diarrhea in hospitalized patients. A severe and increased incidence of C. difficile infection (CDI) is associated predominantly with the NAP1 strain; however, the existence of other severe-disease-associated (SDA) strains and the extensive genetic diversity across C. difficile complicate reliable detection and diagnosis. Comparative genome analysis of 14 sequenced genomes, including those of a subset of NAP1 isolates, allowed the assessment of genetic diversity within and between strain types to identify DNA markers that are associated with severe disease. Comparative genome analysis of 14 isolates, including five publicly available strains, revealed that C. difficile has a core genome of 3.4 Mb, comprising ∼3,000 genes. Analysis of the core genome identified candidate DNA markers that were subsequently evaluated using a multistrain panel of 177 isolates, representing more than 50 pulsovars and 8 toxinotypes. A subset of 117 isolates from the panel had associated patient data that allowed assessment of an association between the DNA markers and severe CDI. We identified 20 candidate DNA markers for species-wide detection and 10,683 single nucleotide polymorphisms (SNPs) associated with the predominant SDA strain (NAP1). A species-wide detection candidate marker, the sspA gene, was found to be the same across 177 sequenced isolates and lacked significant similarity to those of other species. Candidate SNPs in genes CD1269 and CD1265 were found to associate more closely with disease severity than currently used diagnostic markers, as they were also present in the toxin A-negative and B-positive (A-B+) strain types. The genetic markers identified illustrate the potential of comparative genomics for the discovery of diagnostic DNA-based targets that are species specific or associated with multiple SDA strains.
INTRODUCTION
Clostridium difficile is the most common cause of infectious diarrhea in hospitalized patients in the industrialized world (3, 23). With symptoms ranging from self-limited diarrhea to life-threatening fulminant colitis, C. difficile infection (CDI) has affected hundreds of thousands of patients worldwide and substantially burdens health care resources (29). In particular, CDI has caused increased patient morbidity and mortality in hospitals throughout the world since 2003, when outbreaks with increased disease incidence and severity first emerged (62), and from 2004 to 2007 contributed to almost 1,000 deaths in the province of Quebec, Canada (16). Investigations of these and other outbreaks across Canada, the United States, and western Europe led to the recognition of a severe-disease-associated (SDA) strain predominantly responsible for this epidemic (27, 32). This strain has been classified as North American pulse field type 1 (NAP1), ribotype 027, toxinotype III, or restriction-endonuclease type BI (36), which we refer to here as NAP1. Outbreaks have also been associated with other SDA strains, such as the NAP7/toxinotype V/ribotype 078 (NAP7) strain found in cases of human and animal disease (17, 38) and multiple toxin A-negative B-positive (A-B+) pulsotypes and ribotypes responsible for CDI outbreaks in Ireland, the United Kingdom, the United States, and Canada (1, 58).
To date, the monitoring of C. difficile infection in a hospital setting has been a reactive process in which patients are tested after symptoms emerge, usually by employing methods that are time-consuming and expensive and/or lack sufficient sensitivity (11, 26). Improved tests are sought in which all at-risk patients can be tested in a rapid, reliable, and cost-effective manner (45). To address this need, DNA-based diagnostic tests have been previously developed (2, 55, 56, 63), but they rely primarily on targets from previously known genomic regions, such as the genes tcdC (55, 63) and gyrA and gyrB (56). The available tests include numerous commercially available assays (Xpect toxin A/B test, from Remel, Inc., Lenexa, KS; BD GeneOhm Cdiff assay, from BD Diagnostics, San Diego, CA; ProGastro Cd assay, from Prodesse Inc., Waukesha, WI; Cepheid Xpert C. difficile, from Cepheid, Sunnyvale, CA; and illumigene C. difficile, from Meridian Bioscience, Inc.).
With the advent of massively parallel DNA sequencing (MPS) technology (35), bacterial whole-genome sequencing has become rapid and affordable, and there are now sequences from well over 5,000 completed or in-progress microbial genomes available in public databases such as NCBI Entrez. MPS-based genome sequencing has been used in studies of C. difficile and other human pathogens, including a comparative genome analysis of 25 isolates that was performed to provide insight into the molecular evolution of C. difficile (20, 53) and a study of genome comparisons of 3 isolates that was used to identify potential virulence mechanisms in the NAP1 strain (59). These studies have confirmed the mobile nature of the C. difficile genome (54) and that genetic diversity among strains is high. In several studies (20, 22, 53), it has been shown that the NAP7-NAP8 strain type is highly divergent from other strain types and that the C. difficile core genome may be composed of only ∼1,000 genes (20, 22, 53, 58). To date, MPS and comparative genome analyses of C. difficile have not been applied to the search for additional DNA-based diagnostic targets, as has been done for other human pathogens (6, 14, 15, 28).
In this report, we describe the comparative analysis of the genomes of 14 isolates of C. difficile. The genomes of 9 isolates, including 6 eastern Canadian and 3 reference isolates, were sequenced as part of this study, and the genome sequences from 5 additional publicly available isolates were also used in the analyses. Our main objective was to identify DNA targets for use in potentially addressing two major clinical issues: (i) determining whether a given patient is infected with C. difficile and, if so, (ii) determining whether the patient is infected with a strain associated with severe disease. We thus identified DNA-based diagnostic sequences that could be used to detect any isolate of C. difficile, as well as targets able to discriminate between SDA and non-SDA strain types. We also used comparative genome analysis to study the genetic diversity between strain types, estimate the size of the C. difficile core genome, and begin to investigate the existence of additional loci responsible for virulence. Candidate targets identified in the 14-genome analysis were reconfirmed with a larger panel of 177 isolates. Clinical records available for 117 of these isolates were cross-referenced by the use of target alleles to ascertain their association with disease severity.
