Abstract
Sepsis is a major health problem in newborns and children. Early detection of pathogens allows initiation of appropriate antimicrobial therapy that strongly correlates with positive outcomes. Multiplex PCR has the potential to rapidly identify bloodstream infections, compensating for the loss of blood culture sensitivity. In an Italian pediatric hospital, multiplex PCR (the LightCycler SeptiFast test) was compared to routine blood culture with 1,673 samples obtained from 803 children with suspected sepsis; clinical and laboratory information was used to determine the patient infection status. Excluding results attributable to contaminants, SeptiFast showed a sensitivity of 85.0% (95% confidence interval [CI] = 78.7 to 89.7%) and a specificity of 93.5% (95% CI = 92.1 to 94.7%) compared to blood culture. The rate of positive results was significantly higher with SeptiFast (14.6%) than blood culture (10.3%) (P < 0.0001), and the overall positivity rate was 16.1% when the results of both tests were combined. Staphylococcus aureus (11.6%), coagulase-negative staphylococci (CoNS) (29.6%), Pseudomonas aeruginosa (16.5%), and Klebsiella spp. (10.1%) were the most frequently detected. SeptiFast identified 97 additional isolates that blood culture failed to detect (24.7% P. aeruginosa, 23.7% CoNS, 14.4% Klebsiella spp., 14.4% Candida spp.). Among specimens taken from patients receiving antibiotic therapy, we also observed a significantly higher rate of positivity of SeptiFast than blood culture (14.1% versus 6.5%, respectively; P < 0.0001). On the contrary, contaminants were significantly more frequent among blood cultures than SeptiFast (n = 97 [5.8%] versus n = 26 [1.6%]), respectively; P < 0.0001). SeptiFast served as a highly valuable adjunct to conventional blood culture in children, adding diagnostic value and shortening the time to result (TTR) to 6 h.
INTRODUCTION
Pediatric patients with severe trauma and burns, immunodeficiency, malignancy, and prematurity have an increased incidence of septicemia with a high case fatality rate (10 to 50%) (10). Moreover, prolonged hospitalization, broad-spectrum empirical antimicrobial therapy, and supportive care have a strong impact on the cost of care (15, 25).
Oncohematological patients and newborns, particularly preterm infants, are at high risk for severe infections and sepsis due to their deficient and/or immature immunologic defense (5, 7). Rapid detection of the infectious cause and prompt initiation of appropriate antimicrobial treatment are fundamental for the successful treatment of septic patients and for the reduction of antibiotic resistance rates (23, 30).
Blood culture is the current “gold standard” for the detection of bloodstream microbial pathogens; although it allows microbes to be identified and their susceptibility profiles to be tested, it presents several limitations. Lack of rapidity is a major problem: detection of bacterial growth requires approximately 12 to 48 h or more in the case of fastidious bacterial or invasive fungal infection (1, 18). Another remarkable limitation of blood culture is its low sensitivity for previous antibiotic treatment and/or low bacterial concentrations, due to the smaller amount of blood sampled from pediatric patients than from adults (4, 17).
Blood culture may allow the growth of a small quantity of bacteria potentially considered contaminants. Moreover, there is a high risk of contamination by skin saprophytes such as coagulase-negative staphylococci (CoNS) and streptococci, which makes it difficult to implicate them as agents of catheter-associated bacteremia (9). All these issues can be overcome using PCR, because it is based on the direct detection of the microbe without relying on its growth curve or without suffering the bacteriostatic effect of antimicrobial therapy (12, 20). In the present study, we evaluated a commercially available multiplex real-time PCR assay (the LightCycler SeptiFast test) for the direct detection of bacteria and fungi. Results were compared with those obtained from conventional blood cultures, considering both clinical and laboratory data.
MATERIALS AND METHODS
Study site, patients, and tests.
Subjects were recruited from the tertiary care Children's Hospital and Research Institute Bambino Gesù in Rome, Italy, the largest in central and southern Italy. Between May 2007 and May 2009, 2,500 SeptiFast tests were performed on blood samples from 811 subjects, aged 0 to 18 years, as an adjunct diagnostic tool for the diagnosis of sepsis. Wards submitting samples were divided into three groups: (i) intensive care units (ICUs) and surgery department (group a), (ii) oncology, hematology, and neonatology (group b), and (iii) emergency department and pediatrics (group c) (see Table S1 in the supplemental material). The retrospective design of the present study was approved by the Ethics Committee of Children's Hospital and Research Institute Bambino Gesù (study 361/2010, protocol 421CM/vp), which produced dedicated forms for informed consent signed by the children's parents or guardian.
