Abstract
Detection of Staphylococcus aureus isolates with intermediate vancomycin susceptibility (VISA) and heteroresistance (hVISA) remains problematic. The population analysis profile/area under the curve (PAP/AUC) is the gold standard but is cumbersome. We compared the performance of two Etest screening methods (macromethod [MAC] and glycopeptide resistance detection [GRD]) plus brain heart infusion (BHI) agars supplemented with 3 (BHI-V3) or 4 (BHI-V4) mg/liter vancomycin in detecting hVISA and/or VISA phenotypes. Etest hVISA screenings were done in parallel for 485 saved methicillin-resistant S. aureus (MRSA) blood isolates according to the manufacturer's instructions. The PAP/AUC was measured for all isolates according to the modified method. PAP/AUC test isolate/Mu3 ratios of <0.9, 0.9 to 1.3, and >1.3 were considered positive for susceptible MRSA (S-MRSA), hVISA, and VISA, respectively. PAP/AUC revealed seven VISA and 33 hVISA phenotypes. MAC screening was positive for 30 (75.0%) hVISA/VISA and 49 (11.0%) S-MRSA isolates. GRD screening was positive for 28 (70.0%) hVISA/VISA and 63 (14.2%) S-MRSA isolates. Growth on BHI-V3 was noted in all hVISA/VISA and 24 (5.4%) S-MRSA isolates. Growth on BHI-V4 was noted in all VISA and four (12.1%) hVISA isolates. None of the S-MRSA isolates grew on BHI-V4 agar. The sensitivity, specificity, and positive (PPV) and negative (NPV) predictive values were 75.0%, 89.0%, 38.0%, and 97.5% for MAC; 70.0%, 85.8%, 30.8%, and 97.0% for GRD; 100%, 94.6%, 62.5%, and 100% for BHI-V3; and 100, 99.2%, 63.6%, and 100% for BHI-V4 (for detecting VISA). These findings suggest that both Etest screening methods have excellent NPV, but positive results require confirmation. BHI-V3 and BHI-V4 agars provide more precise identification of hVISA and VISA, respectively; they may be reasonable alternatives to PAP/AUC.
INTRODUCTION
Staphylococcus aureus isolates with reduced susceptibility to vancomycin (VA), including those with intermediate susceptibility (VISA), are usually associated with worse treatment outcomes (9, 10, 12, 16). The relevance of isolates with heteroresistance (hVISA), however, remains uncertain (4, 5). Most reported studies include a small number of hVISA and VISA isolates and have questionable power for meaningful analysis (2, 9, 10). Additionally, the variable incidence of hVISA and the use of different testing methods confound the interpretation of these studies (4, 8, 17, 19, 21). Detection of S. aureus isolates with reduced vancomycin susceptibility, including VISA and hVISA isolates, remains problematic (15, 17). The Centers for Disease Control and Prevention recommend an algorithm for detection of S. aureus isolates with reduced susceptibility that was last revised in March 2009 (http://www.cdc.gov/ncidod/dhqp/ar_visavrsa.html). It is based on precise MIC determination and screening on brain heart infusion (BHI) agar supplemented with 6 mg/liter of vancomycin. It appears to be reliable for detecting vancomycin-resistant S. aureus (VRSA) isolates. These methods, however, are inadequate for detection of VISA and hVISA, since the cutoff values for susceptibility have been revised (15). A novel screening agar with 3 mg/liter vancomycin was recently advocated as a screening tool for isolates with intermediate susceptibility (1). The authors reported 100% sensitivity and 65% specificity based on evaluation of 100 isolates with a MIC range of 0.5 to ≥8 mg/liter. An older study, however, reported high false-positive results (7). Mueller-Hinton (MH) agar supplemented with 5 mg/liter teicoplanin (TP) was also examined and was found to have interlaboratory variations (20, 21). BHI agar supplemented with 4 mg/liter vancomycin was studied in a few isolates (3). The gold standard for defining hVISA is the population analysis profile/area under the curve (PAP/AUC) in comparison to a known hVISA control strain (Mu3) (4, 5, 6, 19). This test is not performed in most clinical laboratories. Several screening methods have been advocated for hVISA detection, but their performances have not been compared in a large randomly selected sample of clinical isolates (8, 21). We evaluated the performances of two commonly advocated Etest-based methods for hVISA screening (the macromethod [MAC] and glycopeptide resistance detection [GRD]) and assessed the feasibility of detecting hVISA and VISA by screening BHI agars supplemented with 3 or 4 mg/liter of vancomycin.