MATERIALS AND METHODS
Isolates for whole-genome sequencing.
Within the available hospital collections, six isolates (QCD-32g58, QCD-66c26, QCD-97b34, QCD-37x79, QCD-76w55, and CIP107932) of the predominant SDA strain of C. difficile (NAP1) were selected to emphasize variation across geographic locations and times of isolation. The remaining three of the nine isolates were a NAP2 strain (QCD-63q42), an SDA NAP7 strain (QCD-23m63), and reference strain VPI10463 (ATCC 43255). Isolates were incubated anaerobically at 37°C on 5% Columbia sheep's blood agar in pure culture and identified by standard phenotypic criteria. Genomic DNA from each isolate was extracted using standard commercial column-based extraction (QIAamp DNA Minikit; Qiagen, Mississauga, CA), stored in 1× Tris-EDTA (TE) buffer (pH 8.0), quantitated by spectrophotometry (SmartSpec 3000 UV visible spectrophotometer; Bio-Rad, Mississauga, CA), visualized by 1% agarose gel electrophoresis, and stored at −20°C until further use.
Whole-genome sequencing, gap closure, and comparative genome analysis.
All DNA extractions and sequencing experiments were performed at separate times to minimize the risk of cross-contamination. Isolate QCD-32g58 was sequenced using a first-generation Roche-454 GS system (GS20). The remaining isolates were sequenced later using a second-generation Roche-454 GS system (GS-FLX). Contigs (minimum size, 500 bases) were ordered and oriented based on alignment to the finished genome of C. difficile strain 630 (54). Contigs that could not to be ordered and oriented were placed into separate scaffolds. Assembly improvement was accomplished by designing primers flanking each gap and by performing conventional or long-range PCR (as required by predicted gap size) using the genomic DNA. Bidirectional sequencing of the resulting amplicons was performed with ABI 3730xl systems. Gaps were closed by aligning genomic and gap-directed amplicon sequences with Consed software (18). Genes were predicted using GLIMMER 3.01 software (7). Gene functions were predicted by aligning protein sequences using the NCBI nonredundant (nr) database (e-value < 1.0e-20). A whole-genome multiple alignment was created using Multiz-TBA software (4). DNA variations were catalogued when nucleotide quality (Q) was >63 and fell within alignment blocks of >80% pairwise identity present in all 14 genomes. The bootstrap consensus tree was inferred from 100 replicates and constructed using MEGA 4 software (60). The pairwise genome comparison of QCD-66c26 and QCD-23m63 was computed using lastz software (19), and a histogram showing percent identity was generated using a custom Python script.
Validation via targeted resequencing.
Within available hospital collections, 177 isolates of C. difficile were selected to assess genetic variation for each candidate locus within the NAP1 strain and between strains of various pulsed-field gel electrophoresis (PFGE) types. Of the 177 isolates, 170 (91%) were successfully PCR amplified and DNA sequenced. We also included the 9 isolates from the whole-genome sequencing as sequencing accuracy controls. We also included a species-level control (C. spiroforme [ATCC 29900]) and a phylum-level control (Mycobacterium intracellulare [kindly provided by Marcel Behr, McGill University Health Centre]). Genomic DNA was extracted from isolates by the use of standard lysis-based column extraction, and DNA yields were estimated by spectrophotometry. For each candidate locus, primers were designed using Primer3 software (51) to amplify regions of roughly 700 to 1,000 bp. PCR and bidirectional sequencing of the resulting amplicons were performed using an ABI 377 platform. Sequence chromatograms were base called using Phred software (12, 13), and polymorphisms (Q > 39) were catalogued on the basis of C. difficile VPI10463 analyses.
RESULTS
Genome sequencing and analysis.
We performed whole-genome sequencing and assembly for nine isolates of C. difficile (Table 1). Six isolates were of the predominant SDA strain type (NAP1), and collection dates ranged from 1984 to 2007. The international CIP 107932 reference isolate (isolated in 1984) and the BI-1 isolate (isolated in Minnesota in 1988 [47]) represented NAP1 isolates predating the CDI epidemic of 2003 to 2007. The other four NAP1 isolates (QCD-32g58, QCD-66c26, QCD-37x79, and QCD-97b34) were collected during the CDI epidemic from three locations across Canada. In addition to the six NAP1 isolates, we sequenced and assembled QCD-23m63, an SDA NAP7/toxinotype V/ribotype 078 (NAP7) isolate collected in 2007. The two non-SDA isolates sequenced were international reference strain VPI10463 (ATCC 43255) and a NAP2 isolate (QCD-63q42), collected in 1980 and 2005, respectively.
Table 1.