Inclusion criteria.
Patients were considered for inclusion in the study only if the following decisive factors were met: (i) clinical suggestion of systemic inflammatory response syndrome (SIRS) with suspected bacterial or fungal infection (8), (ii) availability of a filled-out questionnaire with demographic, clinical, and laboratory information (e.g., core temperature, heart and respiratory rates, leukocyte count, systolic blood pressure, risk factors for bloodstream infections, underlying disease/cause of hospitalization, antimicrobial therapy, suspected or proven focus of infection, and concentration of C-reactive protein), and (iii) collection of paired blood samples for SeptiFast (≥1.5 ml) and two blood samples for cultures (0.5 to 10 ml each, depending on whether an aerobic or anaerobic bottle was used, as described below) from a peripheral vein or a central venous line (CVL). Finally, 1,673 paired samples from 803 out of 811 patients tested by SeptiFast and blood culture were studied.
Microbiological techniques.
The LightCycler SeptiFast test M Grade (Roche Molecular Systems, Mannheim, Germany) is an in vitro nucleic acid amplification test for the detection of bacterial and fungal DNA (16S-23S and 18S-5.8S internal transcribed space regions of rRNA genes, respectively) in human blood (the assay is not cleared for diagnostic use in the United States). It allows the identification of more than 20 bacterial and fungal species, as reported in the SeptiFast master list (SML) (Table 1), which cause approximately 90% of all bloodstream infections. The analytical sensitivity of the assay, as indicated by the manufacturer, is between 3 and 100 CFU/ml, depending on the microorganism. Following the manufacturer's instructions, blood sample DNA was extracted with an internal control (IC), provided by the LightCycler SeptiFast kit, in order to exclude false-negative results and was amplified in three individual reactions (Gram-positive bacteria, Gram-negative bacteria, and fungi) on the LightCycler (version 2.0) instrument (Roche Applied Sciences, Mannheim, Germany). PCR products were simultaneously detected by fluorescence and melting temperature (Tm) analysis, using specific hybridization probes and identification software. Regarding blood cultures, bottles added by an antimicrobial removal device (cation-exchange and anion-adsorption resins, 0.6% and 10% wt/vol, respectively) were inoculated with 0.5 to 5 ml (Bactec Peds Plus/F medium; BD Diagnostics, Sparks, MD) and 3 to 10 ml (Bactec Lytic/10 Anaerobic/F medium vials; BD Diagnostics). The bottles were then incubated at 35°C in Bactec 9240/9120 blood culture system (BD Diagnostics) cabinets for 8 days. In case of positivity, Gram staining and culture on solid medium were performed; definitive organism identification and antibiotic susceptibility were determined with accredited routine laboratory methods (Vitek 2 system [bioMérieux, Durham, NC] or Phoenix [BD Diagnostics] system).
Table 1.
SeptiFast master list
| Gram-negative organisms | Gram-positive organisms | Fungi |
|---|---|---|
| Escherichia coli | Staphylococcus aureus | Candida albicans |
| Klebsiella pneumoniae/K. oxytoca | CoNSa | Candida tropicalis |
| Serratia marcescens | Streptococcus pneumoniae | Candida parapsilosis |
| Enterobacter cloacae/E. aerogenes | Streptococcus spp.b | Candida glabrata |
| Proteus mirabilis | Enterococcus faecium | Candida krusei |
| Pseudomonas aeruginosa | Enterococcus faecalis | Aspergillus fumigatus |
| Acinetobacter baumannii | ||
| Stenotrophomonas maltophilia |
The CoNS that can be identified are S. epidermidis, S. haemolyticus, S. hominis, S. pasteuri, S. warneri, S. cohnii, S. lugdunensis, S. capitis, S. caprae, S. saprophyticus, and S. xylosus.
