Abstract
Leptospirosis is considered an underdiagnosed disease. Although several PCR-based methods are currently in use, there is little information on their comparability. In this study, four quantitative real-time PCR (qPCR) assays (SYBR green and TaqMan chemistries) targeting the secY, lfb1, and lipL32 genes were evaluated as diagnostic assays. In our hands, these assays can detect between 102 and 103 bacteria/ml of pure culture, whole-blood, plasma, and serum samples. In three independent experiments, we found a slightly higher sensitivity of the PCR assays in plasma than in whole blood and serum. We also evaluated the specificity of the PCR assays on reference Leptospira strains, including newly described Leptospira species, and clinical isolates. No amplification was detected for DNA obtained from saprophytic or intermediate Leptospira species. However, among the pathogens, we identified sequence polymorphisms in target genes that result in primer and probe mismatches and affect qPCR assay performance. In conclusion, most of these assays are sensitive and specific tools for routine diagnosis of leptospirosis. However, it is important to continually evaluate and, if necessary, modify the primers and/or probes used to ensure effective detection of the circulating Leptospira isolates.
INTRODUCTION
Leptospirosis is a bacterial zoonotic disease caused by pathogenic species of the genus Leptospira (13). The most recent estimates indicate that there are more than 500,000 annual cases of severe leptospirosis worldwide, with a much greater incidence in poor rural populations and urban slum settlements in tropical regions. This neglected disease is expected to become an increasingly important health problem due to predicted global climate changes (5, 12, 23) and expansion of urban slum populations (12, 18).
Leptospirosis is usually transmitted to humans by contact with water that is contaminated with the urine of animal reservoirs. Leptospires penetrate abraded skin or mucous membranes and disseminate into the organism. Infections in the early stage of the disease are similar to influenza-like illnesses and can lead to severe manifestations, such as Weil's disease and severe pulmonary hemorrhage syndrome, for which the fatality rate is more than 10% (15). Early diagnosis is essential because antibiotic treatment is most effective when it is initiated early in the course of the disease (13).
Bacteria are found in the bloodstream in the first few days after exposure. The septicemic phase, or leptospiremia, is followed by an immune phase which is characterized by the appearance of antibodies and the clearance of leptospires from the bloodstream. Culture isolation of causative organisms from biological fluids (blood, cerebrospinal fluid, or urine) takes several weeks, and antibodies can be detectable in the blood by serological methods approximately 1 week after the onset of symptoms. More recently, PCR-based methods have been developed for the diagnosis of leptospirosis (4).
In recent years, several real-time PCR assays have been described, and these assays are now used in many diagnostic and reference laboratories for the detection of leptospires in biological fluids of patients. PCR of blood samples can rapidly confirm the diagnosis in the early phase of the disease (within the first 2 weeks of exposure) and before antibody titers are at detectable levels. However, due to the small amount of leptospira present in blood samples, very sensitive diagnostic tests are required. In addition, the diversity of the genus Leptospira has expanded, with several new species, including pathogens, being described (4) since the first quantitative real-time PCR (qPCR) assays were developed (14, 17, 21). Finally, one of the critical factors for PCR detection is the DNA extraction from the clinical specimen. In this study, we compared the sensitivity and specificity of previously described PCR assays with both SYBR green and TaqMan chemistries (1, 17, 22), as well as the PCR performance of whole-blood, plasma, and serum specimens after DNA extraction with two commercialized kits.
MATERIALS AND METHODS
Strain and culture conditions.
The pathogens Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130, a clinical isolate obtained from a leptospirosis outbreak in Salvador, Brazil (10), and L. borgpetersenii serogroup Mini strain 200801773, isolated from a patient with acute leptospirosis in Mayotte, France (3), were used in this study. Leptospires were cultivated in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (6, 8). Additional reference strains (n = 58) were obtained from the collection maintained by the National Reference Laboratory for Leptospira, which is also a WHO Collaborating Center, at the Institut Pasteur (Paris, France). We also included clinical strains (n = 91) isolated from human and animals of different geographical origins (mainland France and French overseas Territories) since 2007. Other bacterial genomic DNA (Escherichia coli, Borrelia burgdorferi, Borrelia hermsii, Pseudomonas luteola, Plesiomonas shigelloides, Treponema denticola, and Salmonella enterica serovar Mbandaka) were also tested by qPCR assays.