Characteristics of C. difficile isolates used in this study
| Isolate | Yr | PFGE type | Location | Source | Additional characteristic(s) | Genome size (Mb) | Contig no. |
|---|---|---|---|---|---|---|---|
| Isolates sequenced in this study | |||||||
| QCD-66c26 | 2007 | NAP1a | Montreal, Quebec, Canada | 56-yr-old male with severe CDI | BT+h; tcdC δ117; 18-bp deletion | 4.13 | 45 |
| QCD-32g58 | 2004 | NAP1 | Montreal, Quebec, Canada | 70-yr-old male with CDI | BT+; tcdC δ117; 18-bp del | 4.11 | 18 |
| BI-1c | 1988 | NAP1a | Minneapolis, MN | Nonepidemic strain | BT+; tcdC δ117; 18-bp del | 4.4 | 89 |
| CIP 107932 | 1984 | NAP1a | Reims, Marne, France | 28-yr-old female with PMCi | Reference strain for binary toxin | 4.04 | 69 |
| QCD-37x79 | 2005 | NAP1f | London, Ontario, Canada | 67-yr-old patient with severe CDI | BT+; tcdC δ117; 18-bp deletion | 4.33 | 59 |
| QCD-97b34 | 2004 | NAP1g | St. John's, Newfoundland, Canada | 70-yr-old with severe CDI | BT+; tcdC δ117; 18-bp deletion | 4.07 | 74 |
| QCD-63q42 | 2005 | NAP2 | Quebec, Quebec, Canada | 67-yr-old male with severe CDI | Toxinotype 0 | 4.44 | 87 |
| VPI 10463 | 1980b | Reference strain from ATCC | 4.21 | 88 | |||
| QCD-23m63 | 2007 | NAP7 | Montreal, Quebec, Canada | Male with severe CDI | Toxinotype V/ribotype 078 | 3.94 | 80 |
| Publicly available isolates | |||||||
| CD196 | 1985 | NAP1 | Paris, France | Nonepidemic strain | 4.11 | 1 | |
| R20291 | 2006e | NAP1 | Stoke Mandeville, England | Outbreak associated | 4.19 | 1 | |
| Strain 630 | 1980 | Zurich, Switzerland | CDI and PMC | 4.29 | 2 | ||
| NAP07 | 2008d | NAP7 | Unknown | Human feces | 3.90 | 33 | |
| NAP08 | 2008d | NAP8 | Unknown | Human feces | 4.08 | 24 |
Predicted from sequence similarity.
Estimated year of isolation.
N. Razaq et al. (47).
Estimated from date of sequencing.
Estimated from publication.
Subtype b/006.
Subtype a/001.
BT+, binary toxin positive.
PMC, pseudomembranous colitis.
We also used the publicly available genome assemblies of another five isolates, including two additional NAP1 isolates (R20291 and CD196 [59]), NAP7 and NAP8 isolates (Human Microbiome Project [HMP]), and strain 630 (54) (Table 1). Genome assembly details and the NCBI and GenBank accession numbers for all 14 isolates are given in Table 2.
Table 2.
Characteristics of C. difficile genome assemblies used in this study
| Isolate | Technology | Genome assembly status | Genome size (nt)a | No. of contigs | Largest contig (kb) | Contig N80 (kb) | NCBI or GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|---|
| Isolates sequenced in this study | ||||||||
| QCD-66c26 | GS-FLX | Draft | 4,126,050 | 32 | 937.3 | 232.5 | NZ_ABFD00000000 | This study |
| QCD-32g58 | GS-20 | Draft | 4,108,089 | 16 | 1,247.0 | 302.3 | NZ_AAML00000000 | This study |
| BI-1 | GS-FLX | Draft | 4,392,595 | 66 | 356.4 | 59.6 | NZ_ABHE00000000 | This study |
| CIP 1079324 | GS-FLX | Draft | 4,032,580 | 55 | 354.2 | 81.1 | NZ_ABKK00000000 | This study |
| QCD-37x79 | GS-FLX | Draft | 4,329,888 | 45 | 559.0 | 128.7 | NZ_ABHG00000000 | This study |
| QCD-97b34 | GS-FLX | Draft | 4,059,010 | 60 | 366.6 | 76.7 | NZ_ABHF00000000 | This study |
| QCD-63q42 | GS-FLX | Draft | 4,440,437 | 60 | 1,027.2 | 101.2 | NZ_ABHD00000000 | This study |
| VPI 104634 | GS-FLX | Draft | 4,204,780 | 55 | 1293.1 | 138.5 | NZ_ABKJ00000000 | This study |
| QCD-23m63 | GS-FLX | Draft | 3,936,085 | 61 | 440.0 | 93.8 | NZ_ABKL00000000 | This study |
| Publicly available isolates | ||||||||
| CD196 | GS-FLX | Complete | 4,110,554 | 1 | 4,110.5 | 4,110.5 | FN538970 | Stabler et al. (59) |
| R20291 | GS-20 | Complete | 4,191,339 | 1 | 4,191.3 | 4,191.3 | FN545816 | Stabler et al. (59) |
| Strain 630 | ABI377 | Complete | 4,290,252 | 1 | 4,290.2 | 4,290.2 | NC_009089 | Sebaihia et al. (54) |
| NAP07 | GS-FLX | Draft | 3,862,058 | 100 | 269.2 | 35.7 | NZ_ADVM00000000 | Human Microbiome Projectb |
| NAP08 | GS-FLX | Draft | 4,022,033 | 111 | 169.7 | 32.3 | NZ_ADNX00000000 | Human Microbiome Projectb |
nt, number of nucleotides.
The nine genomes sequenced in this study ranged in size from 3.94 Mb (QCD-23m63) to 4.44 Mb (QCD-63q42) (Table 2), which corresponds to the range of genome sizes (3.90 to 4.29 Mb) observed in other C. difficile genome sequencing projects (Table 2). We generated 9 draft genome assemblies in which the contig numbers in the assemblies ranged from 16 (QCD-32g58) to 66 (BI-1). However, the largest assembled contigs were >350 kb for all isolates, and the contig N80 calculation indicated that 80% of the assembled genomes was found in contigs of >59 kb (BI-1) to contigs of >300 kb (QCD-32g58). Given that C. difficile has a typical bacterial gene density (80 to 85%) and typical bacterial gene lengths (500 to 2,000 nucleotides [nt]), our genome assemblies provided contigs that could support synteny analyses combining sequence similarities in the context of information on gene order and orientation. Draft assemblies from the Human Microbiome Project also provided contigs of lengths (contig N80 > 30 kb) useful for gene annotation and the derivation of local synteny relationships.
