Abstract
We noticed that methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) isolates yielded side-scatter (SSC) and fluorescence intensity (FI) differences on flow cytometry (FCM) following incubation in oxacillin broth. The purpose of this study was to determine whether MRSA and MSSA could be reliably differentiated by FCM. S. aureus isolates were incubated in oxacillin-containing Mueller-Hinton broth, stained using the FASTEST total viable organisms kit, and analyzed by FCM in the MicroPRO instrument. SSC versus FI were examined, and gates 1 and 2 were defined to encompass the majority of MSSA and MRSA signal events, respectively. A count ratio (CR) was defined as the ratio of counts in gate 2 to those in gate 1. Initially, 33 isolates were tested after 4 h of incubation for proof-of-concept. Twenty others were then tested after incubation intervals ranging from 30 min to 4 h to determine the earliest possible time for differentiation. Next, 100 separate isolates were tested to determine the best CR cutoff value. Finally, the CR was validated by using an independent cohort of 121 isolates. We noted that MRSA isolates had higher SSC and FI readings than did MSSA isolates after 2 h of incubation. The receiver-operator characteristics curve showed that a CR cutoff of 0.0445 reliably differentiated MRSA from MSSA. In the validation cohort, this cutoff had a sensitivity of 100% and a specificity of 98.7% for identifying MRSA from among S. aureus isolates, following 2 h of incubation. This study demonstrates that MRSA and MSSA can be accurately differentiated by FCM after 2 h of incubation in an oxacillin-containing liquid culture medium.
INTRODUCTION
Several methods have been developed in recent years to detect Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) rapidly. Peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) allows for rapid detection of S. aureus (12) but cannot identify methicillin resistance in S. aureus. Real-time PCR can detect S. aureus and MRSA in as short a period of time as 2 h (16), but the fact that that most of the currently available formats requires batch testing ensures that in reality detection of S. aureus and MRSA by PCR is usually not as rapid as is commonly believed. The BD-GeneOhm assay for the detection of S. aureus and MRSA has been designed to target the insertion site of the staphylococcal cassette chromosome mec (SCCmec) for detection. The advantage of this approach is that it allows for MRSA detection by targeting a single site, thus avoiding the loss of sensitivity that might occur with multiplex PCR. A disadvantage is that mutations at the target site lead to false-negative results, and mutations in the mecA gene itself that result in nonexpression of the mecA gene or deletion of the mecA gene despite the presence of the SCCmec yield false-positive results for MRSA (4, 7). Indeed, surveillance studies for nasal carriage of MRSA in communities have yielded sensitivities and specificities of only ca. 90% (5). The advantage of phenotypic methods is that there is less potential for misclassification based on state of expression of genes. Phenotypic testing for susceptibility using automated systems still require at least 4 to 16 h to provide results. Latex agglutinating assays based on detecting the presence of penicillin-binding protein 2a (PBP2a) are rapid tests that can determine the presence of methicillin resistance when bacteria have been isolated in pure culture in the laboratory but have lower sensitivity with smaller numbers of bacteria (2, 3).
Flow cytometry using the MicroPRO flow cytometer (Advanced Analytical Technologies, Inc., Ames, IA) can count microorganisms accurately and can detect small numbers of microorganisms (17). We hypothesized that it would be possible to differentiate MRSA from MSSA using flow cytometry by comparing counts in paired broth cultures either containing or lacking oxacillin. We found this to indeed be the case, but it would take at least several hours to allow the distinction. Intriguingly, we also noticed that MRSA strains incubated in oxacillin-containing broth had different side-scatter and fluorescence intensity patterns from MRSA strains incubated in oxacillin-deficient culture broths. Similarly, there were differences noted between MRSA incubated in oxacillin-containing broth and MSSA strains regardless of whether or not oxacillin was present in the culture broth. We also noted that this pattern difference was evident at 4 h, long before there was any appreciable difference in cell counts.
We evaluate here whether MRSA and MSSA could be reliably differentiated based on side-scatter and fluorescence intensity patterns on flow cytometry and determined how rapidly this could be accomplished.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial isolates used in the present study were isolated from clinical specimens and identified as per the standard method in our laboratory, which was the Vitek II automated susceptibility system. Fresh overnight bacterial cultures were prepared in Columbia blood agar. An inoculum equivalent to 0.5 McFarland was prepared in saline. The inoculum was further diluted 1/20 in cation-adjusted Mueller-Hinton broth supplemented with 2% NaCl (CAMHB-NaCl) (11, 20). A portion (100 μl) of this diluted suspension was added to each of two tubes, one with 2 ml of CAMHB-NaCl containing 4 μg of oxacillin/ml and the other with 2 ml of CAMHB-NaCl without oxacillin, leaving final suspensions each with an estimated inoculum size of approximately 4 × 105 CFU/ml. These two suspensions were incubated at 37°C.