Blood collection.
Blood was collected from healthy volunteers with no history of leptospirosis. Blood was drawn into Venosafe tubes containing EDTA (Terumo) and BD Vacutainer tubes (BD Diagnostics). Serum was recovered after centrifugation of the Vacutainer tubes at 3,000 rpm for 10 min. This study was part of a protocol approved by the Institut Pasteur (protocol RBM2008-16) and the French Minister for Higher Education & Research (protocol AC-2007-44).
Spiking experiments.
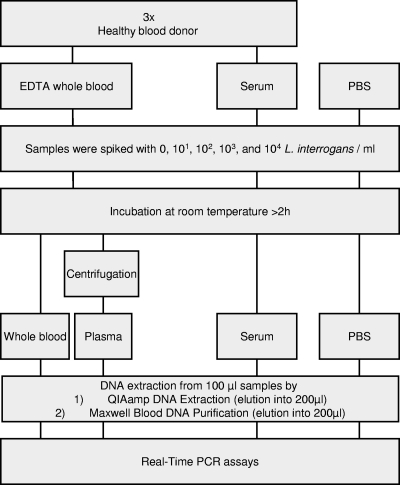
L. interrogans strain Fiocruz L1-130 was grown at 30°C until the optical density reached 0.3 at 420 nm. Exponential-phase bacteria were collected by centrifugation at room temperature and washed by resuspension in physiologically buffered water (PBW), pH 7.6, consisting of 1,840 ml of 0.85% NaCl and 160 ml of Sörensen buffer (0.83% Na2HPO4,12 H2O, and 0.11% KH2PO4). Suspensions of live leptospires in PBW were counted in a Petroff-Hausser counting chamber (Fisher Scientific) and adjusted to 2 × 108 bacteria/ml. Tenfold serial dilutions from 2 × 108 to 2 × 103 bacteria/ml were then performed in PBW. Blood samples were transported and processed for spiking experiments a few hours after collection from volunteers. Bacterial suspensions were spiked into 2 ml of whole blood and 1 ml of serum and PBW so that the final concentrations were 0, 101, 102, 103, and 104 bacteria/ml (see Fig. 2). After 2 h of incubation at room temperature, plasma was collected by removing the supernatant of 1.4 ml of a spiked whole-blood specimen after centrifugation at 1,500 rpm for 5 min.
Fig. 2.
Schematic representation of study design.
Nucleic acid extraction.
Nucleic acids were extracted from 100 μl of spiked whole blood, plasma, serum, or PBW by a Maxwell 16 instrument (Promega) for automated specimen processing by use of a Maxwell blood purification kit, according to the manufacturer's instructions (Promega), and using a QIAamp DNA blood minikit (Qiagen, Inc., Valencia, CA), with elution into 200 μl of 10 mM Tris-Cl–0.5 mM EDTA buffer (pH 9.0). Genomic DNAs were extracted from reference strains and clinical isolates from pure cultures by the Maxwell 16 instrument (Promega) using the Maxwell cell purification kit (Promega). PCR assays were evaluated on calibrated DNAs (2 to 10 ng/μl), except when indicated.
The concentration of DNA was determined spectrophotometrically by measurement of the absorbance at 260/280 nm using a Nanodrop 1000 apparatus (Thermo Fisher Scientific).
qPCR assays.
PCR assays were performed to amplify the lfb1 (17), secY (1), and lipL32 (22) genes of pathogenic Leptospira spp. (Table 1). The CFX96 real-time PCR detection system (Bio-Rad) was used for all qPCRs. SYBR green and TaqMan assays were optimized (primer/probe concentrations, annealing temperature, and incubation time) with the SsoFast EvaGreen supermix (Bio-Rad) and the SsoFast Probe supermix (Bio-Rad), respectively, according to the manufacturer's instructions, using 10-fold serial dilutions of L. interrogans and L. borgpetersenii DNAs. For SYBR green assays, the amplification mixture consisted of 0.4 μM primers (Table 1), 10 μl of SsoFast EvaGreen supermix (Bio-Rad), and 5 μl template DNA in a total volume of 20 μl. Samples were amplified with the following program: initial denaturation at 98°C for 2 min, followed by 40 cycles of denaturation for 5 s at 98°C and annealing/elongation for 30 s at 57°C. For lfb1 assays, PCR was followed by a melting curve analysis. For TaqMan assays, the amplification mixture consisted of 0.7 μM primers (Table 1), 0.15 μM probe, 10 μl of SsoFast Probe supermix (Bio-Rad), and 5 μl template DNA in a total volume of 20 μl. The program was initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation for 5 s at 95°C and annealing/elongation for 30 s at 58°C. In order to detect PCR inhibitors from blood sample DNA extraction, a qPCR assay specifically detecting the human housekeeping gene rnaseP was incorporated (22). All PCRs were run in duplicate, and control reactions without template were included in each assay.