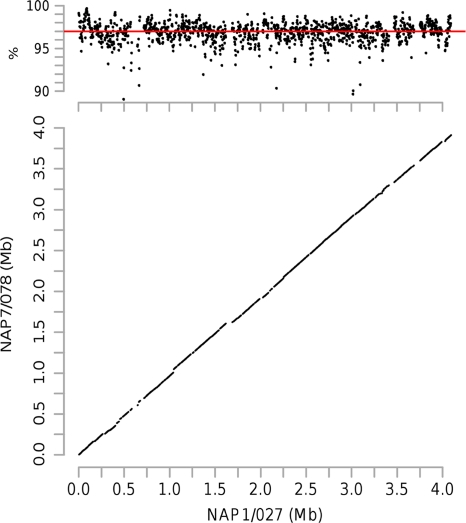
We used whole-genome alignments to determine that a core of 3.4 Mb of orthologous genomic sequence was present in all of the 14 genomes. Within the core genome, the NAP7-NAP8 group of isolates tended to be the most divergent of all the groups, with an average percent identity of 97% compared to the NAP1 group, although this included regions of lower percent identity embedded within the syntenic regions (Fig. 1). The noncore genome sequences represented strain- and isolate-specific insertions and deletions often due to mobile genetic elements and possible extrachromosomal plasmids (data not shown).
Fig. 1.
Percent identity plot (top) and dot plot (bottom) depicting the whole-genome pairwise alignments of a NAP1 isolate (QCD-66c26) and a NAP7 isolate (QCD-23m63). The dot plot in the bottom panel depicts the colinearity of the two genomes, and the percent identity plot in the upper panel depicts the levels of nucleotide-level similarity of the two genomes (the red line indicates average percent identity).
Polymorphism discovery.
Within the 3.4-Mb core genome, we identified 127,442 single nucleotide polymorphisms (SNPs). Although other types of genomic variation, including insertions and deletions, were present, they were not analyzed in this study, as they accounted for less than 3% (3,211) of all instances of nucleotide-level variations. A phylogenetic tree constructed using the SNPs clustered the isolates into three distinct groups: the eight NAP1 isolates, the three NAP7-NAP8 isolates, and the three remaining isolates (ATCC 43255, 630, and QCD-63q42), which we refer here to as the R group (Fig. 2a). Levels of genetic variations among these three groups are greater than the level of variation within any one group (Fig. 2b). The NAP1 isolates differ from the NAP7-NAP8 group at 104,853 SNP positions and differ from the three remaining isolates (R group) at 17,076 SNP positions. The NAP7-NAP8 isolates differ from the R group of isolates at 96,302 SNP positions.
Fig. 2.
(a) Phylogenetic tree of 14 C. difficile genomes constructed using SNP data. Branches corresponding to partitions reproduced in less than 50% of replicate bootstrap determinations are collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. Genomes cluster into 3 distinct groups: NAP1 isolates (red), NAP7-NAP8 isolates (blue), and the 3 remaining isolates (black [R group]). (b) Numbers of polymorphic SNPs observed for the three groups (large boxes), as well as variations observed between isolates within each group (remaining values). SNPs (text at center bottom) that uniquely identify the members of each group are also indicated.
The members of the NAP1 group consisting of 8 isolates are highly identical within the core genome, with identities for any two members ranging from 9 to 62 polymorphic SNP positions (Fig. 2b). We identified 11 nonsynonymous nucleotide substitutions that distinguished members of a subset of Canadian NAP1 isolates that consists of QCD-32g58 (isolated in Quebec in 2004), QCD-66c26 (Quebec, 2007), and QCD-37x79 (Ontario, 2005) as well as the United Kingdom outbreak strain R20291 (Stoke Mandeville) from the others, namely, QCD-97b34 (Newfoundland, Canada, 2004), BI-1 (Minnesota, 1988), CIP107932 (France, 1984), and CD196 (France, 1985). This set of 11 SNPs includes the previously described mutation responsible for resistance to fluoroquinolones (10).
The members of the group of three NAP7-NAP8 isolates are also highly similar to each other, with at most 1,851 SNPs separating QCD-23m63 (NAP 7) from the Human Microbiome Project (HMP) NAP07 isolate. However, the two HMP isolates are very similar, with only 297 SNPs. The number of variations distinguishing the three isolates in the R group is greater than the number of variations within the other two groups, with each of the three isolates in the group having over 11,000 SNPs.
Genetic variations consistently present in members of a single group are candidate DNA markers of potential diagnostic value. We found 10,683 SNPs that distinguish the NAP1 group from the other two groups (Fig. 2b). There were also 89,954 SNPs that separated the NAP7-NAP8 set from all others and 6,342 SNPs that separated the remainder (R group) from the NAP1 and NAP7-NAP8 groups.
The 10,683 SNPs that are specific to the NAP1 group have a genome-wide distribution (Fig. 3a) and include a prominent cluster overlapping the tcdB cytotoxin gene (Fig. 3a). Among the other prominent clusters, 545 SNPs were observed in a 5.8-kb region of 6 genes (CD1265 to CD1270), annotated as a two-component system (TCS) adjacent to an ABC transporter (TCS-ABC) (Fig. 3b). The predicted protein sequences across this TCS-ABC locus showed no premature stop codons, providing preliminary evidence that these remain functional genes despite the increased level of genetic variation. Other prominent clusters include an ABC transporter (CD0336 to CD0337), and a hypothetical protein adjacent to a transcriptional regulator (CD3143 to CD3144).
Fig. 3.
(a) Genome-wide distribution of SNPs that uniquely identify the NAP1 group of isolates. Prominent clusters overlap known genes (e.g., tcdB in PaLoc) and unknown loci (indicated with arrows). (b) Genomic intervals of 5 loci with prominent clusters of NAP1 SNPs. From top to bottom: CD0366-CD0337, tcdB, TCS-ABC, and CD3143-CD3144. Annotations for each genomic interval are indicated as follows (from top to bottom): a scale, the genomic position, the pileup of NAP1 SNPs, and gene annotations (arrows indicate gene direction).
Identification and validation of targets for SDA strains.
