Abstract
Porcine babesiosis is a widespread yet overlooked disease causing economic losses in many regions of the world. To date, the etiological agent of porcine babesiosis has not been molecularly characterized. Here, we provide the first molecular characterization of a piroplasm detected in a symptomatic sow, phylogenetically closely related to the Ungulibabesids. Results pave the way for future molecular epidemiology studies.
TEXT
Babesiosis, caused by intraerythrocytic parasites of the genus Babesia, is one of the most frequently reported infections of free-living and domestic animals. Interest in babesiosis is rising sharply due to its worldwide distribution and public health concerns; indeed, babesiosis is considered an emerging zoonosis of humans (8, 12). Babesiosis is a tick-transmitted disease, with Ixodidae ticks being the major vectors. Although swine babesiosis has been reported in the former Soviet Union, Southern Europe, Africa, and China, Babesia species affecting pigs have not been studied extensively (2, 10, 15, 16). Two porcine species of Babesia are commonly reported, based mainly on the size of the piroplasms detected: Babesia trautmanni, transmitted by Rhipicephalus ticks and characterized by large piroplasms, and the smaller B. perroncitoi, for which the vector still remains unknown (10). Porcine babesiosis is responsible for serious losses and produces antemortem clinical signs that may be similar to those of bovine babesiosis (11). The percentage of parasitized erythrocytes may reach 60%, and abortion associated with fever may appear. Mortality can be significant: 7 out of 56 pigs of a farm died in Italy before therapeutic treatment during an outbreak caused by Babesia trautmanni, and in China, a comparable rate was reported during an outbreak of acute babesiosis caused by B. perroncitoi (6, 9). However, porcine babesiosis remains an overlooked and neglected disease. Outbreaks of babesiosis in pigs have been very rarely described, with the etiological agents still remaining genetically uninvestigated. Consequently, the DDBJ, EMBL, and GenBank sequence databases lack Babesia sequence entries for swine. The lack of molecular data hampers both the development of basic molecular epidemiology studies and the establishment of a role of swine piroplasms, if any, as zoonotic agents.
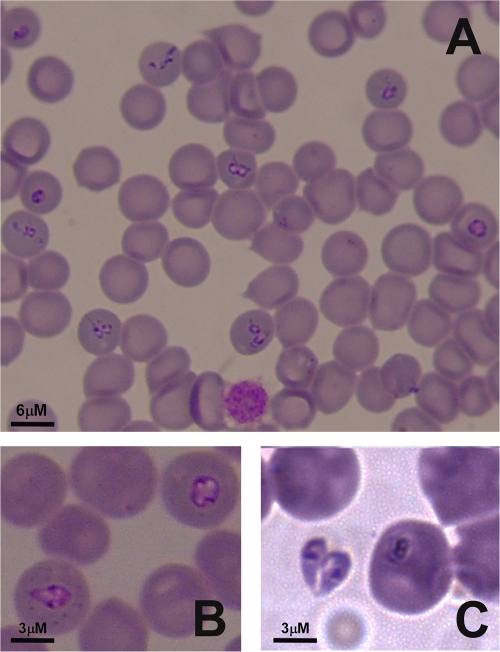
Here, we report the first molecular identification and phylogeny of a Babesia isolate from a symptomatic pig. In summer 2010, EDTA-preserved blood samples and thin blood smears were obtained from a 2.5-year-old pregnant sow reared in a family farm located the region of Anglona (North Sardinia, Italy). The animal showed signs indicative of babesiosis, such as anorexia, depression, lameness, reluctance to move, and high fever, with consequent abortion. Diff-Quick-stained blood smears revealed the presence of babesial inclusions in erythrocytes, with approximately 10% of erythrocytes being parasitized. Smears contained ring-shaped (diameter of 2 to 2.7 μm) and pyriform (1- to 3.3-μm maximum length) protozoa (Fig. 1). Pear-shaped trophozoites occurred either singly (Fig. 1A) or in pairs assembled at their pointed extremities (Fig. 1B). Extracellular forms were also observed (Fig. 1B). The sow recovered after 2 treatments with long-acting oxytetracycline and support therapy (multivitamin complex).
Fig. 1.
(A) Thin blood smears revealing the presence of babesial inclusions in erythrocytes. (B and C) Intraerythrocytic (B) and extracellular (C) forms are shown. Pyriform and ring-shaped protozoa were present, as were groups of small intraerythrocytic parasites (A).
