Abstract
Amplified fragment length polymorphism (AFLP) was employed as a genetic analysis tool for the study of the genetic relatedness of Mycobacterium avium subsp. paratuberculosis isolates harvested from bovine fecal samples and from bovine or human tissues. This analysis revealed genetic differences between these two isolate types that were confirmed through cluster analysis. Dendrogram analysis separated these two isolate types based on the isolation scheme (tissue-associated versus fecal M. avium subsp. paratuberculosis isolates). Further sequence analysis of unique genetic regions from each isolation type revealed no genetic sequence differences. However, Clustal DNA alignments identified AFLP restriction enzyme sites that were undigested in the tissue-associated isolates. AFLP analysis also disclosed that the same AFLP restriction sites were digested in all of the fecal isolates. Sequence analysis further revealed a consensus sequence upstream of the undigested restriction sites for possible methyltransferase recognition in the tissue-associated M. avium subsp. paratuberculosis isolates.
INTRODUCTION
Mycobacterium avium subspecies paratuberculosis is the etiologic agent of a chronic granulomatous enteritis of ruminants known as Johne's disease (12). M. avium subsp. paratuberculosis has also been suspected to be involved in the chronic inflammatory bowel disorder in humans known as Crohn's disease (5, 10, 27). Although prevalent mostly in the bovine host species, this organism has been found to be an infectious agent of numerous mammals and birds alike (6, 9). M. avium subsp. paratuberculosis infections are transmitted through the fecal-oral route by contaminated feed sources, and human cases are suspected to be due to contaminated milk (28).
Many reports have examined the genetic differences and strain diversity among M. avium subsp. paratuberculosis isolates (1, 8, 29–31, 33, 35). These studies report that genetic differences exist between isolates from differing host species, locations, and strain types and that these genetic differences can range from single-nucleotide polymorphisms (SNP) to large genomic rearrangements. Previous data have confirmed that amplified fragment length polymorphism (AFLP) analysis is capable of detecting large polymorphic differences between isolates (29) and can also detect genetic differences due to SNPs at the AFLP restriction sites. Furthermore, the highly sensitive AFLP technique can distinguish between isolates with epigenetic differences at the AFLP restriction sites if the restriction site differences are due to methylation differences (36). Although this information does not allow for the identification of the methylated DNA base responsible for the polymorphism, it serves as a starting point for future epigenetic studies involving M. avium subsp. paratuberculosis isolates. The importance of this information has been demonstrated in multiple previous studies in which DNA methylation has been found to play a key role in the differential regulation of the corresponding gene products and to be able to influence the virulence of the bacterial organism (2, 4, 7, 14, 16–18, 20–22, 24).
In this study, AFLP profiles of M. avium subsp. paratuberculosis isolates from tissue-associated (isolation from infected tissue) and fecal (isolation from bovine fecal samples) sources were analyzed for the distinction of isolate-specific banding patterns. Bioinformatics tools were used to identify host-specific AFLP regions for genetic/epigenetic differences.
MATERIALS AND METHODS
Bacterial isolates.
Four human M. avium subsp. paratuberculosis isolates were obtained from the American Type Culture Collection (ATCC 49164, ATCC 43544, ATCC 43545, and ATCC 43015). Six fecal isolates from Texas Brahman cattle with clinical cases were obtained from Michael Collins (University of Wisconsin School of Veterinary Medicine [UWVM]): isolates T12, T136, T139, T140, T141, and T143. Two bovine M. avium subsp. paratuberculosis isolates were obtained from the ATCC: ATCC 19698 and ATCC 19851. All Texas Brahman cattle isolates and ATCC 19698 were recovered from fecal material (fecal isolates), while all human isolates were recovered from tissue (tissue associated), and ATCC 19851 was isolated from the head of an infected cow (tissue associated). All isolates were identified as M. avium subsp. paratuberculosis by culture requirements, colony morphology, and the presence of IS900 by PCR as described previously (29).
Amplified fragment length polymorphism.
AFLP analysis was performed with the PstI and MseI restriction enzymes as described previously (29) on all isolates except for isolate ATCC 19698, which was used in duplicate as an internal control. In this case, an ATCC 19698 bacterial broth culture was divided into two separate equal aliquots, and the entire AFLP procedure was performed separately on each aliquot.
Cluster analysis.
The BioNumerics computer analysis program, version 4.0 (Applied Maths, Austin, TX), was used for cluster analysis of AFLP-generated data. The Dice method of band scoring was used to create a composite data set from 4 independent AFLP primer sets in order to develop a summary dendrogram.
PCR confirmation of host-specific regions.