To investigate the potential of NAP1 SNPs in the TCS-ABC locus as diagnostic targets associated with disease severity, we selected 2 candidate genes, CD1265 and CD1269, for comparison to 9 putative or known virulence genes (tcdA, tcdB, tcdC, tcdE, codY, fliC, groEL, gyrB, and mviN). These genes were PCR amplified and resequenced using a panel of 177 isolates of C. difficile. Isolates were selected to assess genetic variation for each candidate locus within the NAP1 strain and between strains of various PFGE types. The 177 isolates were collected predominantly from Canadian hospital, provincial, and national reference collections and chosen to sample variations in PFGE types. The isolates were also selected to encompass different collection sites and years of isolation. A second set of international strains facilitated another survey of C. difficile diversity based on toxinotyping (52). A total of 20 contributing sites were represented, including 17 Canadian, 2 American, and one European institution. There were 163 isolates collected in or after 2002, with 22 isolated prior to 2002. Sixty-seven isolates were classified as NAP1, 12 as NAP2, and 109 into more than 50 other pulsovars. Ten isolates were from the reference panel of toxinotyping strains, and 50 were from 9 hospitals in the province of Quebec, Canada. A total of 117 isolates were obtained from The Canadian Nosocomial Infection Surveillance Program (CNISP) and had available patient data. Seventy of the 117 clinical records reported severe CDI, which was defined as death associated with CDI or intensive care units or colectomy due to CDI (all measured at least 30 days after diagnosis).
Our set included 10 A-B+ isolates, all of which were of PFGE type 00065 (NAP9; personal communication) and correlated with toxinotype VIII (22). In the patient data available for 100 of the isolates that were resequenced, SNP genotypes in two genes (CD1269 and CD1265) were seen to be associated with 70% (44/63) of severe-disease cases (comprising the three PFGE types NAP1, NAP7, and NAP9), whereas analysis by existing typing methods, such as PFGE NAP1 classification or determination of deletions in tcdC, accounted for 52% (33/63) or 62% (39/63), respectively, and was limited to two PFGE types (NAP1 and NAP7). For example, the G allele for the SNP at genomic location 1,387,666 in CD1269 (Fig. 4) was associated with more severe-disease cases than the single base deletion at position 117 (Δ117) in tcdC, as revealed by the observation that the A-B+ strains have the G allele but lack the tcdC deletion. In contrast, analysis focusing on the Δ117 deletion in tcdC detected 33 of 63 severe-disease cases, and the larger deletions of at least 18 bp in the 3′ end of tcdC accounted for 39 of 63 of severe-disease cases. Deletions in tcdC were diagnostic markers for two of three strain types previously associated with severe disease (NAP1 and NAP7), whereas analysis of the genotypes for each of 18 SNPs in CD1269 (1/18) and CD1265 (17/18) (the TCS-ABC locus) detected 44 of 63 severe-disease cases and specific alleles were associated with three SDA strain types (NAP1, NAP7, and A-B+ [NAP9/toxinotype VIII]) (Fig. 4). In addition, 5 of the SNPS in CD1265 are triallelic and partition the SDA strains into two groups (NAP1 versus NAP7 and A-B+ [NAP9/toxinotype VIII]) (Fig. 4). For example, the SNP at genomic location 1,383,226 has three alleles, C, T, and A. The A allele is associated with the NAP1 strain, the T allele with the NAP7 and A-B+ strains, and the C allele with all the others.
Fig. 4.
Correlation of disease severity with SNPs from the TCS-ABC locus or with existing diagnostic methodologies. The incidence of severe CDI outcome (SDA column [gray boxes]) was higher for the NAP1 strain and occurred in 7/10 and 2/2 cases for the A-B+ and NAP7 strains (plus one closely related strain, 11ACD0028), respectively (genome-sequenced isolates were not phenotyped and are indicated with an “x”). Molecular markers such as the PFGE type of strain NAP1 and the presence of binary toxin are diagnostic markers for NAP1, with deletions in tcdC additionally capturing the NAP7 strain. Genotypes of SNPs identified in the TCS-ABC locus are identifiers for three SDA strain types (the NAP1 strain, NAP7, and A-B+ strains) and include 5 triallelic SNPs (the third allele is boxed).
Identification and validation of targets for species-level detection.
We selected a set of 12 genes (spoIIIAG, CD0596, CD0117, CD3014, CD0279, CD1795, CD2251, CD0017, cdd1, srlE, prdB, and sspA) that displayed a high level of nucleotide identity across all 14 sequenced genomes as species-wide detection candidates (Table 3). After the panel of 177 isolates was resequenced, sspA was found to be 100% conserved at the nucleotide level for all isolates analyzed (Table 3). The remaining 11 candidates contained a few polymorphisms and did not deviate substantially from what was originally observed in the 14-genome analysis (Table 3). To further investigate whether these gene sequences could be used as DNA-based markers for specific detection of C. difficile (regardless of strain type) and yet remain specific to C. difficile and to no other species, we aligned the genomic sequence from each candidate gene region to sequences in the NCBI nonredundant DNA database. Database hits for all 12 gene regions were well below 90% identity and/or 90% query coverage (Table 3). Furthermore, experiments performed using C. difficile PCR primers for all 12 genes did not lead to amplification products from Clostridium spiroforme and Mycobacterium intracellulare (data not shown).
Table 3.