The Streptococcus species that can be identified are S. agalactiae, S. pyogenes, S. anginosus, S. bovis, S. constellatus, S. cristatus, S. gordonii, S. intermedius, S. milleri, S. mitis, S. mutans, S. oralis, S. parasanguinis, S. salivarius, S. sanguinis, S. thermophilus, S. vestibularis, and viridans group streptococci.
Interpretation of results from blood cultures.
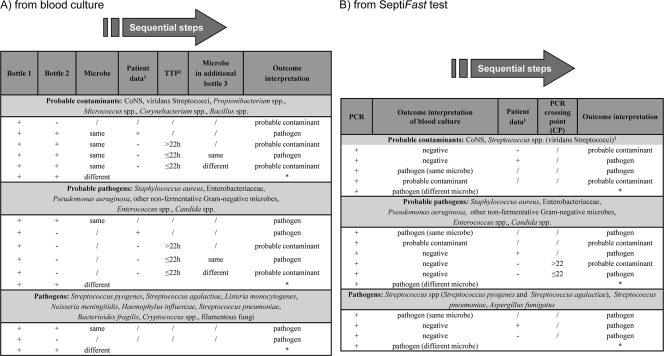
The determination of isolates as contaminants versus pathogens was achieved using standardized algorithms, as described in Fig. 1A, combining microorganism pathogenicity (27) and clinical, laboratory, and microbiological data (8). Positive patient data were considered age-specific vital signs (e.g., core temperature, heart and respiratory rates, and systolic blood pressure) as well as laboratory variables (e.g., leukocyte count, concentration of C-reactive protein, and microbiological evidence of infection focus) in relation to age ranges (8). The condition of sepsis was defined when a SIRS was in the presence of or a result of suspected or proven infection (8), ascertained by the microbiology routine team, which addressed the final interpretation of the results (contaminants versus pathogens) on the basis of type of microbe, time to positivity (TTP), number of positive blood cultures for the same microbe (27), and patient data provided by clinicians.
Fig. 1.
Algorithms for interpretation of blood culture and SeptiFast results (A and B, respectively). *, in case of polymicrobial infections and/or detection of different microorganisms, the same criteria reported above were applied for the assessment of each microorganism as a pathogen or probable contaminant; 1, clinical, laboratory, and microbiological information; 2, TTP, time to positivity; 3, positive result reported by the SeptiFast software.
Interpretation of results from SeptiFast test.
Isolates identified by PCR were considered to be pathogens or contaminants using a standardized algorithm, as reported in Fig. 1B, combining microorganism pathogenicity, interpretation of blood culture results, and clinical, laboratory, and microbiological data. The threshold of the SeptiFast software, based on the bacterial DNA amount, excluded CoNS and streptococci from the positive results and considered them contaminants.
Statistical analysis.
The diagnostic test OpenEpi module was used to calculate the 95% confidence intervals (CIs) for the sensitivity and specificity of the SeptiFast test by considering blood culture to be the “gold standard” reference method. A combined microbiological and clinical algorithm (Fig. 1A and B) was used to confirm positive results by blood culture and SeptiFast as clinically true positive, excluding positive results attributable to contaminants.
The percentage of positive results for each test from paired samples was compared within two-by-two contingency tables using McNemar's test. A P value of <0.05 was considered statistically significant. A conditional logistic regression model was used to test for differences in SeptiFast and blood culture results across medical wards.
RESULTS
Results of 1,673 episodes (paired blood samples tested by blood culture and SeptiFast) were evaluated and are reported in Table 2. Excluding contaminants, 250/1,553 (16.1%) samples gave positive results, 136/250 (54.4%) of which were detected by both methods (not necessarily the same isolates). The rate of positive blood samples was significantly higher by SeptiFast (n = 226/1,553, 14.6%) than blood culture (n = 160/1,553, 10.3%) (P < 0.0001), with a sensitivity of 85.0% (95% CI = 78.7 to 89.7%) and a specificity of 93.5% (95% CIs = 92.1 to 94.7%); the positive predictive value was 60.2% (95% CI = 53.7 to 66.3%), and the negative predictive value was 98.2% (95% CI = 97.3 to 98.8%).
Table 2.