AFLP-generated regions unique to one isolation type were analyzed by agarose gel electrophoresis with ethidium bromide staining. Regions were excised, purified using the Qiagen (Valencia, CA) gel purification kit, and subsequently cloned into chemically competent Escherichia coli cells using the TOPO 2.1 vector according to the manufacturer's protocols (Invitrogen, Carlsbad, CA). Plasmid DNAs of positive transformants were purified with Qiagen's Miniprep kit according to the manufacturer's protocols and were subjected to sequence analysis (Lone Star Labs, Houston, TX). DNA sequence analysis of M. avium subsp. paratuberculosis regions consisted of queries in BLAST searches for sequence alignments against known genomes and for possible sequence homology to known organisms. Sequences were also aligned to the known genomic sequence of M. avium subsp. paratuberculosis K10, and PCR primers external to these regions were derived. PCR primers were designed to amplify regions flanking AFLP-derived regions and AFLP restriction sites for evidence of single-nucleotide polymorphisms (SNP). The PCR conditions used to amplify M. avium subsp. paratuberculosis regions with external primers were as follows: a 20-μl reaction mixture consisting of 100 ng each primer, 10 μl FailSafe PreMix G (Epicenter), 1 U Taq DNA polymerase (Epicenter Technologies, Madison, WI), and 7.6 μl DNA eluted from FTA storage cards (Whatman Inc., Clifton, NJ) according to the manufacturer's protocols. Thermocycler protocols were as follows: initial denaturation at 94°C for 5 min, followed by 45 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. A final elongation was carried out for 10 min at 72°C. Amplicons were viewed on 2% (wt/vol) SeaKem LE agarose gels with ethidium bromide staining.
Data mining of host-specific regions.
PCR products corresponding to isolation-specific regions were used as queries in BLAST searches of the NCBI database and for alignment to the known M. avium subsp. paratuberculosis K10 genomic sequence. Regions of interest were also aligned to each other using MacVector data analysis software in order to locate evidence of consensus sequences used for possible recognition sites for DNA methyltransferases.
AFLP analysis of experimentally infected bovines.
Male Holstein calves, 3 to 4 weeks old and weighing 45 to 55 kg, were fed milk replacer twice daily and water ad libitum. The calves were clinically healthy before the experiment. All experiments were performed under a protocol approved by the Texas A&M University Institutional Animal Use and Care Committee. Calves were fasted for 24 h prior to the nonsurvival surgery; then they were anesthetized and maintained analgesic for the course of the 12-h experiment. In brief, for the preparation and sampling of ligated loops of the ileum and jejunum, anesthesia was induced with propofol (Abbott Laboratories, Chicago, IL), followed by placement of an endotracheal tube and maintenance with isoflurane (Abbott Laboratories, Chicago, IL) for the duration of the experiment. The abdominal wall was clipped and prepared aseptically with chlorhexidine and isopropanol prior to opening. Sterile drapes were used as a barrier. The abdominal wall was opened, and the entire length of the Peyer's patch of the distal jejunum and ileum was exteriorized. The lumen of the ileum and the distal jejunum proximal to the Peyer's patch were flushed with saline to remove any remaining intestinal digest and were manually propelled into the cecum. Six- to 10-cm-long loops of the distal jejunum and ileum were ligated with umbilical tape, leaving a 1- to 2-cm-long area for the interloop. Control loops were injected with 3.0 ml of sterile phosphate-buffered saline (PBS), and infected loops were injected with 3.0 ml of sterile PBS containing 3 × 109 CFU of M. avium subsp. paratuberculosis ATCC 19698. The loops were placed back into the abdominal cavity, and the incision was temporarily closed with Backhaus towel clamps. Intravenous (i.v.) sterile 2.5% dextrose and 0.45% normal physiological saline were used to maintain the circulating blood volume at 5 ml/kg of body weight/h. At 0.5, 1, 2, 4, 8, and 12 h after bacterial inoculation, loops were excised. Samples for bacteriologic culture and DNA extraction were collected as described below. Electrocautery was used to control hemorrhage after the excision of the loops. Throughout the experimental procedure, the calves were monitored for vital signs (blood pressure, heart rate, hydration status, anesthesia depth, and temperature). The calves were euthanized with a rapid overdose (a single bolus at 60 mg/lb i.v.) of pentobarbital sodium after the final 12-h loops were excised.
A 6-mm-diameter biopsy punch was used to collect two tissue samples from Peyer's patches for bacteriology. Intestinal tissue samples were washed three times in PBS, weighed, homogenized in PBS, serially diluted, and plated onto Herrold's egg yolk medium containing amphotericin B, nalidixic acid, and vancomycin (ANV) (Becton Dickinson and Company, Sparks, MD) for incubation at 37°C. The cultures were observed visually weekly for any contamination, and the final CFU counts were recorded on week 16 (19).
The M. avium subsp. paratuberculosis colonies were selected and grown in 7H9 broth containing Mycobactin J, oleic acid-albumin-dextrose-catalase (OADC), and glycerol. DNA was prepared from this bacterial suspension for AFLP studies of host-passaged bacteria.
RESULTS
AFLP analysis of bovine and human isolates.