MATERIALS AND METHODS
All methicillin-resistant S. aureus (MRSA) blood isolates saved at our research laboratory from prior S. aureus bacteremia studies conducted intermittently between 1996 and 2006 were selected (13). They were preserved in skim milk at −80°C until they were tested. The vancomycin MIC was measured by the broth microdilution and Etest methods. Screening for hVISA was done in parallel by the MAC and GRD methods according to the manufacturer's instructions (AB Biodisk). Bacterial suspensions were prepared in Mueller-Hinton broth and diluted to 2.0 and 0.5 McFarland standard for MAC and GRD, respectively. Any visible growth at ≥8 μg VA and TP or ≥12 μg TP only in MAC and ≥8 μg VA or TP in GRD was considered a positive test result. Control strains ATCC 29213 (methicillin-susceptible S. aureus [MSSA]), 700699 (Mu50), and 700698 (Mu3) were included with each run.
PAP/AUC.
The PAP/AUC was measured for all isolates by inoculating serial 10-fold dilutions of the test organism onto increasing concentrations of vancomycin BHI agar (Hardy Diagnostics, Santa Maria, CA). The vancomycin agar concentrations used were 0, 1, 2, 3, 4, and 6 mg/liter. Colony growth at 48 h was measured and graphed as log10 CFU/ml versus the vancomycin concentration (18, 19). Control strains (ATCC 29213 [MSSA], Mu50, and Mu3) were included with each run. The area under the curve was calculated for each sample and compared to the mean Mu3 AUC for each group of PAP. The test isolate AUC/mean Mu3 ratios were calculated. Ratios of 0.9 to 1.3 and >1.3 were considered positive for hVISA and VISA, respectively (5, 19). Growth on BHI agars with 3 (BHI-V3) and 4 (BHI-V4) mg/liter vancomycin was stratified according to hVISA and VISA status. The sensitivities, specificities, and predictive values of the Etest screening methods and BHI-V3 and BHI-V4 agars for distinguishing hVISA and VISA were determined.
RESULTS
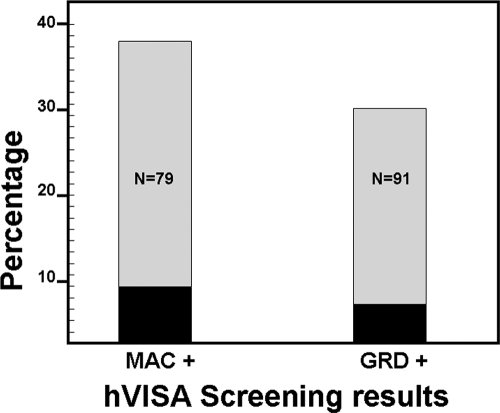
We had 485 saved MRSA blood isolates with vancomycin MICs of 0.5 to 4 mg/liter available for testing. One hVISA screen-negative isolate with a vancomycin Etest MIC of 0.75 mg/liter could not be recovered for additional testing and was excluded. The vancomycin MIC was ≤2 mg/liter for 457 (94.2%) isolates by broth microdilution and for 447 (92.2%) by Etest. Screening for hVISA by both methods was concordantly negative for 376 (77.5%) isolates, concordantly positive for 61 isolates (12.6%), and discordant for 48 (9.9%) isolates. PAP/AUC revealed seven (1.4%) VISA, 33 (6.8%) hVISA, and 445 (91.8%) fully susceptible isolates. MAC screening was positive for 30 (75.0%) hVISA/VISA and 49 (11.0%) fully susceptible isolates. GRD screening was positive for 28 (70.0%) hVISA/VISA and 63 (14.2%) fully susceptible isolates (Fig. 1). MAC-positive GRD-negative screens were more often truly positive than GRD-positive MAC-negative screens (16.7% [odds ratio {OR} = 2.64; 95% confidence interval {CI} = 0.48 to 14.59] versus 3.3% [OR = 0.46; 95% CI = 0.23 to 0.92]; P = 0.1). The sensitivities, specificities, and negative and positive predictive values for hVISA/VISA detection are shown in Table 1.