Results of the SeptiFast test and blood culture per patient episode and per microorganism
| SeptiFast result | No. of samples with the following blood culture result: |
|||
|---|---|---|---|---|
| Positive | Negative | Probable contaminant | Total | |
| Per patient episode | ||||
| Positive | 136 | 90 | 3 | 229 |
| Negative | 24 | 1,303 | 91 | 1,418 |
| Probable contaminant | 0 | 23 | 3 | 26 |
| Total | 160 | 1,416 | 97 | 1,673 |
| Per microorganism | ||||
| Positive | 143 | 97 | 0 | 240 |
| Negative | 27 | 1,303 | 110 | 1,440 |
| Probable contaminant | 0 | 37 | 0 | 37 |
| Total | 170 | 1,437 | 110 | 1,717 |
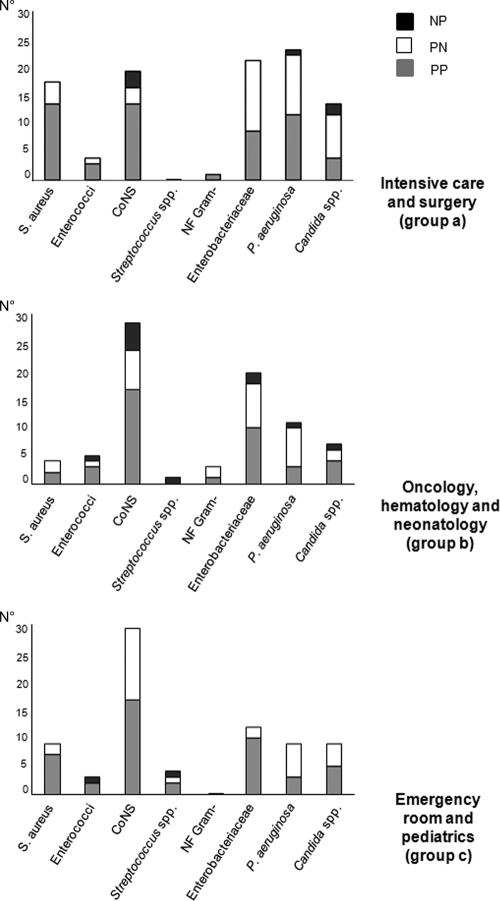
Results for isolates from all episodes are shown in Table 2; the numbers in Table 2 per patient episode are higher than those per microorganism due to the occurrence of polymicrobial infections in 1.3% and 1.0% of episodes for the SeptiFast test and for blood culture, respectively. The same pathogens were detected in 143 (53.6%) out of 267 positive paired samples by the SeptiFast test and blood culture (Table 2). SeptiFast detected an additional 97/267 (36.3%) pathogens in 90 positive samples that blood culture had failed to find. Pathogens exclusively identified by SeptiFast were Pseudomonas aeruginosa (24/97, 24.7%), CoNS (23/97, 23.7%), Klebsiella pneumoniae/K. oxytoca (14/97, 14.4%), and Candida spp. (14/97, 14.4%). Blood culture detected 27/267 (10.1%) additional pathogens in 24 positive samples that SeptiFast failed to find (Table 2); 8/27 (29.6%) of these isolates were not in the SML. Table 3 shows the results obtained by SeptiFast compared to blood culture stratified by pathogens (n = 267) and contaminants (n = 147). Among pathogens, Gram-positive organisms (n = 128/267, 47.9%) were more frequently detected than Gram-negative bacteria (n = 107/267, 40.1%) and fungi (n = 32/267, 12.0%). CoNS were the most frequently detected pathogens (79/267, 29.6%), followed by P. aeruginosa (44/267, 16.5%), and Staphylococcus aureus (31/267, 11.6%). As shown in Table 2, contaminants were significantly more frequent among blood culture results than among SeptiFast results (n = 97/1,673 [5.8%] versus 26/1,673 [1.6%]), respectively; P < 0.0001). For 3 paired cases, positive results for contaminants were obtained by both methods; however, the isolates were not identical (Table 2). Out of 103 contaminants detected by blood culture (excluding those not in the SML), 76 (73.8%) were CoNS. SeptiFast identified 37 contaminants, of which 6 (16.2%) were CoNS, K. pneumoniae/K. oxytoca, and P. aeruginosa (Table 3). Twenty-seven isolates were identified by blood culture and not by SeptiFast: 8 were not in the SML; for 13 isolates (2 Candida albicans isolates, 2 P. aeruginosa isolates, 2 Staphylococcus epidermidis isolates, and 1 isolate each of Candida krusei, Enterococcus faecalis, Enterococcus faecium, K. pneumoniae, Streptococcus bovis, Serratia marcescens, Streptococcus pneumoniae) DNA was not amplified, and 6 Staphylococcus epidermidis isolates were amplified but not reported by the SeptiFast software because of their low concentration. Considering that the risk of developing sepsis is related to both the underlying disease and the clinical interventions, results obtained by the SeptiFast test and blood culture were stratified by groups of medical wards and descriptively summarized (see