TEXT
Wild aquatic birds are the main reservoir of avian influenza viruses (AIV) and play a major epidemiological role in the persistence and spread of infection (7). Given their constant evolution, AIV are a permanent threat to animal and public health and their active surveillance was intensified worldwide during the last decade, notably in France. This report describes the failure to detect atypical H5 virus present in cloacal samples obtained from live common snipes (Gallinago gallinago) in France using 3 European official routine tests recommended by the European (FAO/OIE) Reference Laboratory, namely, one real-time reverse transcriptase PCR (rRT-PCR) assay and two current H5 cleavage site RT-PCR assays (2). The samples studied were part of the 1,813 swabs collected during the 2010 avian influenza active surveillance season; these consisted of 9 cloacal swabs sampled during a common snipe banding program in the Dombes area (eastern France). These birds were all juveniles caught during their western migration in late August. The samples were first shipped to an approved local diagnostic laboratory for screening analyses of pooled samples (two pools of 5 and 4 swabs, respectively). RNA extracts, pooled samples, and the remaining individual samples were subsequently sent to the French National Reference Laboratory for confirmatory and further analyses. The presence of AIV in RNA extracts from the two pools was first determined with an M gene-based rRT-PCR; both were positive (Table 1), but they were negative by the H5- and H7-based rRT-PCR recommended in the official diagnostic manual for avian influenza (8). Afterwards, to determine their subtype, RNAs were reverse transcribed and cDNAs were subjected to partial gene amplification with specific primers to detect all hemagglutinin (HA) and neuraminidase (NA) subtypes, also called PanHA (5) and PanNA (3), respectively. No result was obtained with the PanNA PCR, but the PanHA PCR was positive; resulting HA2 sequences of 550 nucleotides were identified as H5 by BLASTN from GenBank. To confirm this and obtain the cleavage site of the H5 gene, the PanHA and two recommended one-step H5 RT-PCR assays targeting the cleavage site (8) were tested with all 9 individual samples. Of these, only two samples (100212c and 100212i) were positive with PanHA, confirming the first sequences obtained with the pools. However, all individual and pooled samples were negative with the two official H5 cleavage site-specific RT-PCR, so a modified H5 cleavage site PCR was designed by making use of the partial HA2 sequence obtained and tested successfully on the H5-positive individual samples. BLASTN results for cleavage site and partial HA2 sequences confirmed that these were nearest to three H5N2 strains isolated in Korea (4) and Japan between 2004 and 2008. To determine the NA subtype, a specific N2 PCR was designed based on the three previous strains isolated in Asia (A/Korea/GJ54/04, GU351860; A/duck/Tsukuba/168/05, AB472016; A/wild bird/Korea/L60-2/08, GU086247) and it gave positive results with samples 100212c and 100212i. The resulting 327-nucleotide sequence was also close to the aforementioned Korean (4) and Japanese H5N2 sequences. Unfortunately, attempts to grow this virus in specific-pathogen-free hen eggs failed, although standard methods were used (2, 8). This failure may be due to the low viral load and/or the lack of infectivity.
Table 1.
Summary of PCR results for different test samples
| Sample(s) | Result of recommended official testa |
Result of complementary test |
|||||
|---|---|---|---|---|---|---|---|
| M rRT-PCR | H5 rRT-PCR | 2 × one-step H5 cleavage siteb | PanHA | PanNA | Modified cleavage sitec | N2 specific | |
| 100212a (pool of 5) | + (32 ct)e | − | − | + | − | NDd | ND |
| 100212b (pool of 4) | + (29 ct) | − | − | + | − | ND | ND |
| 100212c (individual) | ND | ND | ND | + | − | + | + |
| 100212i (individual) | ND | ND | ND | + | ND | + | + |
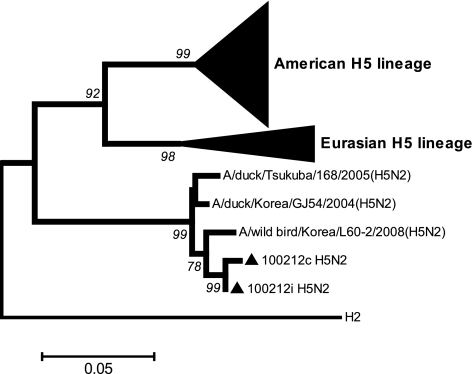
Phylogenic analyses by neighbor joining including the 100 nearest H5 (Fig. 1) and N2 (not shown) sequences indicated that for both genes, these French sequences clustered with the two Korean and Japanese H5N2 strains in a group well distinct from Eurasian and American strains. This is, to our knowledge, the first description in Europe of a virus belonging to this separate lineage; this unique finding in European birds could be either a direct consequence of the virus escaping routine detection tests or due to some of its epidemiological properties. The common ancestor of the Eurasian and American lineages was more recent than the common ancestor of the unusual group and all remaining H5 viruses. Translation of the cleavage site region of the newly described French H5 sequences showed an uncommon PQKETKGLF motif, which was identical to the three Korean and Japanese viruses belonging to the same lineage (Fig. 1), with a K instead of an R at position 6 of this motif (starting from the left), in comparison to the typical low-pathogenicity motif PQRETRGLF (1). The percentage of nucleotide sequence identity between these viruses and the French H5 virus was 97 to 98.3%, whereas this percentage was 99.5% for the two French sequences but only 84.1 to 85.1% for the French H5 virus and the nearest Eurasian or American H5 lineage. The high nucleotide divergence between this unusual lineage and the common Eurasian H5 virus may have caused the observed detection failure of the three H5-specific officially recommended rRT-PCR assays. Indeed, several mismatches between the primers and the probe of these 3 techniques and H5 100212 were observed (see Fig. S1 in the supplemental material). The implication of such a detection failure could be important; indeed, the surveillance of H5 in wild birds could be biased because some H5 viruses may remain undetected. This surveillance is essential to study the evolution of H5 viruses in their adaptation to their host, acquisition of antiviral resistance, and mutation to high pathogenicity. Our observation highlights the need for a constant update of current diagnostic tools in order to maintain their full efficiency as screening methods.