Fig. 1.
Percentages of Etest screen-positive isolates validated as hVISA (gray bars) and VISA (black bars) phenotypes by population profile analysis stratified according to screening results.
Table 1.
Performance of hVISA Etest screening methods
| Screening method | Sensitivity [% (no./total)] (TP/TP + FN)a | Specificity [% (no./total)] (TN/TN + FP) | PPVb [% (no./total)] (TP/TP + FP) | NPVc [% (no./total)] (TN/TN + FN) |
|---|---|---|---|---|
| MAC | 75.0 (30/40) | 89.0 (396/445) | 38.0 (30/79) | 97.5 (396/406) |
| GRD | 70.0 (28/40) | 85.8 (382/445) | 30.8 (28/91) | 97.0 (382/394) |
TP, true positive; TN, true negative; FP, false negative; FN, false negative.
PPV, positive predictive value.
NPV, negative predictive value.
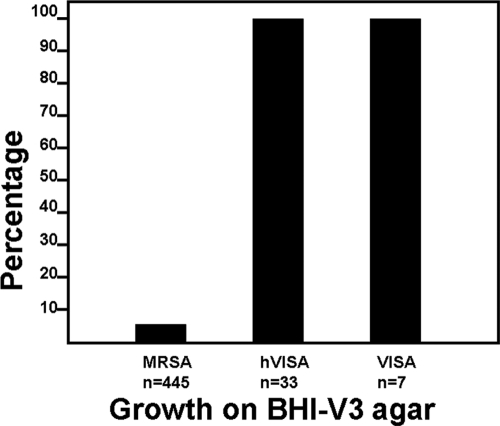
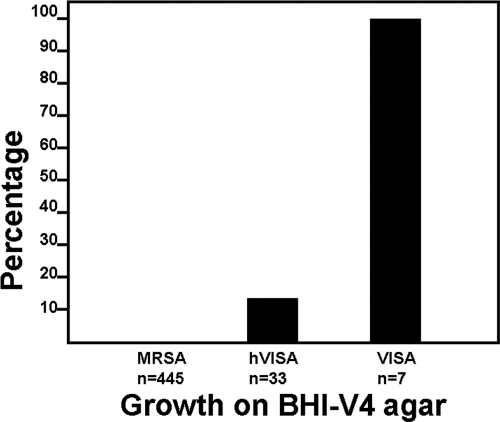
The feasibility of using BHI-V3 and BHI-V4 agars for screening for hVISA and VISA was assessed. On BHI-V3, all VISA, all hVISA, and 24 (5.4%) fully susceptible isolates grew (Fig. 2). The PAP/AUC ratios for the susceptible isolates with growth on BHI-V3 were near the cutoff for hVISA (0.71 to 0.89; median = 0.84), significantly higher than for the inhibited isolates (0.83 ± 0.05 versus 0.51 ± 0.16; P < 0.001). On BHI-V4 agar, growth was noted in all VISA and four (12.1%) hVISA isolates. None of the fully susceptible isolates grew on BHI-V4 (Fig. 3). The hVISA isolates that grew on BHI-V4 agar had PAP/AUC values near the cutoff for VISA (1.18 to 1.29; median = 1.24), significantly higher than the inhibited hVISA isolates (1.24 ± 0.05 versus 0.97 ± 0.07; P < 0.001). The sensitivity, specificity, and predictive values for each agar are shown in Table 2.