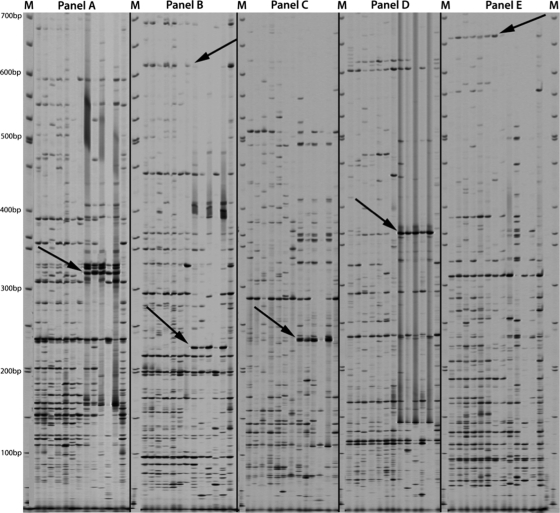
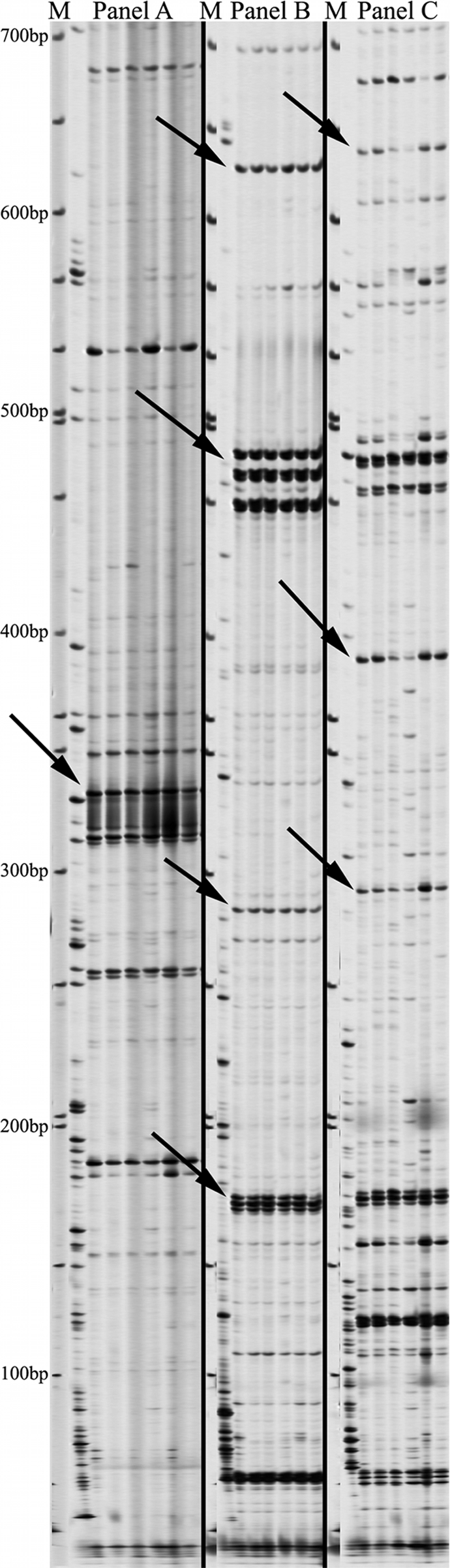
To determine the polymorphic differences between isolates of differing isolation types, AFLP analysis was performed on 4 human and 1 bovine tissue-associated M. avium subsp. paratuberculosis isolate from the ATCC, and the results were compared to those of AFLP analysis of 6 fecal isolates from Texas Brahman cattle. Duplications of ATCC 19698 were used for the normalization of the AFLP protocol. AFLP primers and adapters were used as described previously (29). By comparing these AFLP data, four regions common only to M. avium subsp. paratuberculosis isolates of tissue-associated origin (Fig. 1) were identified. These regions corresponded to particular primer combinations as follows: region 1, primers P-CG and M-AC (330 bp); region 2, primers P-GC and M-AC (220 bp); region 3, primers P-GG and M-AC (240 bp); region 4, primers P-GG and M-AG (365 bp). Along with these tissue-associated isolate-specific regions, two regions that exist only in isolates harvested from fecal material were identified (Fig. 1). These regions correspond to particular primer combinations as follows: region 5, primers P-GC and M-AC (620 bp); region 6, primers P-GG and M-AT (660 bp). The AFLP data suggested a genetic separation of M. avium subsp. paratuberculosis isolates based on colonization in tissue or shedding in the feces.
Fig. 1.
Composite AFLP gel comparison of fecal and tissue-associated M. avium subsp. paratuberculosis isolates. M, molecular weight markers. Isolates in each panel, from left to right, are as follows: fecal isolates T12, T136, T139, T140, T141, and T143; ATCC 19698 (internal control); tissue-associated isolates 19851, 43015, 43544, 43545, and 49164; duplicate internal control (ATCC 19698). (A) AFLP analysis with primers P-CG and M-AC. The arrow points to region 1 (330 bp), found only in tissue-associated isolates. (B) AFLP analysis with primers P-GC and M-AC. The top arrow points to region 5 (620 bp), present in fecal isolates only; the lower arrow points to region 2 (220 bp), present in tissue-associated isolates only. (C) AFLP analysis with primers P-GG and M-AC. The arrow points to region 3 (240 bp), present in tissue-associated isolates only. (D) AFLP analysis with primers P-GG and M-AG. The arrow points to region 4 (365 bp), present in tissue-associated isolates only. (E) AFLP analysis with primers P-GG and M-AT. The arrow points to region 6 (660 bp), present in fecal isolates only.