Table 1.
Primers and probes used in this study
| Primer or probe | Sequence 5′–3′ | Targeta | Reference or source |
|---|---|---|---|
| Primers | |||
| LFB1-F | CATTCATGTTTCGAATCATTTCAAA | lfb1 (LA0322) | 17 |
| LFB1-R | GGCCCAAGTTCCTTCTAAAAG | lfb1 (LA0322) | 17 |
| SecYIVF | GCGATTCAGTTTAATCCTGC | secY (LA0759) | 1 |
| SecYIVR | GAGTTAGAGCTCAAATCTAAG | secY (LA0759) | 1 |
| secY-R | AGTTGAGCCCGCAGTTTTC | secY (LA0759) | 2 |
| secY-F | ATGCCGATCATTTTTGCTTC | secY (LA0759) | 2 |
| LipL32-45F | AAGCATTACCGCTTGTGGTG | lipL32 (LA2637) | 22 |
| LipL32-286R | GAACTCCCATTTCAGCGATT | lipL32 (LA2637) | 22 |
| LipL32-Rb | GAACTCCCATTTCAGCGAT | lipL32 (LA2637) | This study |
| RNAseP3F | CCAAGTGTGAGGGCTGAAAAG | rnaseP | 22 |
| RNAseP3R | TGTTGTGGCTGATGAACTATAAAAGG | rnaseP | 22 |
| Probes | |||
| LipL32-189P | FAM–AAAGCCAGGACAAGCGCCG–BHQ-1b | lipL32 (LA2637) | 22 |
| RNAseP3 | FAM–CCCCAGTCTCTGTCAGCACTCCCTTC–BHQ-1 | rnaseP | 22 |
The gene designation in L. interrogans serovar Lai strain Lai 56601 is given in parentheses.
FAM, 6-carboxyfluorescein; BHQ-1, black hole quencher 1.
Statistics.
Statistical analysis was performed using Prism software (version 5.0c; GraphPad Software, San Diego, CA). Significant differences between group values were determined by one-way analysis of variance (ANOVA) at a P value of ≤0.05.
RESULTS
Several PCR-based methods for detection of Leptospira have been reported in the last decade. This study assessed the relative performance of different quantitative assays (SYBR green and TaqMan chemistries) using different primer sets for the detection of DNA from Leptospira pathogenic species. qPCR-based detection assays targeting the pathogen-specific genes lfb1 (encoding a protein of unknown function) (17), lipL32 (encoding the major surface antigen of pathogenic Leptospira spp.) (22), and the housekeeping gene secY (encoding the preprotein translocase) (1) were compared. An alternative protocol for SYBR green quantification based on the TaqMan qPCR assay targeting the lipL32 gene (22) was also tested (Table 1).
Sensitivity of qPCR assays.
After optimization of the qPCR assays, we performed PCR on serial dilutions of genomic DNAs from L. interrogans and L. borgpetersenii, which are the two main causative agents of leptospirosis, to determine the detection sensitivity of the assays. Except for the failure to amplify secY from some L. borgpetersenii DNAs, all reactions were positive (Table 2). For each PCR assay, the highest dilution (100 bacteria/μl) still yielding a positive signal contained an average of one bacterial or five genome equivalents per PCR (Table 2). Although quantification of bacteria relative to genomic DNA mass is subject to sources of potential error, our data suggest that actively growing leptospires (DNAs were extracted from exponential-phase cultures) may contain multiple copies of the genome per cell, as previously observed in another spirochete (9). In an endpoint dilution experiment (10 fold-dilution of 5 bacteria per reaction mixture), the most sensitive assay was the SYBR green and TaqMan assays targeting lipL32; 7 out of 8 replicates and 4 out of 8 replicates for this dilution were positive by the SYBR green and TaqMan assays, respectively. Leptospiral DNA was amplified from 1 out of 8 diluted samples that were tested with the secY and lfb1 assays (data not shown).