Targets for species-level detection of C. difficile
| Gene or locus | Number of SNPs |
Most similar species by BLASTn analysis (nr) | Query coverage (%) | Maximum identity (%) | |
|---|---|---|---|---|---|
| 14 genomes | Resequencing result | ||||
| sspA | 0 | 0 | Alkaliphilus oremlandii OhILAs | 94 | 74 |
| prdB | 2 | 5 | Clostridium sticklandii | 96 | 76 |
| CD0017 | 2 | 6 | Clostridium botulinum E3 strain Alaska E43 | 83 | 73 |
| cdd1 | 2 | 6 | Leptospira interrogans serovar lai strain 56601 | 39 | 74 |
| srlE | 6 | 6 | Clostridium botulinum B strain Eklund 17B | 97 | 83 |
| CD2251 | 3 | 8 | Trichomonas vaginalis | 13 | 85 |
| CD1795 | 4 | 9 | Polistes sp. MD1 mitochondrion | 16 | 84 |
| CD0596 | 6 | 10 | Brassica rapa subsp. Pekinensis | 22 | 81 |
| CD0117 | 6 | 10 | Alkaliphilus metalliredigens QYMF | 96 | 76 |
| CD3014 | 6 | 10 | Listeria welshimeri serovar 6b strain SLCC5334 | 96 | 86 |
| CD0279 | 8 | 10 | Clostridium acetobutylicum ATCC 824 | 45 | 67 |
| spoIIIAG | 8 | 14 | Mycoplasma mycoides subsp. mycoides SC strain PG1 | 8 | 84 |
DISCUSSION
The primary objectives of this study was to identify DNA markers for C. difficile that could be used to test stool samples of patients and to determine (i) whether they are infected with C. difficile and, if they are infected, (ii) whether the particular strain is associated with severe disease. Our strain selection, which was influenced by the availability of isolates in well-characterized hospital collections, such as the Canadian Nosocomial Infection Surveillance Program, included an emphasis on isolates of the predominant epidemic strain (NAP1) as well as an isolate of another SDA strain (NAP7) previously reported in the literature (17, 38). Our strain choices comprised the majority of SDA strain types (21, 32, 46, 61) as well as two widely used research reference isolates (CIP107932 and VPI10463). Our testing of an extended panel of isolates, from an even wider diversity of strain types than those described here, further demonstrated that we have analyzed C. difficile representing a wide spectrum of naturally occurring genetic diversity.
In agreement with previous studies, we observed variations in genome size that are largely attributable to mobile genetic element activity (20, 53, 54, 59). Mobile genetic elements in C. difficile have been found to carry virulence factors (59); therefore, a more detailed comparative genome analysis of the NAP1 isolates from this study and others might provide further insight into the differential virulence characteristics observed within this strain. We observed 3.4 Mb of conserved sequence present in all 14 genomes, comprising 3,063 genes, a determination that differs substantially from observations in other studies, which estimated a smaller core genome of less than 1,000 genes (20, 22, 53, 58). We do not believe that this is a reflection of strain choices, as our most divergent pairs (NAP1 versus NAP7) were also recognized in other studies as being the most highly divergent (20). Rather, we believe the differences in interpretations of core genome size are due to differences in methodologies and analysis techniques. Given the high quality of the 3 completed genomes and 11 draft genomes used in our analyses, we were able to combine sequence comparisons and syntenic relationships to determine gene orthologs and observed that, compared to NAP1 references, the members of the NAP7-NAP8 group displayed a higher level of genetic diversity, with an average of 97% identity. We believe that in comparative genome hybridization (CGH) studies, for example, where probe mismatches can affect hybridization (41, 49) and where NAP1 or strain 630 genomes have been used as the reference, even a very low number of mismatches between the probe and target DNA can increase the false-negative rate (34, 58). This leads us to suggest that the C. difficile CGH arrays, which were designed using either strain 630 (R group) or NAP1 isolate QCD-32g58, may produce numerous false-negative hybridizations when tested using more a more distantly related strain (e.g., NAP7) and have resulted in the calculation of a smaller core genome size. Other whole-genome sequence-based studies have used differing analytical techniques to determine the core genome size. For example, one study determined the core genome to consist of 622 genes, but only after the researchers stringently considered the nonrecombining genes in the genome (20). Another study estimated the core genome to be comprised of 947 genes (53); however, as it was a gene-centered analysis based on sequence identity without synteny evaluation, that study may have classified genes of lower-than-average sequence identity, such as those present in the NAP7-NAP8 group, as nonorthologous. Our whole-genome multiple-alignment-based approach allowed the identification of orthologous regions of lower sequence identity which might not have been detectable using CGH arrays or might have been classified as below similarity thresholds by the use of a gene-centered approach.
At this time, our catalogue of genetic variation does not include insertions or deletions. Future work may reveal insertions or deletions in SDA strains in addition to those previously identified in tcdC (5, 32, 33, 36). Indeed, whereas deletions in tcdC have previously been hypothesized to lead to increased toxin production (33, 57, 62), findings from a recent study (39) indicated that deletions in tcdC do not predict the biological activity of the PaLoc toxin genes, providing further motivation to identify all sources of genetic variation within the C. difficile genomes that may correlate with disease severity.
There are 64 SNPs that discriminate isolates within the NAP1 strain type, including 14 SNPs that separate eastern Canadian isolates from the others, and these demonstrate the potential of massively parallel sequencing for identification of SNPs suitable for subsequent intrastrain typing and tracking. The SNPs that distinguish the NAP1 strain from all other strains show an uneven genome-wide distribution and cluster in known pathogenicity genes as well as in genes with currently unrecognized roles in CDI. The other genes with clusters of NAP1 SNPs include a general stress gene (CD2599), a carbon starvation gene (CD2600), and an ABC transporter. The most prominent cluster of SNPs that discriminate the epidemic strain was observed in a genetic locus consisting of a two-component system (CD1269 and CD1270) adjacent to an ABC transporter (CD1265 to CD1268). The adjacency of these two loci has been shown to be functionally relevant in Bacillus and is present in other low-GC-content Gram-positive bacteria, including C. difficile (25). The function of the TCS-ABC system has been investigated previously in Bacillus subtilis and includes detoxification of antimicrobial compounds (42). The function of any particular TCS-ABC system is primarily dictated by the protein domains present in the histidine kinase gene (25). Sequence analysis of the histidine kinase gene (CD1270) in this TCS-ABC system suggests that it plays a role in detoxification (data not shown). In silico analysis indicates that this locus is not comprised of pseudogenes and thus may have a functional role. Future experiments are needed to confirm the expression of genes in the TCS-ABC system and investigate their roles in CDI.