Cluster analysis.
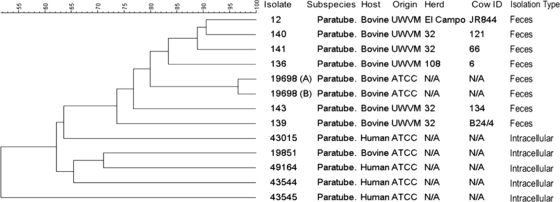
The overall genetic divergence of M. avium subsp. paratuberculosis fecal and tissue-associated isolates was determined by cluster analysis using BioNumerics data analysis software, version 4.0, employing the Dice method with the unweighted-pair group method with arithmetic means (UPGMA). A composite data set analyzing 4 independent AFLP primer sets for each isolate was used to create a dendrogram for the 6 fecal isolates, 5 tissue-associated isolates, and 2 internal controls (ATCC 19698) (Fig. 2). These data revealed a very tight (97% similarity) clustering of the ATCC 19698 internal-control isolates, confirming the very high reproducibility of the AFLP technique. The cluster analysis also revealed clustering of the fecal isolates, as well as clustering of the tissue-associated isolates. These data also provided evidence of multiple strains of M. avium subsp. paratuberculosis within a single bovine herd. Isolates 139, 140, 141, and 143 were all isolated from individual cows in herd 32 from Texas. The cluster analysis revealed a high (89%) similarity of isolates 140 and 141, but isolates 139 and 143 had only 74% similarity to each other and 77% similarity to the other herd 32 isolates, suggesting the possibility of 3 distinct M. avium subsp. paratuberculosis isolates within herd 32. The tissue-associated isolates clustered together despite their different origins (human or bovine).
Fig. 2.
Cluster analysis of AFLP data for fecal and tissue-associated M. avium subsp. paratuberculosis isolates. A comparative dendrogram of fecal and tissue-associated isolates is shown with corresponding percentages of similarity. The isolate number, subspecies designation, host species, origin of the sample, herd, cow identification number (ID), and isolation type are given on the right. N/A, not applicable.
PCR confirmation of isolation-specific regions.
Isolate-specific AFLP bands (regions 1 to 6) were sequenced as described in Materials and Methods. A primer set was designed for the specific region, and PCR analysis was performed, resulting in amplicons for all samples. In contrast to the procedure used in a previous report (29), primer pairs flanking the AFLP regions were created in order to sequence the entire region, including the restriction sites and flanking regions. Analysis of the sequence of each of these products revealed 100% sequence homology to the M. avium subsp. paratuberculosis strain K10 sequence. While this evidence was inconsistent with the differences observed by AFLP analysis, the possibilities of SNPs at the AFLP restriction sites and the genomic divergence of the isolates used in this study from the sequenced K10 genome were then investigated.
PCR results for numerous regions present only in tissue-associated isolates and regions present only in fecal isolates yielded identical amplicons for all isolates tested, regardless of origin. These data suggested that there was no sequence variation event in any of the regions tested. These corresponding PCR products were investigated using SNP analysis at AFLP restriction sites that could lead to different banding patterns on AFLP gels. Sequence analysis of all regions tested revealed no SNPs at either PstI or MseI restriction sites. These data suggested an unknown epigenetic trait as the cause of the AFLP differences observed between tissue-associated and fecal M. avium subsp. paratuberculosis isolates.
Data mining of isolation type-specific regions.
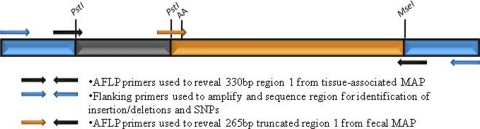
After the possibilities that insertions/deletions and single-nucleotide polymorphisms at restriction sites could account for AFLP differences between tissue-associated and feces-derived M. avium subsp. paratuberculosis isolates were eliminated, the sequences of these regions were analyzed. Upon further examination, two AFLP tissue-associated isolate-specific regions, region 1 (fructose-6-phosphate amidotransferase) and region 3 (a hypothetical protein), revealed a PstI restriction site internal to the AFLP-derived region. Sequence analysis of amplicons obtained from fecal isolates with these primer sets (for region 1, forward primer 5′-GCCCAGATCACCGCCAACTAC-3′ and reverse 5′-CGCATACTTCCTCCCGAACG-3′ were used to amplify a 744-bp product; for region 3, forward primer 5′-GCAACGATTGTCCCAAACCC-3′ and reverse primer 5′-ACAGCACCGACGACGCATTC-3′ were used to amplify a 474-bp product) revealed the same internal PstI sites. To determine if these internal PstI sites were digested in the fecal isolates during the AFLP process, AFLP primer sets were designed to amplify a smaller, truncated region in the fecal isolates, as indicated in Fig. 3.