Fig. 2.
Growth of susceptible MRSA, hVISA, and VISA isolates on BHI agar supplemented with vancomycin (3 mg/liter).
Fig. 3.
Growth of susceptible MRSA, hVISA, and VISA isolates on BHI agar supplemented with vancomycin (4 mg/liter).
Table 2.
Performance of vancomycin-supplemented BHI agars for detecting VISA and hVISA isolates
| Type of agar | Type(s) of isolate | Sensitivity [% (no./total)] (TP/TP + FN)a | Specificity [% (no./total)] (TN/TN + FP) | PPVb [% (no./total)] (TP/TP + FP) | NPVc [% (no./total)] (TN/TN + FN) |
|---|---|---|---|---|---|
| BHI-V3 | hVISA/VISA | 100 (40/40) | 94.6 (421/445) | 62.5(40/64) | 100 (421/421) |
| VISA | 100 (7/7) | 88.1 (421/478) | 10.9 (7/64) | 100 (421/421) | |
| BHI-V4 | hVISA/VISA | 27.5 (11/40) | 100 (445/445) | 100 (11/11) | 93.9 (444/474) |
| VISA | 100 (7/7) | 99.2 (474/478) | 63.6 (7/11) | 100 (474/474) |
TP, true positive; TN, true negative; FP, false negative; FN, false negative.
PPV, positive predictive value.
NPV, negative predictive value.
DISCUSSION
The relevance of hVISA remains uncertain. This uncertainty is due to the lack of standardized testing methods and the variable incidence of hVISA in reported studies (4, 17, 18). PAP, the gold standard for detecting hVISA, is probably unsuitable for clinical laboratories (14, 17). Etest-based methods are suggested as alternate approaches. Both MAC and GRD have been studied and compared, usually in hVISA and VISA isolates. Therefore, their exact sensitivity and specificity in randomly selected clinical isolates are not clearly defined. In our study, we tested a large number of blood isolates; the majority were vancomycin susceptible, with MICs of ≤2 mg/liter. Our findings show good negative predictive values for MAC and GRD, but both tests have a high number of false-positive screens. Based on these findings, we believe that these tests can be used for screening a large number of isolates; however, confirmation of positive screens is warranted.
Our results also show that vancomycin-supplemented BHI agars are useful for detecting hVISA and VISA. Rarely, susceptible isolates with AUC ratios close to the cutoff reading used for defining hVISA grew on BHI-V3 agar, and a few hVISA isolates with AUC ratios near the cutoff value for VISA grew on BHI-V4 agar. Since reduced susceptibility is a continuous process with artificial cutoff values (11), growth of these isolates may be considered a marker of reduced susceptibility. No false-negative results for detecting VISA were noted by any method.
Our findings suggest that BHI-V3 agar provides excellent screening for hVISA but is suboptimal for VISA screening. In comparison, BHI-V4 appears superior for detecting VISA. These observations differ from those of Burnham et al., who report 100% sensitivity and 65% specificity for detecting VISA with BHI-V3 (1). The reason for the difference is unclear but might be related to isolate selection. Burnham et al. selected their isolates based on MIC results and did not perform PAP/AUC. All our tested isolates were confirmed by PAP/AUC.
Finally, PAP/AUC is cumbersome and impractical to perform for a large number of isolates. Our findings suggest that it is reasonable to validate screen-positive isolates and to presume that screen-negative isolates with MICs of ≤2 mg/liter are fully susceptible. Validation of BHI-V3 and BHI-V4 positive results may not be needed.