Before treatments, DNA was extracted from blood samples by using the QIAamp DNA minikit (Qiagen, Italy) according to vendor recommendations. Three DNA aliquots obtained from 3 different blood samples were subjected to Babesia-specific PCR by using primers described previously (1), with minor modifications. Briefly, primers BT1-F and BT2-R, specifically targeting the entire Babesia 18S rRNA gene, were used in a first PCR round. PCR products obtained in the first round were used as the DNA target in 3 heminested PCRs, designed by combining BT1-F with BTH-1R, BT2-R with BT2-F, and BTH-1F with BT2-R. Sequences were obtained by cloning the PCR products obtained from the 3 blood samples into the pCR2.1-TOPO vector (Invitrogen SRL, Milan, Italy) and using an automatic sequencer. At least 3 clones were sequenced for each amplicon. Sequence alignments, obtained with the ClustalW option of BioEdit (7), revealed the presence of an invariable sequence for all the 3 replicates obtained from the 3 distinct DNA samples. Based on sequencing, a consensus sequence was built, containing 1,650 bp of the 18S rRNA gene. Upon performing BLAST analyses, this sequence did not fully match any sequence deposited in the GenBank database but was most closely related (97 to 98% identity with 94 to 100% coverage) to various Babesia sequences found in cattle (Babesia sp. strain Kashi2), in antelope (Babesia occultans and Babesia sp. strain Sable Antelope), and in water buffalo (Babesia orientalis). This Babesia isolate, genetically and geographically distinct from any other previously described species, was tentatively named Babesia sp. strain Suis, based on the host in which this pathogen was detected for the first time.
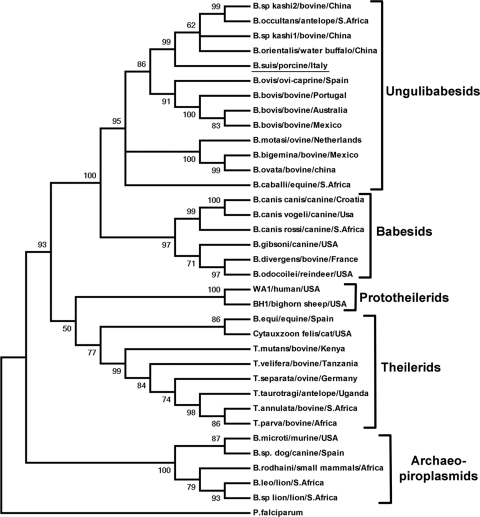
By using the ClustalW option of BioEdit (7), we aligned 1,544 bp of the Babesia sp. Suis 18S rRNA gene sequence to 34 sequences (Table 1) of other members of the Piroplasmida, representative of the 5 groups identified within this order, as defined previously by Criado-Fornelio and coworkers (1). Phylogenetic analyses using both neighbor joining and maximum parsimony were conducted by use of MEGA, version 4 (14), and yielded coinciding trees where Babesia sp. Suis falls into a distinct branch of the Ungulibabesids group, together with B. orientalis, B. occultans, and Babesia sp. Kashi (Fig. 2). The phylogenetic tree is consistent with the topology of previously reported analyses based on 18S rRNA gene sequences of piroplasmids (1). Internal branches of the trees were statistically supported by bootstrapping with 1,000 iterations (4).
Table 1.