Fig. 3.
Graphical representation of the internal PstI site of region 1. Sequence analysis of region 1 revealed an internal PstI site that was not digested in tissue-associated isolates on AFLP gels. New AFLP primers were designed with corresponding 3′ diadenine ends for amplification from fecal M. avium subsp. paratuberculosis (MAP) isolates.
AFLP amplification of M. avium subsp. paratuberculosis fecal and tissue-associated isolates with internally derived AFLP PstI primers with 3′ diadenine ends yielded amplicons of an appropriate size from all fecal isolates, while appropriate amplicons were absent from all tissue-associated isolates (Fig. 4). Corresponding amplicons from fecal isolates were extracted from gels and were subjected to sequencing as described above. Sequence analysis confirmed 100% homology with AFLP region 1 of tissue-associated origin. These data confirmed the presence of an internal PstI restriction site in all fecal and tissue-associated isolates of M. avium subsp. paratuberculosis tested, but this restriction site was digested by the AFLP process only in isolates of fecal origin. An explanation for this occurrence could be a difference in the methylation status of the PstI site between the fecal and tissue-associated groups. These methylation events can inhibit proper PstI digestion, yielding AFLP banding pattern differences.
Fig. 4.
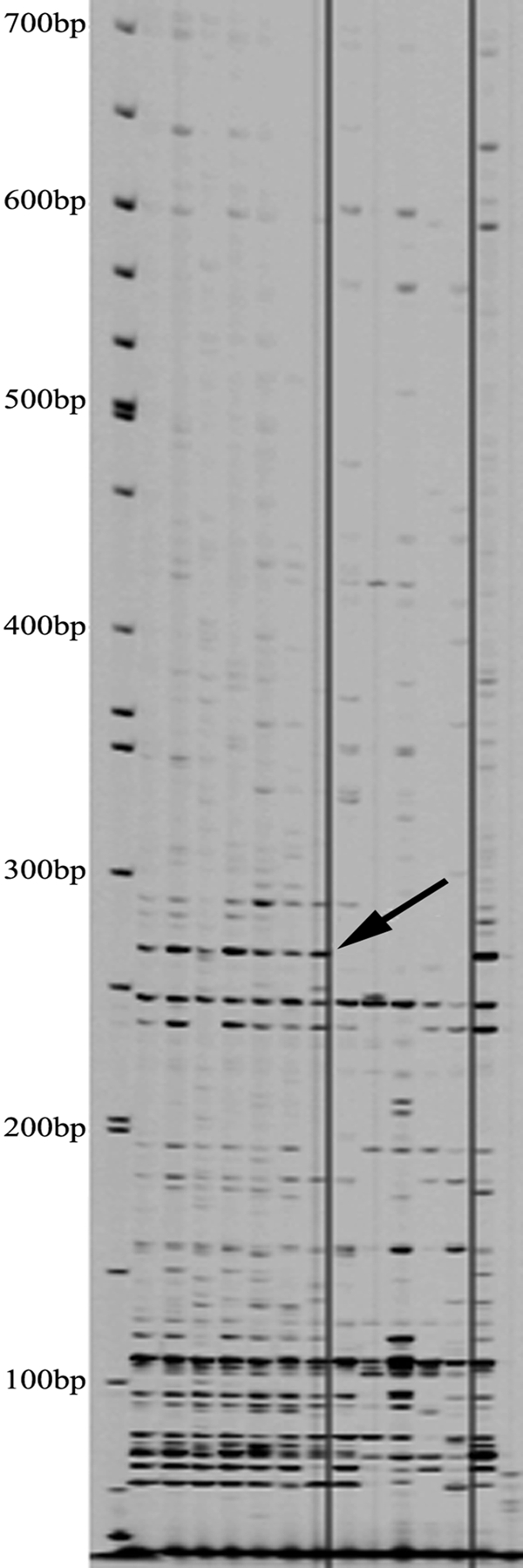
Methylation-specific AFLP gel. A primer set consisting of P-AA and M-AC was used. Isolates are as follows, from left to right: molecular weight marker, fecal isolates (T12, T136, T139, T140, T141, and T143), ATCC 19698, tissue-associated isolates (19851, 43015, 43544, 43545, and 49164), and ATCC 19698. The arrow indicates a truncated AFLP region present in all fecal isolates and absent from all tissue-associated isolates, corresponding to the 3′ end of region 1.
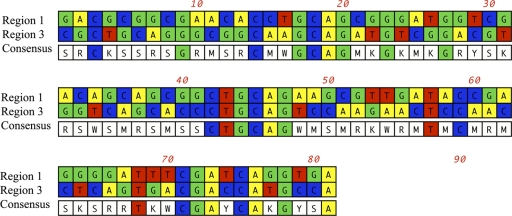
The internal PstI sites of regions 1 and 3 were investigated in detail in order to identify potential methylation sites that might act as inhibitors of restriction of the internal PstI sites in tissue-associated isolates. By aligning the internal PstI sites of regions 1 and 3, a consensus sequence 22 bp upstream of the internal PstI sites, common to both regions, was identified (Fig. 5). The positioning of consensus residues relative to the resistant PstI site suggested that these residues could serve as a binding site for a putative methylase.