Staining protocol.
Viable bacterial cells were labeled by using the FASTEST total viable organism (TVO) kit (Advanced Analytical Technologies, Inc., Ames, IA) as previously described (17). The process involved incubation of the bacteria for 5 min with 100 μl of a proprietary fluorescent nucleic acid dye (excitation wavelength, ∼635 nm; emission spectrum, 665 to 735 nm), followed by incubation for 2 min with 100 μl of a proprietary compound (BRAG3) that can enter only membrane-compromised cells and decrease the fluorescence intensity of such cells, thus only allowing cells with intact membranes (viable cells) to be detected by their fluorescence emission. The processes of addition of reagents, incubation, and counting were automated in the MicroPRO flow cytometer.
Flow cytometry.
The MicroPRO machine was used for flow cytometry. Instrument characteristics include a sheath fluid flow rate of 16 to 19 ml/min, a sample flow rate of 0.1 ml/min, a red diode light source, and fluorescence detection by photomultiplier tubes (detection spectrum of 665 to 735 nm). The cytometer settings were set as follows: fluorescence threshold, 250; scatter threshold, 1; side scatter detector power supply, 70; and fluorescence detector power supply, 135. For analysis, 0.3 ml of broth culture was transferred to a 5-ml sample tube (Fisherbrand 14-956-1D), diluted 10-fold by addition of 2.7 ml of Tris-EDTA buffer, labeled with the FASTEST TVO kit as described above, and a cell count performed in the MicroPRO flow cytometer using a sampling volume of 0.25 ml.
Test of proof-of-concept.
A panel of 33 clinical isolates of S. aureus (17 methicillin susceptible and 16 methicillin resistant) was obtained from the Cleveland Clinic clinical microbiology laboratory. Microorganisms were analyzed by flow cytometry after 4 h of incubation in oxacillin-containing broth. Dot plots of side-scatter versus fluorescence intensity for all microorganisms in the panel were examined to define two overlapping gates, with gate 1 containing most of the signal events corresponding to MSSA and gate 2 corresponding to most of those corresponding to MRSA. A count ratio (CR) was defined as the ratio of signal event counts in gate 2 to that in gate 1. This was done to define a measurable numerical entity to describe signal event pattern differences.
Examining time to differentiation between MRSA and MSSA by flow cytometry.
When it was clear that the CR as crudely defined in the previous section could accurately differentiate MRSA from MSSA, we sought to determine the minimum amount of incubation time necessary. We used 20 clinical strains of S. aureus (10 MRSA and 10 MSSA) isolated from patient specimens for this evaluation. These were incubated in broth culture and analyzed by flow cytometry as described above, after the following specified durations of incubation: 0.25, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, and 4 h. Means of the total signal event count and the CR at each time point were compared between MRSA and MSSA groups.
Determination of cutoff value for CR.
It appeared from these studies that MRSA and MSSA might be reliably differentiated from each other using the CR as early as 1.5 and 2 h, so a larger sample of isolates was tested to determine the optimum duration of incubation and the best cutoff value for the CR. A sample of 100 isolates, which in turn consisted of 50 each of MRSA and MSSA isolates, were selected from clinical isolates in the Cleveland Clinic microbiology laboratory. These were isolates that had been identified as part of routine laboratory workup using the Vitek II automated susceptibility testing system. Each sample was analyzed by flow cytometry after 1.5 and 2 h of incubation. The CRs obtained were used to generate receiver-operating characteristic (ROC) curves for testing at 1.5 and 2 h.
Test validation using an independent sample.
After an appropriate duration of incubation prior to flow cytometry and the optimum cutoff value for the CR were determined from the ROC curves, a cohort of 121 clinical S. aureus isolates were tested. They were incubated for the duration determined to be most appropriate as described above and then analyzed by flow cytometry by using a predetermined cutoff value for the CR, as described in the previous section, to determine the sensitivity and specificity of this flow cytometric method for identification of MRSA among S. aureus isolates.
Discrepant analysis.
Discrepancies in identification of MRSA/MSSA by flow cytometry and automated susceptibility testing by Vitek were resolved by disk diffusion testing using cefoxitin disks, with a zone of inhibition ≤21 mm being considered resistant (11).
Statistical analysis.