Taxonomy, GenBank accession numbers, hosts, regions of isolation, and percentages of sequence identity of Babesia sp. Suis with the 33 piroplasms used as operational taxonomic units in the phylogenetic analysis
| Species | GenBank accession no. | Host | Region | % sequence identity with Babesia sp. Suisa |
|---|---|---|---|---|
| Plasmodium falciparum | M19172.1 | Human | Africa | 64.5 |
| Babesia ovis | AY150058.1 | Goat | Spain | 94.3 |
| Babesia bovis | AY150059.1 | Cattle | Portugal | 85.9 |
| Babesia canis canis | AY072926.1 | Dog | Croatia | 91.8 |
| Babesia canis rossi | DQ111760.1 | Dog | South Africa | 92.6 |
| Babesia canis vogeli | AY371198.1 | Dog | United States | 92.3 |
| Babesia equi | AY150064.2 | Horse | Spain | 88.7 |
| Babesia sp. Kashi2 | AY726557.1 | Cattle | China | 98.0 |
| Babesia occultans | EU376017.1 | Antelope | South Africa | 98.0 |
| Babesia sp. Kashi1 | AY726556.1 | Cattle | China | 96.3 |
| Babesia orientalis | AY596279.1 | Water buffalo | China | 97.7 |
| Theileria annulata | M64243.1 | Cattle | South Africa | 88.5 |
| Theileria mutans | AF078815.1 | Cattle | Kenya | 88.3 |
| Theileria separata | AY260175.1 | Sheep | Germany | 88.7 |
| Theileria velifera | AF097993.1 | Cattle | Tanzania | 89.5 |
| Theileria taurotragi | L19082.1 | Antelope | Uganda | 88.9 |
| Theileria parva | L02366.1 | Cattle | Africa | 88.6 |
| Babesia motasi | AY260180.1 | Texel sheep | Netherlands | 93.8 |
| Babesia bigemina | X59605.1 | Cattle | Mexico | 93.3 |
| Babesia ovata | AY603400.1 | Cattle | China | 94.1 |
| Babesia caballi | Z15104.1 | Horse | South Africa | 94.1 |
| Babesia gibsoni | DQ184507.1 | Dog | United States | 93.4 |
| Babesia divergens | FJ944826.1 | Cattle | France | 92.7 |
| Babesia microti | U09833.1 | Mouse | United States | 86.9 |
| Babesia sp. strain WA1 | AY027816.1 | Human | United States | 87.8 |
| Piroplasmida sp. strain BH1 | AF158708.1 | Bighorn sheep | United States | 87.1 |
| Babesia bovis | M87566.1 | Cattle | Australia | 84.5 |
| Babesia bovis | L31922.1 | Cattle | Mexico | 86.0 |
| Babesia odocoilei | AY237638.1 | Reindeer | United States | 92.8 |
| Babesia sp. strain Dog | AY534602.1 | Dog | Spain | 86.6 |
| Babesia leo | AF244911.1 | Lion | South Africa | 86.9 |
| Babesia sp. strain Lion | AF244914 | Lion | South Africa | 87.1 |
| Cytauxzoon felis | AY679105.1 | Cat | United States | 87.1 |
| Babesia rodhaini | M87565.1 | Small mammals | Africa | 86.3 |
Percentages of identity were evaluated by pairwise alignment and by the sequence identity matrix option of BioEdit (8).
Fig. 2.
Coinciding neighbor-joining and maximum parsimony trees showing the evolutionary relationships of Babesia sp. Suis and the other 34 taxa used in this study. The Ungulibabesids are spread in 2 branches, one of which has Babesia sp. Suis as the most ancestral species. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. All positions containing gaps and missing data were eliminated from the data set (complete-deletion option). There were a total of 1,512 positions in the final data set.
So far, only two Babesia species in pig have been reported, B. trautmanni and B. perroncitoi, but none of them have been characterized at the molecular level. Moreover, literature on porcine babesiosis is relatively scarce. Notably, babesiosis is of economic importance for pigs, as it can be responsible for serious losses following infection, producing signs not unlike those described for cattle (11).
In this study we report the first molecular characterization of a piroplasm in pig. This species, tentatively named Babesia sp. Suis, was detected in a sow from a family farm located in North Sardinia showing symptoms typical of porcine babesiosis, including abortion. Interestingly, an outbreak of porcine babesiosis characterized by a high mortality rate was previously reported in the same area in 1993 (9). However, the lack of molecular data in that previous study and the impossibility of obtaining suitable samples for molecular comparisons render it impossible to verify the homology of Babesia sp. Suis with that responsible for the 1993 outbreak. The sizes of the piroplasms described in our study ranged from 2 to 2.7 μm and from 1 to 3.3 μm in ring-shaped (diameter) and pyriform (maximum-length) forms, respectively. These sizes could be indicative of a coinfection with both “small” and “large” piroplasms. However, direct sequencing of amplicons obtained by PCR always resulted in one type of 18S sequence, suggesting the presence of a unique species variable in size. Biometrical polymorphisms within B. ovis and B. canis were previously reported (5, 13), suggesting that biometrical analysis alone is not sufficient for taxonomical studies of Babesia but should always be accompanied by molecular studies. The absence of a vacuole in the ring forms observed would argue for this isolate being B. trautmanni. This is also supported by the fact that Rhipicephalus is the best-represented tick genus in Sardinia (3). However, the lack of information on the B. trautmanni 18S rRNA gene hampers taxonomical comparisons.