Fig. 5.
Alignment of sequences of regions 1 and 3, with internal PstI sites aligned at position 42. Region 3 has an AFLP-digested PstI site at position 3. Region 1 has an AFLP-digested PstI site at position 15. The consensus sequence shows evidence of a 4-bp stretch common to both regions, starting at position 17. The internal AFLP PstI primer with a 3′ diadenine end is located at position 48 of region 1.
AFLP analysis of experimentally infected bovines.
To determine if host selective pressures could alter AFLP banding patterns, bovine ileal loop analysis was performed. ATCC 19698 was isolated from bovine ileal loops at 30 min and at 1, 2, 4, 8, and 12 h. DNA from these isolates was analyzed by AFLP, and the results were compared with those for the ATCC 19698 inoculum (Fig. 6). The AFLP banding patterns demonstrated clear differences between the inoculum and the tissue-associated isolates obtained at all time points. These banding pattern differences further supported our hypothesis of a host pressure-induced epigenetic trait, unique to tissue-associated M. avium subsp. paratuberculosis, which inhibits digestion by PstI, causing AFLP banding pattern differences. This was evident in that the inoculum isolate had a banding pattern different from those of the isolates recovered at later time points (Fig. 6A to C, arrows). Moreover, this finding was further supported by the fact that the tissue-associated isolates from all time points, each representing an independent DNA isolation, had banding patterns very similar to each other. These banding pattern differences were not likely caused by DNA rearrangement, due to the very short period within the host cell (30 min). All of the time points fall far short of the 24-h M. avium subsp. paratuberculosis generation time.
Fig. 6.
AFLP gel of tissue-associated isolates. The composite gel image shows results for tissue-associated isolates obtained at 30 min and at 1, 2, 4, 8, and 12 h. M, molecular weight markers. AFLP primers P-GC and M-CG (A), P-CT and M-GG (B), and P-CT and M-TG (C) were used. Bands within each panel, from left to right, represent the inoculum and isolates recovered at 30 min and at the 1-, 2-, 4-, 8-, and 12-h time points. Arrows indicate regions present in all tissue-associated time point isolates and absent from the inoculum.
DISCUSSION
In this study, M. avium subsp. paratuberculosis isolates that were isolated from tissue (tissue associated) and fecal samples were differentiated by the use of AFLP. Distinguishing genetic variations were evident upon examination of AFLP-amplified DNA and were unique to isolates of specific origins (tissue associated or fecal) (Fig. 1). These genetic variations were not confined to one isolation type of M. avium subsp. paratuberculosis; on the contrary, isolation-specific regions were found to be present in fecal and tissue-associated isolates alike. These data confirmed previously published results suggesting a differentiation among M. avium subsp. paratuberculosis isolates from different hosts (3, 13).
Since verification of the AFLP data suggested a genetic separation of M. avium subsp. paratuberculosis tissue-associated and fecal isolates, cluster analysis was performed. This analysis confirmed the AFLP observations that the banding patterns of fecal and tissue-associated isolates were genetically distinct. The cluster analysis demonstrated the reproducibility of AFLP by the clustering of the duplicate sample of ATCC 19698 to a similarity of 97%. It is also noteworthy that the ATCC 19698 isolate itself fell in the middle of the fecal cluster of isolates (Fig. 2). This was anticipated, because ATCC 19698 was a fecal isolate of M. avium subsp. paratuberculosis. Another important observation from the cluster analysis was the uniqueness of three different bovine M. avium subsp. paratuberculosis strains from herd 32. Cluster analysis revealed that isolates 140 and 141 clustered closely together, while isolates 139 and 143 diverged from one another, showing only 74% similarity to each other and 77% similarity to the other herd 32 isolates (Fig. 2). Taken together, these data suggested that herd 32 was infected with 3 strains of M. avium subsp. paratuberculosis. This observation correlates with the findings of Möbius et al., in which a number of genetically distinct M. avium subsp. paratuberculosis isolates were recovered from single herds in Germany (25). Dendrogram analysis further revealed a clustering of the human M. avium subsp. paratuberculosis isolates as a less similar group of isolates than the bovine isolates. These data were reasonable in view of the fact that the human isolates were recovered from regions all over the world over a period of decades, in contrast with the bovine isolates, which, with the exception of ATCC 19698, were isolated recently exclusively from herds in Texas. These data validated previous work in which multiplex PCR of IS900 integration loci (MPIL) revealed that human M. avium subsp. paratuberculosis isolates exhibit greater genetic diversity than bovine isolates (26).