Table 2.
Mean CT of 10-fold serial dilutions of L. interrogans and L. borgpetersenii of real-time PCR assays
| No. of leptospires/μl | DNA concn (ng/μl) | No. of gDNAb copies/μl | Mean CTa |
|||
|---|---|---|---|---|---|---|
| SYBR green |
TaqMan, lipL32 | |||||
| lfb1 | secY | lipL32 | ||||
| L. interrogans | ||||||
| 105 | 2.5 | 500,000 | 20.19 | 20.21 | 19.15 | 20.46 |
| 104 | 0.25 | 50,000 | 23.16 | 23.21 | 22.53 | 22.60 |
| 103 | 0.025 | 5,000 | 26.41 | 26.50 | 25.70 | 26.27 |
| 102 | 0.0025 | 500 | 30.09 | 30.30 | 29.49 | 28.94 |
| 101 | 0.00025 | 50 | 34.18 | 32.19 | 32.67 | 34.15 |
| 100 | 0.000025 | 5 | 36.82 | 38.50 | 38.37 | 36.45 |
| L. borgpetersenii | ||||||
| 105 | 2.5 | 500,000 | 19.92 | NA | 19.88 | 18.94 |
| 104 | 0.25 | 50,000 | 22.52 | NA | 22.92 | 22.02 |
| 103 | 0.025 | 5,000 | 26.11 | NA | 26.26 | 25.80 |
| 102 | 0.0025 | 500 | 29.34 | NA | 29.38 | 29.31 |
| 101 | 0.00025 | 50 | 33.28 | NA | 33.04 | 32.90 |
| 100 | 0.000025 | 5 | 36.81 | NA | 36.19 | 36.37 |
Mean of two independent experiments done in duplicate. NA, no amplification.
Based on the size of the genome of L. interrogans strain Fiocruz L1-130 (4.6 Mb); 1 genome is ∼5 fg. gDNA, genomic DNA.
Specificity of the qPCR assays.
Today, 20 species have been described in the genus Leptospira. This includes 9 pathogenic species (L. interrogans, L. kirschneri, L. kmetyi, L. borgpetersenii, L. santarosai, L. noguchii, L. weilii, L. alexanderi, and L. alstoni), 6 saprophytic species (L. biflexa, L. wolbachii, L. meyeri, L. vanthielii, L. terpstrae, and L. yanagawae), and 5 intermediate species (L. inadai, L. broomii, L. fainei, L. wolffii, and L. licerasiae), which are species of unclear pathogenicity forming a group distinct from pathogens and saprophytes by 16S rRNA sequence analysis (4).
We tested the specificities of the qPCR assays using reference strains from the 20 Leptospira species (Table 3). None of the saprophytic and intermediate strains tested by qPCR assay showed positive amplification. All 9 pathogenic species, except L. kmetyi, were amplified using the four qPCR assays. L. kmetyi, which was isolated from soil in Malaysia (20), is clustered to the pathogenic group using 16S rRNA analysis (4). The pathogen-specific genes lfb1 and lipL32 were amplified from L. kmetyi but not secY (Table 3). After amplification and sequencing with primers secY-R and secY-F (Table 1), we were able to amplify the secY gene from L. kmetyi. secY sequence analysis revealed four primer mismatches that could account for the absence of amplification (Fig. 1). No amplification was detected in DNA obtained from bacteria other than Leptospira.
Table 3.