Table S1 in the supplemental material); in particular, the histograms in Fig. 2 show the concordance of results of blood culture versus SeptiFast for major pathogens and their distribution among medical wards. S. aureus, P. aeruginosa, Enterobacteriaceae, and Candida spp. prevail in ICUs and surgery wards, while CoNS are found mainly in the hematology, oncology, neonatology, emergency department, and pediatric wards. The distributions of enterococci, Streptococcus spp., and nonfermentative (NF) Gram-negative organisms are similar among the three groups. Of interest, CoNS, P. aeruginosa, Enterobacteriaceae, and Candida spp. were often identified exclusively by SeptiFast; S. aureus was never detected exclusively by blood culture.
Table 3.
Isolates detected by SeptiFast plus blood culture or by only one testa
| Species group and species | Total no. of positive blood samples (% of all positive blood samples) | No. of samples |
Concordance (%) | No. of samples with: |
|||
|---|---|---|---|---|---|---|---|
| SeptiFast positive/blood culture positive | SeptiFast positive/blood culture negative | SeptiFast negative/blood culture positive | SeptiFast contaminants | Blood culture contaminants | |||
| Gram-positive bacteria (128 pathogens and 108 contaminants) | |||||||
| Bacillus spp.b | 0 | 0 | 0 | 0 | 0 | 2 | |
| Corynebacterium spp.b | 0 | 0 | 0 | 0 | 0 | 2 | |
| Clostridium spp.b | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Enterococcus faecalis | 8 (3.0) | 7 | 0 | 1 | 87.5 | 1 | 2 |
| Enterococcus faecium | 4 (1.5) | 1 | 2 | 1 | 25.0 | 1 | 1 |
| Micrococcus spp.b | 0 | 0 | 0 | 0 | 0 | 1 | |
| Staphylococcus aureus | 31 (11.6) | 23 | 8 | 0 | 74.2 | 5 | 3 |
| CoNS | 79 (29.6) | 48 | 23 | 8 | 60.8 | 6 | 76 |
| Streptococcus pneumoniae | 2 (0.7) | 1 | 0 | 1 | 50.0 | 1 | 0 |
| Streptococci other than S. pneumoniae | 3 (1.1) | 1 | 1 | 1 | 33.3 | 1 | 6 |
| Gram-negative bacteria (107 pathogens and 35 contaminants) | |||||||
| Abiotrophia defectiveb | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Acinetobacter baumannii | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Citrobacter spp.b | 1 (0.4) | 0 | 0 | 1 | 0 | 2 | |
| Enterobacter aerogenes/E. cloacae | 12 (4.5) | 7 | 5 | 0 | 58.3 | 3 | 3 |
| Escherichia coli | 6 (2.2) | 5 | 1 | 0 | 83.3 | 3 | 5 |
| Klebsiella pneumoniae/K. oxytoca | 27 (10.1) | 12 | 14 | 1 | 44.4 | 6 | 1 |
| Burkholderia cepaciab | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Proteus mirabilis | 2 (0.7) | 1 | 1 | 0 | 50.0 | 0 | 0 |
| Pseudomonas aeruginosa | 44 (16.5) | 18 | 24 | 2 | 41.0 | 6 | 0 |
| Pseudomonas oryzihabitansb | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Salmonella spp.b | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Serratia marcescens | 7 (2.6) | 4 | 2 | 1 | 57.1 | 3 | 0 |
| Stenotrophomonas maltophilia | 4 (1.5) | 2 | 2 | 0 | 50.0 | 0 | 1 |
| Fungi (32 pathogens and 4 contaminants) | |||||||
| Candida albicans | 12 (4.5) | 4 | 6 | 2 | 33.3 | 1 | 1 |
| Candida famatab | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
| Candida krusei | 4 (1.5) | 2 | 1 | 1 | 50.0 | 0 | 0 |
| Candida parapsilosis | 13 (4.9) | 6 | 7 | 0 | 46.2 | 0 | 2 |
| Candida tropicalis | 1 (0.4) | 1 | 0 | 0 | 100 | 0 | 0 |
| Geotrichum capitatumb | 1 (0.4) | 0 | 0 | 1 | 0 | 0 | |
The total number of isolates tested was 414, which included 267 pathogens and 147 contaminants.