AFLP analysis not only revealed the genetic diversity of M. avium subsp. paratuberculosis isolates from fecal and tissue-associated samples but also identified 6 genetic regions specific to one isolation type or the other. In this study, the genetic basis for the isolation-specific variations was investigated. Sequence analysis, PCR analysis, and SNP analysis of the tissue-associated isolate-specific regions revealed sequence homology to the M. avium subsp. paratuberculosis K10 isolate and no discernible difference from any fecal isolate. However, upon examination of regions 1 and 3, an internal PstI restriction site refractory to restriction during the AFLP process was apparent. Regions 1 and 3 corresponded to AFLP bands unique to tissue-associated isolates. Their apparent uniqueness was not, however, based on any differences in the DNA sequence. Therefore, we hypothesized that an epigenetic event such as DNA methylation at this internal PstI site might be a reason for the unique AFLP banding patterns. An AFLP primer with a 3′ diadenine end was created for the amplification of the truncated 3′ region of region 1 in fecal isolates (Fig. 3). These AFLP data revealed the presence of an amplicon corresponding in size to this truncated region in the fecal isolates that was absent from the tissue-associated isolates, suggesting the possibility of a tissue-associated isolate-specific methylation event of the internal PstI site of region 1, inhibiting AFLP digestion of tissue-associated isolate DNA. The idea that mycobacteria contain methylated DNA that will inhibit restriction enzymatic activity is not new. Although many studies have proven the existence of DNA methylation in mycobacteria, no study has proven where these methylation events take place or revealed the particular methyltransferases required for these events (15, 34).
Support for the hypothesis of tissue-associated isolate-specific methylation of the internal PstI site in region 1 was found in the presence of a consensus sequence 22 bp upstream of both internal PstI sites in regions 1 and 3. Although the methyltransferase has not yet been identified, a putative enzyme recognition sequence, 5′-GCAGnnGnnnGnnnnnnnnnnnnnnCTGCAG-3′, was observed. The presence of a methylase recognition sequence 22 bp upstream of the methylation site is not uncommon. For example, the restriction enzyme BsgI of Bacillus sphaericus recognizes a site 14 bp from the restriction site (5′-GTGCAGnnnnnnnnnnnnnnnn^-3′). Additionally, the restriction enzyme EcoP15I of E. coli P15 binds 25 bp upstream of the restriction site (5′-CAGCAGnnnnnnnnnnnnnnnnnnnnnnnnn^nn-3′). In the case of BsgI, as with the M. avium subsp. paratuberculosis site, the recognition sequence does not constitute a perfect palindrome. These examples demonstrate the possibility of a tissue-associated isolate-specific methylase as the explanation for the differences evident in the AFLP comparisons of the banding patterns of M. avium subsp. paratuberculosis tissue-associated and feces-derived isolates. Although the evidence of DNA methylation at these sites is somewhat circumstantial, investigations into the nature of DNA methylation and the specific methyltransferase involved are currently under way. The preliminary results of this research suggest a lack of 5-methyl-cytosine at these sites based on sodium bisulfite modification analysis. This is not surprising in view of previous reports of certain mycobacterial isolates lacking 5-methyl-cytosine residues and suggestions that N6-methyl-adenine is the predominant methylated DNA base in mycobacteria (34). Current measures for the detection and analysis of N6-methyl-adenine at these locations are under way.
It has been well established that DNA modifications can lead to the survival of bacteria and/or modifications in the virulence of bacteria (14, 17, 21, 23, 24, 32, 34). Although confirming evidence for a role of PstI methylation in the survival of tissue-associated isolates or the virulence of M. avium subsp. paratuberculosis has yet to be shown, the evidence for such an effect is accumulating. DNA methylation has been proven to play a role in the differences in transcription and therefore gene product levels in bacteria (11, 14, 17, 21). These data, coupled with the discovery of a consensus sequence that could be a recognition sequence for a DNA methyltransferase, support the contention that the methylation profiles of M. avium subsp. paratuberculosis tissue-associated and feces-derived isolates are unique and that these methylation patterns may influence the transcription levels of the methylated genes. This contention is also supported by the bovine ileal loop AFLP analysis, which revealed differences in banding patterns between tissue-associated isolates and the original inoculum. A possible explanation for our detection of these differences could be that the tissue-associated bacteria bearing the epigenetic modification were not detectable in the original inoculum but were selected during infection. An alternate explanation could be the presence of a tissue-associated isolate-specific methylation trait that is displayed only after the host cell internalizes the bacterium. Once the bacterium is internalized, the site is methylated, inhibiting enzymatic restriction of PstI and yielding the genetic differences observed by AFLP analysis in the M. avium subsp. paratuberculosis tissue-associated isolates. The latter explanation posits a host-dependent epigenetic trait, which would therefore be reversible. It is our opinion that the former is more likely the correct explanation. This is further supported by the fact that the epigenetic patterns are stable after the ileal loop subculturing, suggesting a state independent of host pressures. These data support the proposed interpretation that the epigenetic trait common to M. avium subsp. paratuberculosis tissue-associated isolates inhibits PstI digestion, and thus, the data support the presence of an epigenetic trait expressed solely in M. avium subsp. paratuberculosis tissue-associated bacteria.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Alexander D. C., Turenne C. Y., Behr M. A. 2009. Insertion and deletion events that define the pathogen Mycobacterium avium subsp. paratuberculosis. J. Bacteriol. 191:1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allamane S., et al. 2000. Bacterial DNA methylation and gene transfer efficiency. Biochem. Biophys. Res. Commun. 276:1261–1264 [DOI] [PubMed] [Google Scholar]
