Abstract
In this article, we describe a chronic case of rhinofacial mucormycosis caused by Mucor irregularis, formerly known as Rhizomucor variabilis var. variabilis, a rare mycotic agent in humans. The infection caused progressive destruction of the nasal septum and soft and hard palate, leading to collapse of the nose bridge and an ulcerative gaping hole. The mucoralean mold cultured from a nasal biopsy specimen was determined by multilocus DNA sequence data to be conspecific with M. irregularis.
CASE REPORT
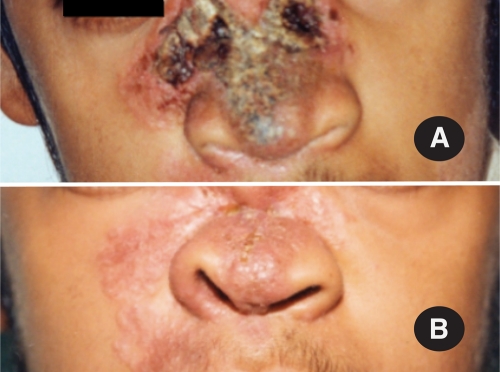
An 18-year-old male patient from south India presented to the Jawaharlal Nehru Medical College, Belgaum, India, in April 2002 with a 12-year history of an infection of the face and nose. At the age of 6 years, he developed a small, hypopigmented, rubbery, elevated lesion over the cheek, which gradually increased in size. He consulted local physicians on several occasions but without any relief. At the age of 15 years, he noticed a small ulcerative lesion in the hard palate, which gradually spread to the soft palate and nasal septum. Over the following 3 years, the infection gradually destroyed the hard palate, resulting in palatal perforation. In addition, progressive destruction of the nasal septum led to collapse of the nasal bridge. An ulcer formed on the nasal bridge, which resulted in a gaping hole of approximately 1 cm in diameter (Fig. 1). At this stage, he was treated empirically with antituberculosis drugs for a period of 6 months at a medical center without any success.
Fig. 1.
Patient showing nasal lesions caused by Mucor irregularis NRRL 32535 (Hema 8791) before (A) and after (B) 6 months of treatment with fluconazole. Given that fluconazole is inactive against mucoralen fungi, we speculate the infection resolved independently, possibly because this isolate cannot grow at 37°C.
Subsequently, the patient was examined at the Jawaharlal Nehru Medical College in April 2002. His hemogram and chest X ray were within normal limits. Skin slit smear examination for Hansen' bacilli, VDRL test, Treponema pallidum hemagglutination (TPHA) for syphilis, tests for tuberculosis (i.e., IgA, IgM, and IgG), including PCR, as well as an enzyme-linked immunosorbent assay (ELISA) test for HIV, were all negative. In addition, examination of acid-fast-stained slides of a skin and palate biopsy specimen for Mycobacterium tuberculosis and Mycobacterium leprae were negative. The epidermis appeared normal in thickness; the dermis showed few granulomas formed by epithelioid cells, ill-formed Langhans' giant cells, and lymphocytes.
Even though a mucoralean fungus was detected in the nasal biopsy (see below), and these fungi have been shown to be broadly resistant to fluconazole (16), this antifungal was used instead of amphotericin B or posaconazole, because of their prohibitive cost. Hence, the patient was hospitalized and treated with 200 mg fluconazole intravenously twice a day for 7 days, which was well tolerated. Dead tissue and crusts were debrided daily until the area looked healthy. The patient was discharged from the hospital after 1 week of treatment and placed on oral fluconazole therapy at 200 mg/day for a period of 30 days followed by 150 mg/daily for another 30 days as a maintenance dose. In addition, a fluconazole solution (2.0 mg/ml) was applied locally four times/day over the eroded area for a period of 3 months. At the time of a 6 month-checkup, the wound had largely healed (Fig. 1B) and reconstructive surgery was planned. However, the patient refused to undergo reconstructive surgery and decided to undergo treatment with homeopathic medicines. He was lost for a further follow-up.