Bacterial strains used for real-time PCR assaysa
| Species | Strain | qPCR assay result |
|||
|---|---|---|---|---|---|
| SYBR green |
TaqMan, lipL32 | ||||
| lfb1 | secY | lipL32 | |||
| Pathogens | |||||
| L. interrogans | Fiocruz L1-130 | + | + | + | + |
| L. kirschneri | Moskva V | + | + | + | + |
| L. noguchii | CZ 214 K | + | + | + | + |
| L. borgpetersenii | M84 | + | + | + | + |
| L. weilii | Celledoni | + | + | + | + |
| L. santarosai | LT821 | + | + | + | + |
| L. alexanderi | L 60 | + | + | + | + |
| L. alstoniib | 79601 | + | + | + | + |
| L. kmetyi | Bejo-Iso 9 | + | − | + | + |
| Intermediates | |||||
| L. wolffii | Khorat-H2 | − | − | − | − |
| L. licerasiae | VAR010 | − | − | − | − |
| L. inadai | LT64-68 | − | − | − | − |
| L. fainei | BUT6 | − | − | − | − |
| L. broomii | 5399 | − | − | − | − |
| Saprophytes | |||||
| L. wolbachii | CDC | − | − | − | − |
| L. meyeri | Veldrat | − | − | − | − |
| L. biflexa | Patoc 1 | − | − | − | − |
| L. vanthieliic | WaZ Holland | − | − | − | − |
| L. terpstraed | LT 11-33 | − | − | − | − |
| L. yanagawaee | Sao Paulo | − | − | − | − |
| Other bacteria | |||||
| B. burgdorferi | B31 | − | − | − | − |
| B. hermsii | HS1 | − | − | − | − |
| E. coli | XL10 | − | − | − | − |
| T. denticola | ATCC 35405 | − | − | − | − |
| P. luteola | LAM | − | − | − | − |
| P. shigelloides | 24-78 | − | − | − | − |
Results of qPCR assays with DNA extracted from pure cultures (>2 ng/μl) and pathogenic status of Leptospira spp. are indicated.
Genomospecies 1.
Genomospecies 3.
Genomospecies 4.
Genomospecies 5.
Fig. 1.
Nucleotide sequence polymorphisms of secY and lipL32. Alignment of secY (A) and lipL32 (B) nucleotide sequences of L. interrogans serovar Copenhageni strain Fiocruz L1-130 (FioL1-130), L. kmetyi strain Bejo-Iso 9 (L. kmetyi), L. borgpetersenii strain 200801773 (Lborg1773), and L. borgpetersenii strain 200801929 (Lborg1929). Primer and probe sequences are represented with a black background (L. interrogans strain Fiocruz L1-130 serves as the template DNA for the published sequences of primers and probe). Nucleotides with a white background within the black background indicate mismatched positions.
We also tested the detection ability of the qPCR assays for typing Leptospira strains on additional reference strains belonging to pathogens (see data in the supplemental material) and 91 clinical strains (including 44 L. interrogans isolates, 22 L. borgpetersenii isolates, 17 L. kirschneri isolates, 5 L. weilii isolates, and 3 L. santarosai isolates). PCR primers and probes targeting lfb1 and lipL32 were found to be specific for our clinical isolates (data not shown). However, in our hands, as previously observed (Table 2), the Leptospira-specific secY gene-targeted PCR with primers secYIVF and secYIVR was not capable of detecting three L. borgpetersenii isolates from Mayotte (Indian Ocean) (3). After amplification with another set of primers (Table 1), alignment of the secY sequences showed identical sequences between the three strains and revealed mismatches to both of the primer sequences. This includes two mismatches located at the 3′ end of the reverse primer that may interfere with amplification (Fig. 1A). Since secY is shared by all species of the genus Leptospira, including saprophytes and intermediates, the use of degenerated primers resulted in the amplification of some species other than pathogens (data not shown). By using secYIVF in combination with secY-R (instead of secYVIR), the pathogenic strains with negative results (L. kmetyi and the L. borgpetersenii isolates) had threshold cycle (CT) values of 20 to 22 (for 1 ng of DNA per reaction mixture). To further determine the specificity of the assay, genomic DNA of the 20 Leptospira species was tested. None of the nonpathogenic species was amplified by secYIVF and secY-R (data not shown).
Performance of the PCR assays was evaluated on calibrated DNAs (1 ng/μl) extracted from a set of clinical isolates from Mayotte which are representative members of new genotypes (3). Of them, the same three strains that did not amplify with the qPCR assay targeting secY showed mean CT values higher than those for other isolates by using the TaqMan assay targeting lipL32 (data not shown). Mismatches in the probe and/or primers may be the cause of this discrepancy. Strains which gave these abnormal CT results were further evaluated by sequencing analysis. There were two mistmatches in the reverse primer, including one mismatch located at the 5′ end of the primer that makes it likely to interfere with amplification; one mismatch in the 3′ end of the reverse primer; and two mismatches in the TaqMan probe (Fig. 1B). This may prevent optimal amplification and accurate quantification for these isolates. Those samples with discordant results had significantly lower CT values with primer LipL32-45F and reverse primer LipL32-Rb, which does not include the 3′ end nucleotide of primer LipL32-286R. We did not detect variations in target sequences of lfb1 primers among the sequenced PCR products (data not shown).