- 3. Amonsin A., et al. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bart A., van Passel M. W., van Amsterdam K., van der Ende A. 2005. Direct detection of methylation in genomic DNA. Nucleic Acids Res. 33:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barta Z., Csipo I., Mekkel G., Zeher M., Majoros L. 2004. Seroprevalence of Mycobacterium paratuberculosis in patients with Crohn's disease. J. Clin. Microbiol. 42:5432–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beard P. M., et al. 2001. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 39:1517–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casadesús J., Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castellanos E., et al. 2009. Discovery of stable and variable differences in the Mycobacterium avium subsp. paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl. Environ. Microbiol. 75:676–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corn J. L., Manning E. J., Sreevatsan S., Fischer J. R. 2005. Isolation of Mycobacterium avium subsp. paratuberculosis from free-ranging birds and mammals on livestock premises. Appl. Environ. Microbiol. 71:6963–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. François B., Krishnamoorthy R., Elion J. 1997. Comparative study of Mycobacterium paratuberculosis strains isolated from Crohn's disease and Johne's disease using restriction fragment length polymorphism and arbitrarily primed polymerase chain reaction. Epidemiol. Infect. 118:227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. García-Del Portillo F., Pucciarelli M. G., Casadesús J. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 96:11578–11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris N. B., Barletta R. G. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris N. B., Payeur J. B., Kapur V., Sreevatsan S. 2006. Short-sequence-repeat analysis of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium isolates collected from animals throughout the United States reveals both stability of loci and extensive diversity. J. Clin. Microbiol. 44:2970–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heithoff D. M., Sinsheimer R. L., Low D. A., Mahan M. J. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967–970 [DOI] [PubMed] [Google Scholar]
- 15. Hemavathy K. C., Nagaraja V. 1995. DNA methylation in mycobacteria: absence of methylation at GATC (Dam) and CCA/TGG (Dcm) sequences. FEMS Immunol. Med. Microbiol. 11:291–296 [DOI] [PubMed] [Google Scholar]
- 16. Herman J. G., Graff J. R., Myohanen S., Nelkin B. D., Baylin S. B. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U. S. A. 93:9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heusipp G., Falker S., Schmidt M. A. 2007. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297:1–7 [DOI] [PubMed] [Google Scholar]
- 18. Ivascu C., et al. 2007. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas. Int. J. Biochem. Cell Biol. 39:1523–1538 [DOI] [PubMed] [Google Scholar]
- 19. Khare S., et al. 2009. Early phase morphological lesions and transcriptional responses of bovine ileum infected with Mycobacterium avium subsp. paratuberculosis. Vet. Pathol. 46:717–728 [DOI] [PubMed] [Google Scholar]
- 20. Low D. A., Casadesús J. 2008. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11:106–112 [DOI] [PubMed] [Google Scholar]
- 21. Low D. A., Weyand N. J., Mahan M. J. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marinus M. G., Casadesús J. 2009. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33:488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mashhoon N., Pruss C., Carroll M., Johnson P., Reich N. 2006. Selective inhibitors of bacterial DNA adenine methyltransferases. J. Biomol. Screen. 11:497–510 [DOI] [PubMed] [Google Scholar]
- 24. Mehling S., Lavender H., Clegg S. 2007. A Dam methylation mutant of Klebsiella pneumoniae is partially attenuated. FEMS Microbiol. Lett. 268:187–193 [DOI] [PubMed] [Google Scholar]
- 25. Möbius P., Luyven G., Hotzel H., Kohler H. 2008. High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J. Clin. Microbiol. 46:972–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motiwala A. S., et al. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naser S. A., Ghobrial G., Romero C., Valentine J. F. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039–1044 [DOI] [PubMed] [Google Scholar]
- 28. O'Mahony J., Hill C. 2002. A real time PCR assay for the detection and quantitation of Mycobacterium avium subsp. paratuberculosis using SYBR Green and the Light Cycler. J. Microbiol. Methods 51:283–293 [DOI] [PubMed] [Google Scholar]
- 29. O'Shea B., et al. 2004. Amplified fragment length polymorphism reveals genomic variability among Mycobacterium avium subsp. paratuberculosis isolates. J. Clin. Microbiol. 42:3600–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paustian M. L., Kapur V., Bannantine J. P. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paustian M. L., et al. 2008. Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ratel D., Ravanat J. L., Berger F., Wion D. 2006. N6-methyladenine: the other methylated base of DNA. Bioessays 28:309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Semret M., et al. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Srivastava R., Gopinathan K. P., Ramakrishnan T. 1981. Deoxyribonucleic acid methylation in mycobacteria. J. Bacteriol. 148:716–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stevenson K., et al. 2009. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol. 9:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu M., Li X., Korban S. S. 2000. AFLP-based detection of DNA methylation. Plant Mol. Biol. Rep. 18:361–368 [Google Scholar]