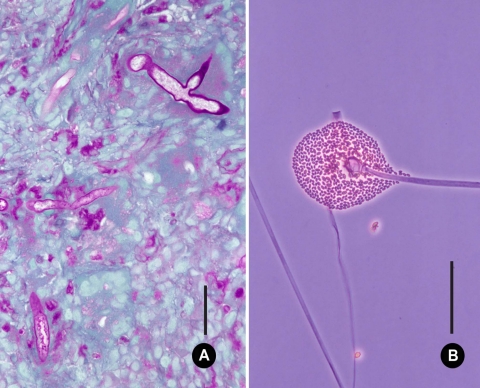
Nasal biopsy tissue taken during the examination was submitted to the Microbiology Service for histological and mycological examination and culture. Sections stained by hematoxylin and eosin (H&E) and periodic acid-Schiff stain (PAS) showed focal squamous metaplasia and ulceration of the respiratory epithelium. The subepithelium showed areas of necrosis, pleomorphic cellular infiltrates with plasma cells, and scattered histiocytes in the vascular granulation tissue. Numerous granulomas consisting of multinucleated giant cells, macrophages, and lymphocytes were also present. Sections stained by PAS showed broad, ribbon-like, often folded, nonseptate hyphae characteristic of a mucoralean fungus (Fig. 2A).
Fig. 2.
Microscopic features of Mucor irregularis NRRL 32535. (A) Section of nasal biopsy tissue showing broad, nonseptate, hyphal elements stained with periodic acid Schiff's reagent. Bar, 25 μm. (B) Sporangiophore, columella, and sporangiospores. Bar, 250 μm.
Direct KOH examination of the biopsy tissue showed broad, hyaline, nonseptate hyphae. A portion of the biopsy tissue was cultured on Sabouraud dextrose agar containing chloramphenicol. Duplicate plates were incubated at 28°C and 37°C. After 4 days of incubation, several whitish to lemon-yellow colonies were visible on plates incubated at 28°C. No growth was observed at 37°C. A subculture of the isolate (Hema 8791, where Hema is the senior author's isolate code) was grown on potato dextrose agar (PDA) and malt extract agar (MEA). Duplicate plates were incubated at 25 and 36°C in the dark. Colonies on PDA and MEA were fast-growing, white at first, although they became yellow to gray-ochraceous after 7 days at 25°C, and measured 40 to 45 mm in diameter. Growth at 36°C was slow, with colonies reaching 20 to 23 mm in diameter after 10 days. No growth was observed at 37 and 40°C. Microscopically, hyphae were broad, hyaline, nonseptate, and branched. Sporangiophores were erect, up to 12.0 to 14.0 μm wide, and unbranched at first, later branching sparingly. Sporangia were globose, yellowish, and up to 70 to 75 μm in diameter with ellipsoidal to spherical columellae. Sporangiospores were ellipsoidal and often flattened on one side, measuring 6.0 to 8.5 μm by 2.5 to 4.5 μm (Fig. 2B). Although this analysis indicated Hema 8791 resembled Mucor hiemalis morphologically (3), it differed from this species in that the maximum growth temperature documented for M. hiemalis is 30°C (8), whereas the temperature maximum for Hema 8791 was 36°C.
A subculture of Hema 8791 was submitted to the NCAUR-ARS-USDA for multilocus DNA sequencing, where it was accessioned in the ARS Culture Collection (http://nrrl.ncaur.usda.gov/) as NRRL 32535. Mycelium obtained by culturing in yeast-malt broth was freeze-dried prior to extraction of total genomic DNA. Methods for extracting genomic DNA, PCR amplification, and DNA sequencing followed published protocols (13, 14, 19). DNA sequence data from NRRL 32535 (Hema 8791) was obtained from domains D1 and D2 of the nuclear ribosomal large subunit (28S) ribosomal DNA (rDNA) (14), the nuclear ribosomal small subunit (18S) rDNA (20), and the translation elongation factor (EF-1α) gene (14), using Platinum TAQ DNA polymerase Hi-Fi (Invitrogen Life Technologies, Carlsbad, CA) in an ABI 9700 thermocycler with the following cycling parameters: 1 cycle of 90 s at 94°C; 40 cycles of 30 s at 94°C, 30 s at 52°C, and 3 min at 68°C; and finally 1 cycle for 5 min at 68°C followed by a 4°C soak. PCR products were purified using Montage PCR96 cleanup filter plates (Millipore Corp., Billerica, MA) and then sequenced using ABI BigDye chemistry version 3.1 in a 9700 thermocycler with the following cycling parameters: 1 cycle of 15 s at 96°C; 40 cycles of 15 s at 96°C, 10 s at 50°C, and 4 min at 60°C followed by a 4°C soak. Sequencher version 4.1.2 (Gene Codes, Ann Arbor, MI) was used to edit and align sequence chromatograms, after which they were exported and used to query the GenBank database (www.ncbi.nlm.nih.gov/BLAST). BLAST searches of the GenBank nucleotide database revealed that the 18S rDNA, partial 28S rDNA, and partial EF-1α gene sequences of NRRL 32535 (Hema 8791) showed 100% identity to sequences of the ex type strain of Rhizomucor variabilis var. variabilis NRRL 28773 (CBS 103.93) (GenBank accession no. AF113435, AF113476, and AF157284). This species was recently reclassified as Mucor irregularis (3).