Species differentiation may be possible on the basis of melting temperature (Tm) variability among the lfb1 amplification products (17). For all L. interrogans strains (n = 70) except two, a single melting peak at 81°C (±0.5°C) was observed and demonstrated the ability to differentiate between L. interrogans, which is the most common pathogenic species, and other pathogenic species by melting curve analysis (see the data in the supplemental material). Two isolates of L. interrogans with discrepant results (serovar Kremastos strain 2414VB and serogroup Pyrogenes strain Nouméa 8) revealed a Tm of 83.5°C instead of 81°C. L. kirschneri isolates (n = 29) exhibited a Tm of 82.50°C (±0.5°C). Differentiation of L. interrogans and L. kirschneri with a peak Tm difference of 1°C may therefore be difficult. Other pathogenic Leptospira spp. (L. weilii, L. santarosai, L. kmetyi, L. alexanderi, and L. noguchii) exhibited higher Tms, but they may be difficult to distinguish among each other according to their melting temperatures (see the data in the supplemental material).
Detection and quantification of Leptospira in human blood samples.
To test the sensitivity of the qPCR assays for diagnosis, blood samples were isolated from healthy blood donors according to standard procedures, and these samples were artificially spiked with known concentrations of L. interrogans (Fig. 2). The aim of this study was also to evaluate commercially available DNA extraction kits, which should provide high DNA yields and reduce inhibition to improve the results that can be obtained with qPCR assays.
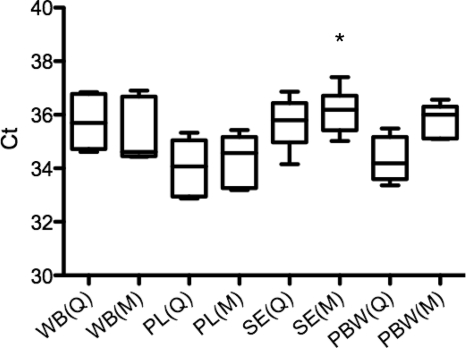
Total DNA was extracted using the Qiagen and Promega kits (manually and automatically, respectively) and quantified spectrophotometrically. These two methods exhibited similar performance in DNA extraction from whole blood (9.3 ± 0.65 ng/μl), plasma (2.45 ± 0.37 ng/μl), and serum (1.65 ± 0.37 ng/μl). The two kits similarly yielded pure and high-quality DNA (A260/A280 ratio > 1.7 for purified DNAs). Quantification of leptospires was assessed using the TaqMan probe targeting lipL32. For samples spiked with 103 L. interrogans bacteria/ml, the DNA extracted with the two kits produced positive results for all samples in each of the three experiments (Fig. 3). PBW spiked with leptospires had CT values (34.33 ± 0.85 and 35.77 ± 0.64) similar to the CT values obtained for spiked blood specimens (from 34.36 ± 0.97 to 36.19 ± 0.85), suggesting little inhibition of the assay by blood samples. Since the Promega kit used was designed for blood extraction (Maxwell blood DNA purification kit), this kit was less efficient in DNA purification from L. interrogans diluted in PBW than from L. interrogans in other samples (Fig. 3). One-way ANOVA analysis of the CT values for the spiked samples showed a significant inhibition of nucleic acid in serum specimens extracted by the Qiagen kit in comparison to the inhibition of other nucleic acid extracts prepared by any method (n = 6 for each sample, P < 0.05) (Fig. 3). For lower concentrations of 102 bacteria/ml, plasma was the only blood fraction from which DNA was extracted to produce positive results (data not shown).
Fig. 3.
Box plot of the mean CT for DNA extracted from whole blood, plasma, serum, and PBW spiked with 103 L. interrogans/ml. Bacteremia was assessed using qPCR assay with the TaqMan probe targeting lipL32. A one-way ANOVA (Dunnett's multiple comparison test; *, P < 0.05) was used to compare the mean CT of DNA extracted from whole blood (WB), plasma (PL), serum (SE), and PBW using the Qiagen (Q) and Maxwell (M) DNA purification kits. Each box represents the interquartile spread between the first and third quartiles (25th and 75th percentiles). The line inside the box is the median, and the lines extending from the box represent minimum and maximum values.