The SigmaPlot 11.0 software (Systat Software Inc., San Jose, CA) was used to calculate the means and confidence intervals (CIs) of CRs of MRSA and MSSA at various time points for the cohort used to determine the earliest time the two could be reliably differentiated and to graphically depict these findings. The same software was also used to generate the ROC curves and calculate the associated areas and specificities and sensitivities.
RESULTS
Differentiation of MRSA and MSSA in the test of proof-of-concept.
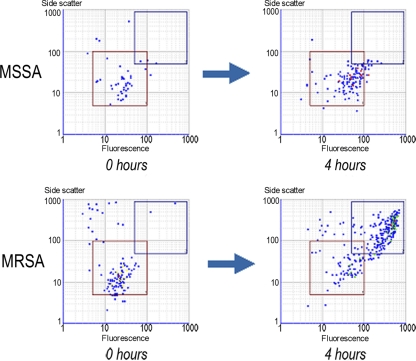
MRSA and MSSA could be differentiated with 100% accuracy using the CR in the small proof-of-concept sample when the two gates were defined as follows. For gate 1, the parameters were as follows: fluorescence intensity low, 5; fluorescence intensity high, 100; side scatter low, 5; and side scatter high, 100. For gate 2, the parameters were as follows: fluorescence intensity low, 50; fluorescence intensity high, 900; side scatter low, 50; and side scatter high, 900. Representative dot plots with diagrammatic representation of gates are shown (Fig. 1). The CRs for individual MRSA and MSSA isolates were determined. An arbitrary cutoff 0.1 accurately differentiated MRSA from MSSA. The results are presented in Table 1, suggesting that flow cytometry after a short period of incubation in oxacillin-containing broth could potentially differentiate MRSA from MSSA.
Fig. 1.
Representative dot plots of MRSA and MSSA strains on flow cytometry after incubation in oxacillin-containing broth for 4 h, with gate 1 (red) and gate 2 (blue) highlighting the differences in signal events (the x axis represents fluorescence intensity, and the y axis represents side scatter).
Table 1.
CRs for the sample of MRSA and MSSA isolates in the proof-of-concept study as tested by flow cytometry after 4 h of incubation in oxacillin-containing broth
| Group | No. of isolates |
||
|---|---|---|---|
| CR |
Total | ||
| ≥0.1 | <0.1 | ||
| MRSA | 16 | 0 | 16 |
| MSSA | 0 | 17 | 17 |
| Total | 16 | 17 | 33 |
Time to differentiation of MRSA and MSSA by flow cytometry.
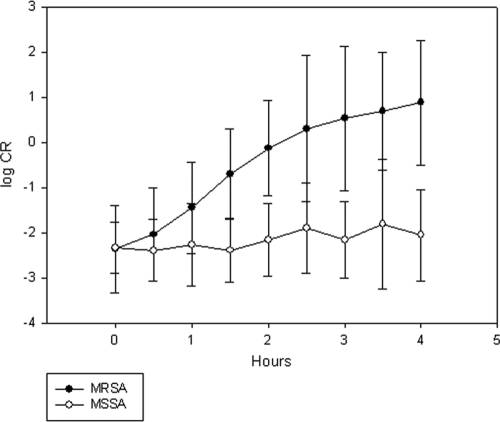
Total counts and CR (ratio of counts in gate 2 to those in gate 1) of 10 MRSA and MSSA isolates each were compared after various durations of incubation in oxacillin-containing broth. By up to 4 h of incubation there was no significant difference in the mean TVO counts for the MRSA and MSSA groups. The mean CR for the MRSA group, however, became increasingly greater than that of the MSSA group with increasing duration of incubation. Beyond 1.5 h of incubation there was clear separation of the mean CR between the MRSA and MSSA groups (Fig. 2).
Fig. 2.
Comparison of mean CR for 5 MRSA and 5 MSSA strains after increasing durations of incubation (log10 scale). Error bars represent 2 SD on each side of the mean.
Determination of cutoff value for the CR.
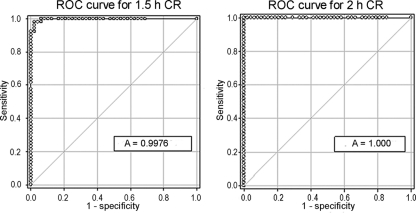
One hundred isolates (50 MRSA and 50 MSSA) were analyzed by flow cytometry after incubation in oxacillin-containing broth as described above for 1.5 and 2 h, and the CRs were determined. ROC curves were generated to determine the ability of the CR to differentiate MRSA from MSSA (Fig. 3). The areas under the ROC curves at 1.5 and 2.0 h were 0.9976 (95% CI = 0.9930 to 1.0002) and 1.000 (95% CI = 1.000 to 1.000), respectively. At 2 h a CR cutoff 0.0445 had 100% sensitivity and a 100% specificity for differentiating MRSA from MSSA in this cohort.