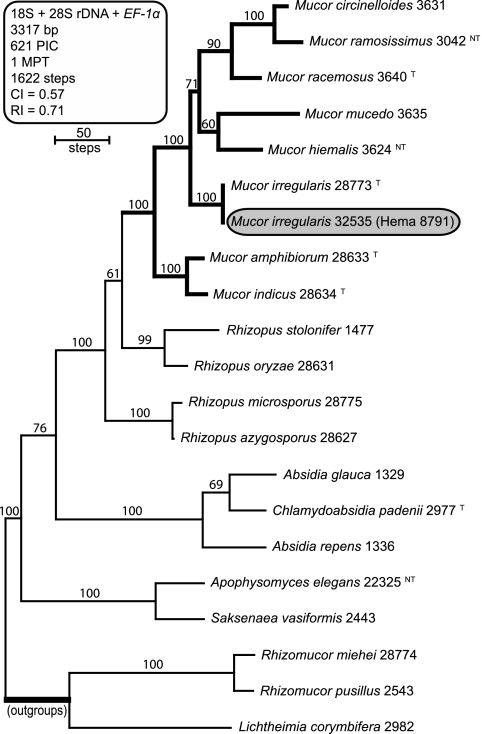
Sequences of NRRL 32535 were added to a three-locus data set, comprising previously published mucoralean sequences (14, 19), to infer its evolutionary relationship to the most common human-pathogenic members of the Mucorales. Maximum parsimony phylogenetic analysis of the combined three-locus data set (18) indicated that NRRL 32535 (Hema 8791) and the ex type strain of Rhizomucor variabilis NRRL 28773 (CBS 103.93) were conspecific (Fig. 3). Furthermore, our results and those of Álvarez et al. (2) indicate that Rhizomucor is polyphyletic, as presently circumscribed, given that R. variabilis is distantly related to the type species of Rhizomucor, Rhizomucor pusillus (Fig. 3). Alverez et al. (3) recently transferred R. variabilis to Mucor as M. irregularis. The latter epithet was chosen because the ex type strain of M. variabilis NRRL 3209 (CBS 564.66) is distantly related to the Mucor clade (data not shown).
Fig. 3.
Bootstrap consensus phylogram depicting phylogenetic relationships of Mucor irregularis NRRL 32535 (Hema 8791) to other clinically important members of the Mucorales. The phylogram is the single-most-parsimonious tree (MPT) inferred from a three-locus data set comprising the nuclear ribosomal small subunit 18S rDNA, and portions of the nuclear ribosomal large subunit 28S rDNA and translation elongation factor (EF-1α) gene. Note that M. irregularis NRRL 32535 and NRRL 28773, the ex type strain (T) of Rhizomucor variabilis var. variabilis, appear to be conspecific. Numbers above internodes represent bootstrap support based on 1,000 maximum parsimony pseudoreplicates of the data. Sequences of Rhizomucor miehei, R. pusillus, and Lichtheimia corymbifera were used to root the phylogram. PIC, parsimony informative characters; CI, consistency index; RI, retention index; T, ex type strain; NT, ex neotype strain.
Antifungal susceptibility tests (with single drugs and two-drug combinations) were performed on both isolates according to the CLSI M-38A2 protocol and checkerboard titrations (7). Mucor irregularis NRRL 28773 and NRRL 32535 were susceptible to amphotericin B (<0.12 μg/ml), posaconazole (0.5 and 0.25 μg/ml), and itraconazole (0.5 μg/ml) and nonsusceptible to echinocandins (>4.0 μg/ml), fluconazole (>256 μg/ml), and voriconazole (8.0 μg/ml). The combination of amphotericin B with caspofungin was synergistic: ΣFICi = 0.25 and 0.31, respectively, where ΣFICi is the summation fractional inhibitory concentration index. These findings do not support treatment of mucormycoses with the latter three antifungals, as previously noted (16), and they strongly suggest that the use of fluconazole in the present case report did not have a direct impact on the clinical outcome of the infection.