Microorganisms not in SML.
Fig. 2.
Concordance of results of blood culture and SeptiFast test related to clinical wards (NP, SeptiFast negative/blood culture positive; PN, SeptiFast positive/blood culture negative; PP, SeptiFast plus blood culture positive). NF Gram−, nonfermentative Gram-negative organisms. Numbers on the y axis refer to the number of isolates.
The higher rate of positive results by SeptiFast than blood culture was consistent across medical wards for the test by medical ward interaction term in a conditional logistic regression model (P = 0.6647).
Antimicrobial treatment may significantly impact the sensitivity of blood culture. Therefore, SeptiFast and blood culture results were analyzed for patients who either had received or not received antimicrobial treatment at the time of sampling (Table 4). SeptiFast showed a markedly and significantly higher rate of positive results than blood culture among specimens taken from patients receiving antibiotic therapy (14.1% versus 6.5%, respectively; P < 0.0001); this observation was apparent across all medical specialties.
Table 4.
Results obtained by SeptiFast and blood culture among patients receiving or not receiving antimicrobial therapy
| Ward and antimicrobial therapy | No. of samplesa |
|||
|---|---|---|---|---|
| SeptiFast/blood culture positive | SeptiFast positive/blood culture negative | SeptiFast negative/blood culture positiveb | Total | |
| Intensive care, surgery | ||||
| Yes | 26 | 34 | 4 | 64 |
| No | 31 | 6 | 2 | 39 |
| Oncology, hematology, neonatology | ||||
| Yes | 15 | 23 | 6 | 44 |
| No | 25 | 6 | 5 | 36 |
| Emergency department, pediatrics | ||||
| Yes | 9 | 26 | 1 | 36 |
| No | 37 | 2 | 1 | 40 |
The total numbers of pathogens from patients under antimicrobial therapy/patients not under antimicrobial therapy for the SeptiFast-positive plus blood culture-positive, SeptiFast-positive and blood culture-negative, and SeptiFast-negative and blood culture-positive groups were 50/93, 83/14, and 11/8, respectively (total, 144/115).
Including microorganisms not in the SML.
DISCUSSION
PCR-based methods have been reported to reduce the time to result (TTR) from more than 40 to 23 h compared to blood culture; however, these approaches still depend on the previous growth of organisms in culture (21). In contrast, the SeptiFast test is culture independent and delivers results in less than 6 h by allowing direct detection of organisms from whole blood. Moreover, in this study, we observed a markedly and significantly higher rate of positivity by the SeptiFast test than blood culture, paired with a lower number of contaminants. The rate of positive results was significantly higher by SeptiFast (14.6%) than by blood culture (10.3%) (P < 0.0001). Among those pathogens that SeptiFast identified in addition to those identified by blood culture, P. aeruginosa, CoNS, Klebsiella spp., and Candida spp. were the most frequently detected. In a cohort of newborns with sepsis, other authors evaluated amplification of the bacterial 16S rRNA gene for the early diagnosis of bloodstream infection: compared to blood culture, the sensitivity was 66.7% and the specificity was 87.5% (19). These percentages are markedly lower than the 85% and 93.5%, respectively, that we observed in the present study for the SeptiFast test compared to blood culture, reinforcing the evidence of its higher performance than previous PCR-based methods. Remarkably, SeptiFast identified additional pathogens (S. aureus, CoNS, Enterobacter aerogenes/E. cloacae, K. pneumoniae/K. oxytoca, P. aeruginosa, Candida albicans, and Candida parapsilosis) in 36.3% (97/267) of specimens that gave negative results by blood culture. Indeed, in many of these cases, a high rate of SeptiFast positivity was associated with high crossing-point values, indicating a low concentration of pathogens. Thus, infections caused in particular by Enterobacteriaceae, P. aeruginosa, and Candida spp. may be missed when blood culture alone is used, and the combined use of blood culture and SeptiFast may therefore improve patient care, i.e., in ICUs and surgery, oncology, hematology, and neonatology wards. Since patients in these medical wards are often undergoing empirical antimicrobial therapy to prevent bloodstream and other infections, we also investigated the role of the SeptiFast test in patients receiving antimicrobial treatment (2, 6). We observed a significantly higher rate of positive results by SeptiFast than blood culture in patients undergoing antimicrobial therapy, as previously reported for patients affected by infectious endocarditis (3). The finding was ascertained for patients belonging to all clinical groups. It seems to represent a step forward in the management of patients under antimicrobial treatment, regardless of the particular illness affecting them. However, a slight spectrum bias might be introduced, considering that several samples are more likely to be drawn for critically ill patients belonging to surgery wards and ICUs under prolonged antimicrobial treatment.