Fig. 3.
ROC curves for the count ratio (CR) for differentiating MRSA from MSSA by flow cytometry after 1.5 h (1.5 h CR) and 2 h (2 h CR) of incubation in oxacillin-containing broth.
Performance of the CR in the validation cohort.
The CR of 0.0445 was tested in a validation cohort of 121 S. aureus isolates (46 MRSA and 75 MSSA) that were incubated in oxacillin-containing broth for 2 h and analyzed by flow cytometry. The validation cohort consisted of S. aureus isolated from patients more than 6 months after the microorganisms that comprised the initial test cohort were obtained and consisted of isolates from blood, respiratory, and wound/abscess cultures. There was excellent correlation between the results of flow cytometry and conventional susceptibility testing (Table 2). One isolate identified as MSSA by conventional testing was identified as MRSA by flow cytometry. Discrepancy resolution testing demonstrated that this isolate had a zone of inhibition of 32 mm on testing with disk diffusion using cefoxitin and was confirmed to be MSSA. In the validation cohort, flow cytometry with a CR cutoff 0.0445 was 100% sensitive and 98.7% specific in differentiating MRSA from MSSA after 2 h of incubation in oxacillin-containing broth.
Table 2.
Performance of the CR for differentiating MRSA from MSSA in the validation cohort
| Group | No. of isolates |
||
|---|---|---|---|
| CR |
Total | ||
| ≥0.0445 | <0.0445 | ||
| MRSA | 46 | 0 | 46 |
| MSSA | 1 | 74 | 75 |
| Total | 47 | 74 | 121 |
DISCUSSION
This study demonstrates that flow cytometry can be used to differentiate MRSA from MSSA after incubation for 2 h in an oxacillin-containing liquid culture medium. These findings were validated using an independent second cohort of MRSA and MSSA isolates, which further supports our findings. Differentiation at 2 h is too early for separation based on differential rates of growth, as confirmed by our experiments that demonstrated no substantial difference in viable bacterial cell counts when MRSA and MSSA isolates were incubated in an oxacillin-containing medium for up to 4 h (data not shown). However, MRSA, but not MSSA, incubated in oxacillin, undergoes some changes in the cell that confer increased fluorescence intensity and side-scatter properties to the cell.
The nature of these cellular changes is unknown, and an investigation of possible hypotheses was outside the scope of the present study. Nevertheless, some comments are in order. It is very reasonable to expect that the process of incubating bacteria in the presence of cell wall-active antibiotics will induce changes in the morphology of the bacteria. An important question from a clinical perspective is whether these changes can be effectively measured to accurately differentiate susceptible from resistant bacteria before they can be differentiated on the basis of cell viability. Antibiotics have previously been shown to induce membrane potential and membrane permeability changes in bacteria (6, 8). Thus, live bacteria adapting to stress are probably changing in measurable ways. It has also been shown that the magnitude of cell-associated fluorescence does not necessarily correlate with cell viability (10, 18). Previous reports have described successful determination of susceptibility of S. aureus to penicillin and oxacillin in 90 min to 4 h, based on measurable drug-induced membrane potential changes using membrane-potential sensitive dyes in small numbers of microorganisms (13, 19). The task of differentiating bacteria by flow cytometry based on susceptibility is a difficult one because of challenges in differentiating single bacteria from clumps of bacteria, the dependence of fluorescence signals on size, and variations in metabolic patterns between species and within organisms of the same species under suboptimal culture conditions (such as antibiotic exposure) (15). These difficulties notwithstanding, and regardless of the nature of the cellular changes induced by antibiotic exposure, the important point from a clinical perspective is that these changes are measurable and the methodology described here reliably differentiates MRSA from MSSA much sooner than would be possible in conventional ways. Our study isolates came from patients at a tertiary referral center and were spread out in time over several months, suggesting that this assay is able to differentiate MRSA from MSSA among many different strains of S. aureus.
Latex agglutination assays targeting PBP2a can accurately detect methicillin resistance in 15 min in staphylococci isolated in culture (1, 2, 9, 21). These assays would be faster than flow cytometry for determining the presence of methicillin resistance in S. aureus isolated in pure culture on solid media. The sensitivity of latex agglutination is lower with smaller inocula, and PBP2a latex agglutination assays have lower sensitivity when used directly in positive blood culture bottles (3, 14). The promise of flow cytometry is that it can detect far fewer microorganisms and that the technology could potentially be developed to be able to identify and determine resistance of microorganisms directly in body fluids. A test based on detection of a PBP2a protein could mistakenly identify MSSA as MRSA if the bacterium is susceptible to methicillin but carries a mecA gene producing a product with impaired function (1).