An alarming increase in mucormycoses in India has been reported by several Indian Medical centers (4–6, 9, 10, 12, 17). According to Chakrabarti et al. (6, 7), the rise in number of patients with invasive mucormycosis may be due to the increasing population of diabetics in India. It is estimated that more than 30 million diabetics reside in India alone; a large number of them remain undiagnosed and uncontrolled due to deficiencies in the health care system. Previous retrospective surveys have reported that rhino-orbito-cerebral infection is the most common clinical form of mucormycosis, followed by cutaneous, disseminated, and gastrointestinal infections (4, 6, 9, 10). In spite of advances made in diagnosing mucormycoses among Indian patients, the number of etiologic agents isolated in culture and identified to genus level remains disappointedly small. We reviewed the Indian literature comprising 434 cases reported over a span of 40 years (from 1967 to 2007) and found that the causal agent was cultured and identified to genus in only 80 cases (18.4%). In the remaining 354 cases (81.6%), the diagnosis was based on histopathological and/or direct microscopic examination of the biopsy/autopsy tissue. In an earlier retrospective review of 461 Indian cases of mucormycosis, the etiologic agent was identified to the genus in only 172 (37.3%) cases (9). The etiologic agents identified in order of frequency were Rhizopus oryzae, Apophysomyces elegans, Saksenaea vasiformis, Cunninghamella bertholletiae, and Lichtheimia corymbifera (formerly Absidia corymbifera). Among the rare causal agents, Mucor was reported in only 11 cases, and none of these were identified to the species. The identification reported herein was supported by morphological data and confirmation by molecular phylogenetic analysis of a three-locus DNA sequence data set. In the present case, the Mucor infection in our immunocompetent patient progressed slowly over a period of 6 to 7 years. We speculate that it did not disseminate to the cerebral region due to the inability of the isolate to grow at 37°C.
The pathogen now recognized as Mucor irregularis (3, 15) was originally described as Rhizomucor variabilis in 1991 based on an isolate causing a primary cutaneous infection on the hand of a woman from Jiangsu Province, China (22). As in the present case report, the patient lacked any underlying disease or immunodeficiency. Zheng and Chen (22) reported their isolate showed optimum growth between 24 and 30°C, with a maximum growth temperature of 38°C. Additional cases of primary cutaneous mucormycosis have been described recently from three adjacent provinces of eastern China (11, 21). Rhizomucor variabilis var. regularior was described by Zheng and Chen in 1993 (23) based on an isolate from a cutaneous lesion of an immunocompetent patient. The first report of R. variabilis var. regularior from a palate biopsy specimen was recently published involving a 14-year-old female leukemic posthematopoietic cell transplant patient from the United States (1). A recent study by Álvarez et al. (3), however, has demonstrated that R. variabilis var. regularior and R. variabilis var. variabilis are not conspecific. Moreover, these authors reported that R. variabilis var. regularior may be nested within M. circinellioides. Prior to the present study, M. irregularis (R. variabilis var. variabilis) had not been reported as an etiologic agent of cutaneous infections in India. However, in light of present findings that R. variabilis is phylogenetically much more closely related to Mucor than Rhizomucor, it would be worthwhile to examine reports of Mucor species causing mycotic infections in India and elsewhere to better understand the spectrum and genetic diversity of the species involved.
Nucleotide sequence accession numbers.
Nucleotide sequences for Mucor sp. NRRL 32535 (Hema 8791) were deposited in GenBank under accession no. HQ332515 (18S rDNA), HQ332516 (28S rDNA D1/D2 region), and HQ332517 (partial EF-1α gene).
Acknowledgments
We thank Stacy Sink for generating the DNA sequence data reported in this study and Nathane Orwig for collecting the DNA sequence data in the National Center for Agricultural Utilization Research (NCAUR) DNA core facility.
Mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Abuali M. M., et al. 2009. Rhizomucor variabilis var. regularior and Hormographiella aspergillata infections in a leukemic bone marrow transplant recipient with refractory neutropenia. J. Clin. Microbiol. 47:4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Álvarez E., et al. 2009. Spectrum of zygomycete species identified in clinically significant specimens in the United States. J. Clin. Microbiol. 47:1650–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Álvarez E., et al. 2011. Two new species of Mucor from clinical samples. Med. Mycol. 49:62–72 [DOI] [PubMed] [Google Scholar]
- 4. Andrews G., Kurien M., Anandi V., Ramakrishna B., Raman R. 1996. Nasosinusal fungal granuloma—clinical profile. Singapore Med. J. 37:470–474 [PubMed] [Google Scholar]
- 5. Chakrabarti A., et al. 2001. Ten years' experience in zygomycosis at a tertiary care centre in India. J. Infect. 42:261–266 [DOI] [PubMed] [Google Scholar]
- 6. Chakrabarti A., et al. 2006. The rising trend of invasive zygomycosis in patients with uncontrolled diabetes mellitus. Med. Mycol. 44:335–342 [DOI] [PubMed] [Google Scholar]
- 7. Chaturvedi V., et al. 2008. Multilaboratory testing of antifungal combinations against a quality control isolate of Candida krusei. Antimicrob. Agents Chemother. 52:1500–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Hoog G. S., Guarro J., Gené J., Figueras M. J. 2002. Atlas of clinical fungi, 2nd ed Centraalbureau voor Schimmelcultures, Utrecht, Netherlands [Google Scholar]
- 9. Diwakar A., et al. 2007. Zygomycosis—a case report and overview of the disease in India. Mycoses 50:247–254 [DOI] [PubMed] [Google Scholar]
- 10. Kulkarni N. S., Bhide A. R., Wadia R. S. 2005. Rhinocerebral mucormycosis: an analysis of probable mode of spread and its implication in an early diagnosis and treatment. Indian J. Otolaryngol. Head, Neck Surg. 57:121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu X.-L., et al. 2009. Primary cutaneous zygomycosis caused by Rhizomucor variabilis: a new endemic zygomycosis? A case report and review of 6 cases from China. Clin. Infect. Dis. 49:e39–e43 [DOI] [PubMed] [Google Scholar]
- 12. Mohindra S., Mohindra S., Gupta R., Bakshi J., Gupta S. K. 2007. Rhinocerebral mucormycosis: the disease spectrum in 27 patients. Mycoses 50:290–296 [DOI] [PubMed] [Google Scholar]
- 13. O'Donnell K., Cigelnik E., Nirenberg H. I. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493 [Google Scholar]
- 14. O'Donnell K., Lutzoni F. M., Ward T. J., Benny G. L. 2001. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93:286–296 [Google Scholar]
- 15. Schell W., O'Donnell K., Alspaugh J. A. 2011. Heterothallic mating in Mucor irregularis and first isolate of the species outside of Asia. Med. Mycol. [Epub ahead of print.] doi:10.3109/13693786.2011.568975 [DOI] [PubMed] [Google Scholar]
- 16. Sun Q. N., Fothergill A. W., McCarthy D. I., Rinaldi M. G., Graybill J. R. 2002. In vitro activities of posaconazole, itraconazole, voriconazole, amphotericin B, and fluconazole against 37 clinical isolates of zygomycetes. Antimicrob. Agents Chemother. 46:1581–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sundaram C., et al. 2005. Cerebral zygomycosis. Mycoses 48:396–407 [DOI] [PubMed] [Google Scholar]
- 18. Swofford D. L. 2002. PAUP: phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 19. Voigt K., Cigelnik E., O'Donnell K. 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 37:3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 21. Zhao Y., et al. 2009. Primary cutaneous mucormycosis caused by Rhizomucor variabilis in an immunocompetent patient. Mycopathologia 168:243–247 [DOI] [PubMed] [Google Scholar]
- 22. Zheng R.-Y, Chen G.-Q. 1991. A non-thermophilic Rhizomucor causing human primary cutaneous mucormycosis. Mycosystema 4:45–57 [Google Scholar]
- 23. Zheng R.-Y., Chen G.-Q. 1993. Another non-thermophilic Rhizomucor causing human primary cutaneous mucormycosis. Mycosystema 6:1–12 [Google Scholar]