Enzyme-based strategies for the detection of C. difficile have limited sensitivity and specificity (11). To address this, alternative gene targets, such as the tpi housekeeping gene, have been investigated (8). While this assay is specific to C. difficile, it requires the additional procedure of restriction fragment length polymorphism (RFLP) to achieve its specificity, making it less suitable in a clinical setting. Our genome analysis has identified numerous highly conserved genes that may be suitable for PCR-based specific detection of C. difficile. The 12 candidate genes displayed a high level of sequence conservation across the 14 genomes as well as a diverse population of 177 isolates, representing major PFGE types and toxinotypes. Moreover, our in silico analysis showed that many of these candidates have low sequence identity to other bacterial species and, upon further experimental validation and development, may define a more rapid and specific clinical detection assay.
The development of genetic markers to track severe-disease-causing strains has previously relied on variations in known pathogenicity genes, such as the detection of deletions in tcdC (63). While deletions in tcdC are diagnostic markers for the NAP1 strain and the SDA NAP7 strain, they are not diagnostic markers for the A-B+ isolates (NAP9/toxinotype VIII) that have been found in food animals (44) and retail meats (50). Using comparative genome analysis with candidate gene resequencing, we have identified numerous SNPs that detect these three SDA strain types, and this has in turn led to an increase of almost 20% in the detection of severe-disease-causing cases. However, strains containing these SNPs do not always lead to CDI, and severe CDI can be observed in cases of infection with non-SDA strains. Part of this phenomenon may be attributable to host factors that alter disease severity, such as variation in immune response (30, 31) and exposure to certain antibiotics (32, 40, 43) or acid-reducing agents (9, 32, 43). Alternatively, as our resequencing of candidate regions was limited, the resequencing of additional genes or loci may uncover SNPs that are markers for additional severe-disease-causing isolates. Also, severe disease or outbreak occurrences and numbers of outbreak infections may result from a constellation of genetic loci, such as those responsible for sporulation (37) or gut survival, and investigation of these other loci may identify SNPs that account for additional severe-disease cases.
This report demonstrates the utility of massively parallel DNA sequencing to identify clinically relevant diagnostic markers of C. difficile. As the cost of whole-genome sequencing continues to decrease, we envision this approach being applied to the study of other human pathogens.
ACKNOWLEDGMENTS
We thank the DNA sequencing teams at the Genome Center at Washington University School of Medicine and the McGill University and Genome Quebec Innovation Centre for the sequencing of the C. difficile isolates. We thank Manon Lorange for the isolation of C. difficile QCD-63q42. We also thank Marcel Behr for providing C. spiroforme and Mycobacterium intracellulare control DNAs. We thank the Human Microbiome Project (HMP) and the HMP DACC for prepublication data release of the C. difficile NAP07 (ADVM00000000.1) and NAP08 (ADNX00000000.1) sequences.
This study was funded by Genome Canada and Genome Quebec (K.D.) and the NHGRI (E.R.M.). V.F. was the recipient of a Canadian Institutes of Health Research Doctoral Research Award. M.T.O. was the recipient of an AMMI Canada/CIHR/CFID/Bayer Healthcare research fellowship.
Footnotes
Published ahead of print on 20 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. al-Barrak A., et al. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 25:65–69 [PubMed] [Google Scholar]
- 2. Barbut F., Braun M., Burghoffer B., Lalande V., Eckert C. 2009. Rapid detection of toxigenic strains of Clostridium difficile in diarrheal stools by real-time PCR. J. Clin. Microbiol. 47:1276–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbut F., et al. 1996. Prevalence and pathogenicity of Clostridium difficile in hospitalized patients. A French multicenter study. Arch. Intern. Med. 156:1449–1454 [PubMed] [Google Scholar]
- 4. Blanchette M., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curry S. R., et al. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45:215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai J., et al. 2011. Multiple-genome comparison reveals new loci for mycobacterium species identification. J. Clin. Microbiol. 49:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhalluin A., et al. 2003. Genotypic differentiation of twelve Clostridium species by polymorphism analysis of the triosephosphate isomerase (tpi) gene. Syst. Appl. Microbiol. 26:90–96 [DOI] [PubMed] [Google Scholar]
- 9. Dial S., Alrasadi K., Manoukian C., Huang A., Menzies D. 2004. Risk of Clostridium difficile diarrhea among hospital inpatients prescribed proton pump inhibitors: cohort and case-control studies. CMAJ 171:33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drudy D., Kyne L., O'Mahony R., Fanning S. A. 2007. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg. Infect. Dis. 13:504–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eastwood K., Else P., Charlett A., Wilcox M. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J. Clin. Microbiol. 47:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 13. Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 14. Feng Y., et al. 2011. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J. Clin. Microbiol. 49:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia Pelayo M. C., et al. 2009. A comprehensive survey of single nucleotide polymorphisms (SNPs) across Mycobacterium bovis strains and M. bovis BCG vaccine strains refines the genealogy and defines a minimal set of SNPs that separate virulent M. bovis strains and M. bovis BCG strains. Infect. Immun. 77:2230–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilca R., et al. 2008. Surveillance des diarrhées associées à Clostridium difficile au Québec: bilan du 22 août 2004 au 18 août 2007. Institut National de Santé Publique du Québec, Québec, Canada: http://www.inspq.qc.ca/pdf/publications/745_Cdifficile_bilan2004-2007.pdf [Google Scholar]
- 17. Goorhuis A., et al. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction Ribotype 078. Clin. Infect. Dis. 47:1162–1170 [DOI] [PubMed] [Google Scholar]