DISCUSSION
Leptospirosis is often diagnosed late, due to its wide spectrum of symptoms, ranging from a flu-like syndrome to renal failure, that mimic the clinical presentations of many other diseases, such as dengue and malaria. The diagnosis of leptospirosis is also challenging, as culture of Leptospira and seroconversion require weeks (4). qPCR assay is therefore highly useful for early diagnosis of leptospirosis. However, Leptospira is found in the bloodstream within the first week of illness and in relatively low numbers. It has been shown that a density of 104 leptospires/ml was a critical threshold for the vital prognosis of patients (19, 24). Blood samples also have to be collected by 2 days after the start of antibiotherapy, since antibiotics quickly remove Leptospira from the blood.
The sensitivity of the PCR assays has been determined in the studies described in the original publications (1, 17, 22). The limit of detection was repeatedly determined to be approximately 1 leptospire per PCR mixture for pure cultures (103 bacteria/ml). Under similar conditions, our lower limit of detection for each assay was comparable to those in these published studies. We then evaluated which of the blood fractions (whole blood, plasma, and serum) and nucleic acid extraction kits had the most efficient recovery for clinical samples. In our spiking experiments with blood samples, qPCR assays would allow specific and sensitive detection of 103 bacteria per ml. Leptospira spiked at 102 bacteria/ml was better detected in plasma, which corresponds to the fraction collected after centrifugation of the tube with whole blood. For DNA extraction from plasma, the use of both a larger sample volume (500 μl instead of 100 μl) and a smaller elution volume (50 μl instead of 200 μl), resulting in a 10-fold increase in DNA concentration, increased the likelihood of a positive result (data not shown).
Recent reports have already compared different blood fractions for detection of leptospiral DNA in clinical specimens (11, 22). A detection limit of 101 to 102 leptospires/ml of blood was usually determined (1, 22). Serum PCR was found to be less sensitive than PCR performed on other fractions for detection of Leptospira DNA (11, 22). In our hands, serum was also found not to be the best fraction for detecting leptospires, whatever the kit of DNA extraction used. Because leptospires can be detected in the clot, spiking experiments before clotting, which was not the case in our study, would decrease the number of leptospires in serum (22).
The most sensitive and specific qPCR assays for the detection of pathogenic Leptospira species are the SYBR green and TaqMan assays targeting lfb1 and lipL32, respectively. Because detection of Leptospira by rapid and less expensive methods than the TaqMan assay using SYBR green could be an important diagnostic tool, we also modified the TaqMan protocol targeting lipL32 established by Stoddard et al. (22). This SYBR green assay targeting lipL32 showed similar performance. The melting curve analysis of the assay targeting lfb1 provides useful additional information, as it can differentiate between L. interrogans and other pathogenic species (see the data in the supplemental material). Our results indicate that the qPCR assay targeting secY was not 100% specific for the detection of pathogenic Leptospira. The lack of sequence homology in newly described species and exotic isolates, such as some clinical isolates from Mayotte (3), can lead to false-negative results or decreased sensitivity. For example, mismatches located in the 3′-end region of primers, such as the one identified for the reverse primers of secY and lipL32 (Fig. 1), are well-known to be exceptionally detrimental to PCR priming. These mismatches are likely to be the cause of the underquantification of some strains in blood samples. This was confirmed by modifying the set of primers used for the amplification of secY and lipL32. The PCR assays may also not be suitable tools to screen for so-far-undescribed pathogenic strains. Failure to detect some isolates upon diagnosis can significantly impact patient care. Further sequence analysis of target genes of circulating Leptospira strains will need to be undertaken to evaluate the impact of nucleotide mismatches on PCR diagnosis. The redesign of the probes and primers should improve the sensitivity in detecting pathogenic Leptospira isolates.
To increase specificity for the saprophytes and the so-called intermediates, PCR assays targeting the 16S rRNA genes (7, 16) can also be used. This may facilitate the identification of previously undescribed species and/or strains unrelated to pathogens. Recently, intermediate species have thus been implicated in long-term renal carriage by asymptomatic individuals (7).