In conclusion, phylogenetic analysis allowed us to place Babesia sp. Suis in a distinct ancestral branch of the Ungulibabesids group. The host tropism of its most close relatives (Babesia sp. Kashi, B. occultans, Babesia sp. Sable Antelope, and B. orientalis), and their geographical distribution indicate that Babesia sp. Suis represents a porcine-specific pathogen. This first molecular characterization paves the way for investigating a possible role of porcine piroplasms as zoonotic agents and establishes a milestone for future molecular epidemiology studies. More data are needed to assess the clinical relevance, the geographical distribution, and the tick vector associated with this Babesia sp.
Nucleotide sequence accession number.
The partial 18S rRNA nucleotide sequence of Babesia sp. Suis was deposited in GenBank under accession number HQ437690.
Acknowledgments
This work was supported by Misura P5 Biodiversitá Animale (Regione Autonoma della Sardegna). Rosanna Zobba was supported by the program Promozione della Ricerca Scientifica e dell'Innovazione Tecnologica in Sardegna, LR 7/2007, PO Sardegna FSE 2007-2013 (Regione Autonoma della Sardegna).
We thank Paola Rapelli for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Criado-Fornelio A., Martinez-Marcos A., Buling-Saraña A., Barba-Carretero J. C. 2003. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe. Part II. Phylogenetic analysis and evolutionary history. Vet. Parasitol. 114:173–194 [DOI] [PubMed] [Google Scholar]
- 2. Dipeolu O. O., Otesile E. B., Fagbemi B. O., Adetunji A. 1983. Studies on blood parasites of pigs in Nigeria: pathogenicity of Babesia trautmanni in experimentally infected pigs. Zentralbl. Veterinarmed. B 30:97–102 [DOI] [PubMed] [Google Scholar]
- 3. Di Todaro N., Piazza C., Otranto D., Giangaspero A. 1999. Ticks infesting domestic animals in Italy: current acarological studies carried out in Sardinia and Basilicata regions. Parassitologia 41:39–40 [PubMed] [Google Scholar]
- 4. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 5. Fukumoto S., et al. 2000. Morphological changes of Babesia gibsoni grown in canine red blood cell-substituted severe combined immune deficiency mice. J. Parasitol. 86:956–958 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y., et al. 1997. The cure of acute Babesiosis perroncitoi in swine. Trop. Anim. Health Prod. 29:64–65 [DOI] [PubMed] [Google Scholar]
- 7. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 8. Kjemtrup A. M., Conrad P. A. 2000. Human babesiosis: an emerging tick-borne disease. Int. J. Parasitol. 30:1323–1327 [DOI] [PubMed] [Google Scholar]
- 9. Ligios C., Scala A. 1993. An outbreak of pig babesiosis in Sardinia. Soc. Ital. Sci. Vet. 47:1379–1383 (In Italian.) [Google Scholar]
- 10. Penzhorn B. L. 2006. Babesiosis of wild carnivores and ungulates. Vet. Parasitol. 138:11–21 [DOI] [PubMed] [Google Scholar]
- 11. Pumell R. E. 1981. Babesiosis in various hosts, p. 25–63 In Ristic M., Kreier J. P. (ed.), Babesiosis. Academic Press, New York, NY [Google Scholar]
- 12. Setty S., Khalil Z., Schori P., Azar M., Ferrieri P. 2003. Babesiosis. Two atypical cases from Minnesota and a review. Am. J. Clin. Pathol. 120:554–559 [DOI] [PubMed] [Google Scholar]
- 13. Shayan P., Hooshmand E., Nabian S., Rahbari S. 2008. Biometrical and genetical characterization of large Babesia ovis in Iran. Parasitol. Res. 103:217–221 [DOI] [PubMed] [Google Scholar]
- 14. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 15. Uilenberg G. 2006. Babesia—a historical overview. Vet. Parasitol. 138:3–10 [DOI] [PubMed] [Google Scholar]
- 16. Yin H., Lu W., Luo J. 1997. Babesiosis in China. Trop. Anim. Health Prod. 29:11–15 [DOI] [PubMed] [Google Scholar]