The use of molecular assays bears the danger of detecting contaminant DNA that could interfere with the presence of DNA from true pathogens (13). Actually, the presence of low concentrations of CoNS and Streptococcus species DNA may reflect contamination during the work flow at different stages, and therefore, it is not considered a reliable result by the SeptiFast software. In this regard, contamination rates for these isolates were significantly lower for SeptiFast (7/399, 1.8%) than for blood culture (82/399, 20.6%).
Despite several advantages of the PCR-based methods, a potential limitation of these molecular assays is the detection of DNA from dead microorganisms, resulting in clinically false-positive results. However, a recent report on the SeptiFast detection of pathogen DNA in blood has convincingly related the DNA presence to the actual infection status of the patient (22). There is no doubt that interpretation of results of the molecular assays should be assessed in a broader context that also accounts for other laboratory data and clinical conditions of the patient. Without consideration of those microorganisms not covered by the SML, SeptiFast missed 4.8% (19/399) of isolates that were interpreted to be pathogens. That proportion apparently highlights the potential true pitfall of the test. Indeed, a complete lack of amplification was observed in 3.3% (13/399) of these cases, possibly due to genetic variability or mutations of the target site, inappropriate sample preparation, and/or inhibition of PCR. We do not think that low-level bacteremia caused the lack of amplification, since the TTP of the respective blood culture samples was lower than the annual average calculated for the same microorganisms in our laboratory (data not shown). In the remaining 1.5% (6/399) of the missed cases (e.g., all S. epidermidis isolates in blood culture), the SeptiFast software reported negative results, despite amplification of bacterium-specific DNA (threshold). These samples were obtained from patients in groups a and b; all were carriers of central venous lines, for which bacterial colonization is commonly observed (11). The pathogens identified were characteristic for the respective medical ward and risk factors, and the representative nature of the pediatric cohort described here reinforces the significance of the SeptiFast results. Timely initiation of targeted antimicrobial therapy remains a crucial step to reduce morbidity and mortality in children affected by sepsis, e.g., full-term critical newborns affected by congenital immunodeficiency; preterm neonates; and pediatric patients suffering from malignancies, severe trauma, transplantation procedures, and burns. Furthermore, rapid detection of the infectious cause and prompt initiation of appropriate antimicrobial treatment are essential to reduce antibiotic resistance rates. Besides a 6-h TTR directly from whole blood and an increased sensitivity compared to blood culture, this study highlights the possibility that this real-time-based detection method may facilitate early patient-tailored specific antimicrobial treatment, especially in preterm newborns and full-term neonates affected by sepsis, for whom empirical treatment is generally abused in neonatal ICUs. Furthermore, this assay may dramatically reduce the number/definition of contaminant bacteria compared to that with blood culture, an issue which is of crucial importance in pediatric patients. However, there is no way to ensure that the entire sample set is collected according to guidelines (e.g., skin cleansing and blood culture inoculum) at the patient's bedside, leading to an inadequate criterion bias, although this is more representative of actual clinical practice.