In conclusion, all four molecular assays are reliable methods for the detection and quantification of pathogenic Leptospira in clinical samples. The tests that show better performance on local isolates in the individual laboratory should be selected. Multicenter comparison should also be carried out to validate the usefulness of each test as a diagnostic tool. It is recommended that more than one technique be used to increase the diagnostic sensitivity and specificity; this can be performed by developing real-time multiplex PCR assays. The use of more than one target may be important to distinguish between true- and false-positive PCR results.
This study provides the first description of nucleotide mismatches in the currently used PCR assays and highlights that further sampling and sequencing of circulating Leptospira isolates are required to optimize the PCR for diagnosis of leptospirosis.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge L. Collet for the isolation of Leptospira isolates from Mayotte and the critical reading of the manuscript and M. Cornet, F. X. Weill, and J. C. Fenno for supplying non-Leptospira DNA. The Platform ICAReB (Investigation Clinique et Accès aux Ressources Biologiques) at the Institut Pasteur is thankfully acknowledged for providing blood samples.
This work was funded by the Institut Pasteur and the Institut de Veille Sanitaire (InVS) from the French Ministry of Health.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Ahmed A., Engelberts M. F., Boer K. R., Ahmed N., Hartskeerl R. A. 2009. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS One 4:e7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed N., et al. 2006. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bourhy P., et al. 2010. Isolation and characterization of new Leptospira genotypes from patients in Mayotte (Indian Ocean). PLoS Negl. Trop. Dis. 4:e724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cerqueira G. M., Picardeau M. 2009. A century of Leptospira strain typing. Infect. Genet. Evol. 9:760–768 [DOI] [PubMed] [Google Scholar]
- 5. Dufour B., Moutou F., Hattenberger A. M., Rodhain F. 2008. Global change: impact, management, risk approach and health measures—the case of Europe. Rev. Sci. Tech. 27:529–550 [PubMed] [Google Scholar]
- 6. Ellinghausen H. C., McCullough W. G. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45–51 [PubMed] [Google Scholar]
- 7. Ganoza C. A., et al. 2010. Asymptomatic renal colonization of humans in the Peruvian Amazon by Leptospira. PLoS Negl. Trop. Dis. 4:e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson R. C., Harris V. G. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 94:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitten T., Barbour A. G. 1992. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics 132:311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ko A. I., Galvao Reis M., Ribeiro Dourado C. M., Johnson W. D., Jr., Riley L. W. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354:820–825 [DOI] [PubMed] [Google Scholar]
- 11. Kositanont U., et al. 2007. Detection and differentiation between pathogenic and saprophytic Leptospira spp. by multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 57:117–122 [DOI] [PubMed] [Google Scholar]
- 12. Lau C. L., Smythe L. D., Craig S. B., Weinstein P. 2010. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 104:631–638 [DOI] [PubMed] [Google Scholar]
- 13. Levett P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levett P. N., et al. 2005. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 54:45–49 [DOI] [PubMed] [Google Scholar]
- 15. McBride A. J., Athanazio D. A., Reis M. G., Ko A. I. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18:376–386 [DOI] [PubMed] [Google Scholar]
- 16. Merien F., Amouriaux P., Perolat P., Baranton G., Saint Girons I. 1992. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J. Clin. Microbiol. 30:2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merien F., et al. 2005. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett. 249:139–147 [DOI] [PubMed] [Google Scholar]
- 18. Reis R. B., et al. 2008. Impact of environment and social gradient on leptospira infection in urban slums. PLoS Negl. Trop. Dis. 2:e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Segura E. R., et al. 2005. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin. Infect. Dis. 40:343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slack A. T., et al. 2009. Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int. J. Syst. Evol. Microbiol. 59:705–708 [DOI] [PubMed] [Google Scholar]
- 21. Smythe L. D., et al. 2002. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect. Dis. 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stoddard R. A., Gee J. E., Wilkins P. P., McCaustland K., Hoffmaster A. R. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 64:247–255 [DOI] [PubMed] [Google Scholar]
- 23. Storck C. H., Postic D., Lamaury I. P. J. M. 2008. Changes in epidemiology of leptospirosis in 2003-2004, a two El Niño Southern Oscillation period, Guadeloupe archipelago, French West Indies. Epidemiol. Infect. 136:1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Truccolo J., Serais O., Merien F., Perolat P. 2001. Following the course of human leptospirosis: evidence of a critical threshold for the vital prognosis using a quantitative PCR assay. FEMS Microbiol. Lett. 204:17–321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.