Fig. 1.
The phylogenetic tree of the partial HA2 sequence of H5 was inferred using the neighbor-joining construct method with 548 nucleotides. Evolutionary distances were computed by the Kimura 2-parameter method. The American and Eurasian H5 lineages are represented by 21 Eurasian and 76 American sequences isolated from 1959 to 2007. The tree is rooted with H2 (CY042265). The Japanese, Korean, and French sequences are available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession no. AB558466, GU351859, GU086229, CY084425, and CY004427 (February 2011).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited under accession numbers CY084425 to CY084428.
Supplementary Material
Acknowledgments
This work was supported by EPIZONE (Network of Excellence for Epizootic Disease Diagnosis and Control) and by a Avian Influenza Research Fund (FRIA) grant supplied by the food department of the Ministry of Agriculture, Nutrition, Fishing and Rural Affairs.
We are grateful to A. Henry, K. Ogor, C. Guillou-Cloarec, C. Allée, and M. O. Le Bras for their excellent assistance, the diagnostic departmental laboratory of Ain (LDA01) staff for preliminary AIV detection, and J. C. Laporte and the departmental technical staff of the ONCFS of Ain (SD 01) for bird catching and sampling in the field.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 6 April 2011.
The authors have paid a fee to allow immediate free access to this article.
Contributor Information
Eric Niqueux, Anses, Ploufragan/Plouzané Laboratory, National Reference Laboratory for Avian Influenza and Newcastle Disease, Avian and Rabbit Virology, Immunology and Parasitology Unit, B.P. 53, 22440 Ploufragan, France.
Jean Hars, ONCFS French Wildlife and Hunting Agency, Research Department, Wildlife Sanitary Unit, 38610 Gières, France.
Véronique Jestin, Anses, Ploufragan/Plouzané Laboratory, National Reference Laboratory for Avian Influenza and Newcastle Disease, Avian and Rabbit Virology, Immunology and Parasitology Unit, B.P. 53, 22440 Ploufragan, France.
REFERENCES
- 1. Anonymous 2007. One Step RT PCR for detection of H5 & H7 avian influenza & cleavage site sequencing. Avian Influenza-European Community Reference Laboratory, Weybridge, United Kingdom: http://defra.gov.uk/vla/science/docs/sci_ai_vi545.pdf [Google Scholar]
- 2. European Commission 2006. Manual for avian influenza as provided for in council directive 2005/94/EC. Official J. Eur. Union L237/18 http://eur-lex.europa.eu/LexUriServ.do?uri=OJ:L:2006:010:0016:0016:EN:PDF
- 3. Gall A., et al. 2009. Rapid and highly sensitive neuraminidase subtyping of avian influenza viruses by use of a diagnostic DNA microarray. J. Clin. Microbiol. 47:2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim H. R., et al. 2010. Characterization of H5N2 influenza viruses isolated in South Korea and their influence on the emergence of a novel H9N2 influenza virus. J. Gen. Virol. 91:1978–1983 [DOI] [PubMed] [Google Scholar]
- 5. Phipps L. P., Essen S. C., Brown I. H. 2004. Genetic subtyping of influenza A viruses using RT-PCR with a single set of primers based on conserved sequences within the HA2 coding region. J. Virol. Methods 122:119–122 [DOI] [PubMed] [Google Scholar]
- 6. Starick E., Romer-Oberdorfer A., Werner O. 2000. Type- and subtype-specific RT-PCR assays for avian influenza A viruses (AIV). J. Vet. Med. B Infect. Dis. Vet. Public Health 47:295–301 [DOI] [PubMed] [Google Scholar]
- 7. Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Organization for Animal Health (OIE) 2008. Avian influenza, p. 465–481 In Manual of diagnostic tests and vaccines for terrestrial animals. World Organization for Animal Health (OIE), Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.