In conclusion, since Etest screening methods for detecting hVISA and VISA are suboptimal, and since population analysis is not practical, screening for isolates with reduced susceptibility on vancomycin-supplemented BHI agars provides a reliable alternative. These agars can be made commercially available so that testing methods can be standardized and the relevance of these isolates can be better defined. We recommend BHI-V3 for hVISA screening and BHI-V4 for VISA detection.
ACKNOWLEDGMENT
The study was supported by the St. John Hospital Medical Education Fund.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Burnham C.-A. D., Weber C. J., Dunn M., Jr 2010. Novel screening agar for detection of vancomycin-nonsusceptible Staphylococcus aureus. J. Clin. Microbiol. 48:949–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charles P. G. P., Ward P. B., Johnson P. D. R., Howden B. P., Grayson L. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448–451 [DOI] [PubMed] [Google Scholar]
- 3. Chesneau O., Morvan A., El Solh N. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887–890 [DOI] [PubMed] [Google Scholar]
- 4. Falagas M. E., Makris G. C., Dimopoulos G., Matthaiou D. K. 2008. Heteroresistance: a concern of increasing clinical significance? Clin. Microbiol. Infect. 14:101–104 [DOI] [PubMed] [Google Scholar]
- 5. Howden B. P., Davies J. K., Johnson P. D. R., Stinear T. P., Grayson L. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khosrovaneh A., et al. 2004. Frequency of reduced vancomycin susceptibility and heterogeneous subpopulation in persistent or recurrent methicillin resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 38:1328–1330 [DOI] [PubMed] [Google Scholar]
- 7. Kosowska-Shick K., et al. 2008. Incidence and characteristics of vancomycin-nonsusceptible strains of methicillin-resistant Staphylococcus aureus at Hershey Medical Center. Antimicob. Agents Chemother. 52:4510–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leonard S. N., Rossi K. L., Newton K. L., Rybak M. J. 2009. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63:489–492 [DOI] [PubMed] [Google Scholar]
- 9. Lodise T. P., et al. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52:3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maor Y., et al. 2009. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J. Infect. Dis. 199:619–624 [DOI] [PubMed] [Google Scholar]
- 11. Mohr J. F., Murray B. E. 2007. Point: vancomycin is not obsolete for the treatment of infections caused by methicillin resistant Staphylococcus aureus. Clin. Infect. Dis. 44:1536–1542 [DOI] [PubMed] [Google Scholar]
- 12. Moise P. A., Sakoulas A., Forrest A., Schentag J. J. 2007. Vancomycin in vitro bactericidal activity and its relationship to efficacy in clearance of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 51:2582–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Musta A. C., et al. 2009. Vancomycin MIC plus heteroresistance and outcome of methicillin-resistant Staphylococcus aureus bacteremia: trends over 11 years. J. Clin. Microbiol. 47:1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rybak M. J., et al. 2008. Characterization of vancomycin heteroresistant Staphylococcus aureus (hVISA) from the Detroit metropolitan area over a 22-year period (1986–2007). J. Clin. Microbiol. 46:2950–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swenson J. M., et al. 2009. Accuracy of commercial and reference susceptibility testing methods for detecting vancomycin-intermediate Staphylococcus aureus. J. Clin. Microbiol. 47:2013–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soriano A., et al. 2008. Influence of vancomycin minimal inhibitory concentration on the treatment of methicillin resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46:193–200 [DOI] [PubMed] [Google Scholar]
- 17. Voss A., et al. 2007. A multi-center blinded study on the efficiency of phenotypic screening methods to detect glycopeptides intermediately susceptible Staphylococcus aureus (GISA) and heterogeneous GISA (h-GISA). Ann. Clin. Microbiol. Antimicrob. 6:9 doi:10.1186/1476-0711-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walsh T. R., et al. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wootton M., et al. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399–403 [DOI] [PubMed] [Google Scholar]
- 20. Wootton M., MacGowan A. P., Walsh T. R., Howe R. A. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yusof A., et al. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptides-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J. Clin. Microbiol. 46:3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]