- 18. Gordon D. 2003. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinformatics 11.2.1–11.2.43 doi:10.1002/0471250953.bi1102s02 [DOI] [PubMed] [Google Scholar]
- 19. Harris R. S. 2007. Improved pairwise alignment of genomic DNA. Ph.D. thesis Pennsylvania State University, University Park, PA [Google Scholar]
- 20. He M., et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hubert B., et al. 2007. A portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of C. difficile-associated disease in Quebec. Clin. Infect. Dis. 44:238–244 [DOI] [PubMed] [Google Scholar]
- 22. Janvilisri T., et al. 2009. Microarray identification of Clostridium difficile core components and divergent regions associated with host origin. J. Bacteriol. 191:3881–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson S., Gerding D. N. 1998. Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 26:1027–1034 [DOI] [PubMed] [Google Scholar]
- 24. Jordan S., Hutchings M. I., Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 25. Joseph P., Fichant G., Quentin Y., Denizot F. 2002. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 4:503–513 [PubMed] [Google Scholar]
- 26. Killgore G., et al. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuijper E. J., Coignard B., Tull P. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12:2–18 [DOI] [PubMed] [Google Scholar]
- 28. Kuroda M., et al. 2010. Genome-wide single nucleotide polymorphism typing method for identification of Bacillus anthracis species and strains among B. cereus group species. J. Clin. Microbiol. 48:2821–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kyne L., Hamel M. B., Polavaram R., Kelly C. P. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346–353 [DOI] [PubMed] [Google Scholar]
- 30. Kyne L., Warny M., Qamar A., Kelly C. P. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357:189–193 [DOI] [PubMed] [Google Scholar]
- 31. Kyne L., Warny M., Qamar A., Kelly C. P. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 342:390–397 [DOI] [PubMed] [Google Scholar]
- 32. Loo V. G., et al. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 33. MacCannell D. R., et al. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 44:2147–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machado H. E., Renn S. C. 2010. A critical assessment of cross-species detection of gene duplicates using comparative genomic hybridization. BMC Genomics 11:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margulies M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDonald L. C., et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 37. Merrigan M., et al. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192:4904–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mulvey M. R., et al. 2010. Hypervirulent Clostridium difficile strains in hospitalized patients, Canada. Emerg. Infect. Dis. 16:678–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murray R., Boyd D., Levett P. N., Mulvey M. R., Alfa M. J. 2009. Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of toxin B or toxin A. BMC Infect. Dis. 9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muto C. A., et al. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26:273–280 [DOI] [PubMed] [Google Scholar]
- 41. Naiser T., Kayser J., Mai T., Michel W., Ott A. 2008. Position dependent mismatch discrimination on DNA microarrays—experiments and model. BMC Bioinformatics 9:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohki R., et al. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135–1144 [DOI] [PubMed] [Google Scholar]
- 43. Pépin J., et al. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254–1260 [DOI] [PubMed] [Google Scholar]
- 44. Pirs T., Ocepek M., Rupnik M. 2008. Isolation of Clostridium difficile from food animals in Slovenia. J. Med. Microbiol. 57:790–792 [DOI] [PubMed] [Google Scholar]
- 45. Planche T., et al. 2008. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect. Dis. 8:777–784 [DOI] [PubMed] [Google Scholar]
- 46. Quesada-Gómez C., et al. 2010. Emergence of Clostridium difficile NAP1 in Latin America. J. Clin. Microbiol. 48:669–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Razaq N., et al. 2007. Infection of hamsters with historical and epidemic BI types of Clostridium difficile. J. Infect. Dis. 196:1813–1819 [DOI] [PubMed] [Google Scholar]
- 48. Renn S. C., et al. 2010. Using comparative genomic hybridization to survey genomic sequence divergence across species: a proof-of-concept from Drosophila. BMC Genomics 11:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rennie C., et al. 2008. Strong position-dependent effects of sequence mismatches on signal ratios measured using long oligonucleotide microarrays. BMC Genomics 9:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodriguez-Palacios A., Staempfil H. R., Duffield T., Weese J. S. 2007. Clostridium difficile in retail ground meat, Canada. Emerg. Infect. Dis. 13:485–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rozen S., Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 52. Rupnik M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 32:541–555 [DOI] [PubMed] [Google Scholar]
- 53. Scaria J., et al. 2010. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One 5:e15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sebaihia M., et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature Genetics 38:779–786 [DOI] [PubMed] [Google Scholar]
- 55. Sloan L. M., Duresko B. J., Gustafson D. R., Rosenblatt J. E. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spigaglia P., Carattoli A., Barbanti F., Mastrantonio P. 2010. Detection of gyrA and gyrB mutations in Clostridium difficile isolates by real-time PCR. Mol. Cell Probes 24:61–67 [DOI] [PubMed] [Google Scholar]
- 57. Spigaglia P., Mastrantonio P. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stabler R. A., et al. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stabler R. A., et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 61. Walkty A., et al. 2010. Molecular characterization of moxifloxacin resistance from Canadian Clostridium difficile clinical isolates. Diagn. Microbiol. Infect. Dis. 66:419–424 [DOI] [PubMed] [Google Scholar]
- 62. Warny M., et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 63. Wolff D., Bruning T., Gerritzen A. 2009. Rapid detection of the Clostridium difficile ribotype 027 tcdC gene frame shift mutation at position 117 by real-time PCR and melt curve analysis. Eur. J. Clin. Microbiol. Infect. Dis. 28:959–962 [DOI] [PubMed] [Google Scholar]