Flow cytometry is an alternative to molecular methods. It can be easily designed to identify only viable cells, which is an advantage over PCR which cannot easily differentiate live from dead cells. Specifically for methicillin resistance testing in S. aureus, it would not be expected to suffer from inaccuracies arising from mutation, nonexpression or deletion of the mecA gene from the SCCmec. Flow cytometry has not found wide application in clinical microbiology yet, but we demonstrate here its potential.
ACKNOWLEDGMENT
This study was partially supported by Advanced Analytical Technologies, Inc., Ames, IA.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Bressler A. M., et al. 2005. Correlation of penicillin binding protein 2a detection with oxacillin resistance in Staphylococcus aureus and discovery of a novel penicillin binding protein 2a mutation. J. Clin. Microbiol. 43:4541–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cavassini M., Wenger A., Jaton K., Blanc D. S., Bille J. 1999. Evaluation of MRSA-Screen, a simple anti-PBP 2a slide latex agglutination kit, for rapid detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:1591–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chapin K. C., Musgnug M. C. 2004. Evaluation of penicillin binding protein 2a latex agglutination assay for identification of methicillin-resistant Staphylococcus aureus directly from blood cultures. J. Clin. Microbiol. 42:1283–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donnio P.-Y., et al. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farley J. E., et al. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gant V. A., Warnes G., Phillips I., Savidge G. F. 1993. The application of flow cytometry to the study of bacterial responses to antibiotics. J. Med. Microbiol. 39:147–154 [DOI] [PubMed] [Google Scholar]
- 7. Huletsky A., et al. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mason D. J., Allman R., Stark J. M., Lloyd D. 1994. Rapid estimation of bacterial antibiotic susceptibility with flow cytometry. J. Microsc. 176:8–16 [DOI] [PubMed] [Google Scholar]
- 9. Miller M. B., Meyer H., Rogers E., Gilligan P. H. 2005. Comparison of conventional susceptibility testing, penicillin-binding protein 2a latex agglutination testing, and mecA real-time PCR for detection of oxacillin resistance in Staphylococcus aureus and coagulase-negative Staphylococcus. J. Clin. Microbiol. 43:3450–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mortimer F. C., Mason D. J., Gant V. A. 2000. Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescent probes. Antimicrob. Agents Chemother. 44:676–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clinical Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Oliveira K., Procop G. W., Wilson D., Coull J., Stender H. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ordonez J. V., Wehman N. M. 1993. Rapid flow cytometric antibiotic susceptibility assay for Staphylococcus aureus. Cytometry 14:811–818 [DOI] [PubMed] [Google Scholar]
- 14. Qian Q., Venkataraman L., Kirby J. E., Gold H. S., Yamazumi T. 2010. Direct detection of methicillin resistance in Staphylococcus aureus in blood culture broth by use of a penicillin binding protein 2a latex agglutination test. J. Clin. Microbiol. 48:1420–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shapiro H. M. 2001. Multiparameter flow cytometry of bacteria: implications for diagnostics and therapeutics. Cytometry 43:223–226 [DOI] [PubMed] [Google Scholar]
- 16. Shrestha N. K., Tuohy M. J., Hall G. S., Isada C. M., Procop G. W. 2002. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J. Clin. Microbiol. 40:2659–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shrestha N. K., Tuohy M. J., Procop G. W. 2006. Comparison of flow cytometry and quantitative culture for counting microorganisms, abstr. O-075. Abstr. 106th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 18. Suller M. T., Lloyd D. 1999. Fluorescence monitoring of antibiotic-induced bacterial damage using flow cytometry. Cytometry 35:235–241 [DOI] [PubMed] [Google Scholar]
- 19. Suller M. T., Stark J. M., Lloyd D. 1997. A flow cytometric study of antibiotic-induced damage and evaluation as a rapid antibiotic susceptibility test for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 40:77–83 [DOI] [PubMed] [Google Scholar]
- 20. Thornsberry C., McDougal L. K. 1983. Successful use of broth microdilution in susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J. Clin. Microbiol. 18:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamazumi T., Furuta I., Diekema D. J., Pfaller M. A., Jones R. N. 2001. Comparison of the Vitek gram-positive susceptibility 106 card, the MRSA-Screen latex agglutination test, and mecA analysis for detecting oxacillin resistance in a geographically diverse collection of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 39:3633–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]