One of the major limitations of this study was the retrospective design and the fact that the chart review was focused on the relationship between microbiological outcomes and groups (typology) of pediatric patients. However, this experience can be exploited as a pilot study in future prospective studies on age-related pediatric populations.
Overall, our study aims at integrating the entire group of studies on the SeptiFast assay, which appears to be an advantageous addition to blood culture for the diagnosis of bloodstream infection and improving the management and outcome of pediatric patients (3, 14, 16, 17, 22, 24, 26, 28, 29). We need to go further and address additional issues to completely evaluate the contribution of this real-time assay to the management and treatment of pediatric patients, especially under critical conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria Teresa D'Urbano and Silvia Tredici from the Unit of Microbiology, Children's Hospital and Research Institute Bambino Gesù, Rome, Italy, for their technical advice. We especially thank the physicians and departments involved in this study and the statistical support of John Duncan. We also acknowledge Andreina Santoro for her careful English revision of the manuscript.
Oliver Liesenfeld is an employee of Roche Molecular Diagnostics, manufacturer of the SeptiFast test used in this study. Beatrice Pizzorno is an employee of Roche Diagnostics Italy, distributor of the SeptiFast test in Italy. All the rest of us declare an absence of a conflict of interest.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 6 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Beekmann S. E., Diekema D. J., Chapin K. C., Doern G. V. 2003. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J. Clin. Microbiol. 41:3119–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carrigan S. D., Scott G., Tabrizian M. 2004. Toward resolving the challenges of sepsis diagnosis. Clin. Chem. 50:1301–1314 [DOI] [PubMed] [Google Scholar]
- 3. Casalta J. P., et al. 2009. Evaluation of the LightCycler SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 28:569–573 [DOI] [PubMed] [Google Scholar]
- 4. Connell T. G., Rele M., Cowley D., Buttery J. P., Curtis N. 2007. How reliable is a negative blood culture result? Volume of blood submitted for culture in routine practice in a children's hospital. Pediatrics 119:891–896 [DOI] [PubMed] [Google Scholar]
- 5. Danai P. A., Moss M., Mannino D. M., Martin G. S. 2006. The epidemiology of sepsis in patients with malignancy. Chest 129:1432–1440 [DOI] [PubMed] [Google Scholar]
- 6. Dellinger R. P., et al. 2008. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock. Crit. Care Med. 36:296–327 [DOI] [PubMed] [Google Scholar]
- 7. Fanaroff A. A., et al. 2007. Trends in neonatal morbidity and mortality for very low birthweight infants. Am. J. Obstet Gynecol. 196:147.e1–147.e8 [DOI] [PubMed] [Google Scholar]
- 8. Goldstein B., Giroir B., Randolph A., and the members of the International Consensus Conference on Pediatric Sepsis 2005. International Pediatric Sepsis Consensus Conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr. Crit. Care Med. 6:2–8 [DOI] [PubMed] [Google Scholar]
- 9. Hall K. K., Lyman J. A. 2006. Updated review of blood culture contamination. Clin. Microbiol. Rev. 19:788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jafari H. S., McCracken G. H., Jr 1992. Sepsis and septic shock: a review for clinicians. Pediatr. Infect. Dis. J. 11:739–748 [DOI] [PubMed] [Google Scholar]
- 11. Kirchhoff L. V., Sheagren J. N. 1985. Epidemiology and clinical significance of blood cultures positive for coagulase-negative staphylococcus. Infect. Control 6:479–486 [DOI] [PubMed] [Google Scholar]
- 12. Klaschik S., et al. 2004. Detection and differentiation of in vitro-spiked bacteria by real-time PCR and melting-curve analysis. J. Clin. Microbiol. 42:512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klaschik S., Lehmann L. E., Raadts A., Hoeft A., Stuber F. 2002. Comparison of different decontamination methods for reagents to detect low concentrations of bacterial 16S DNA by real-time-PCR. Mol. Biotechnol. 22:231–242 [DOI] [PubMed] [Google Scholar]
- 14. Lamoth F., et al. 2010. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J. Clin. Microbiol. 48:3510–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leibovici L., et al. 1997. Septic shock in bacteremic patients: risk factors, features and prognosis. Scand. J. Infect. Dis. 29:71–75 [DOI] [PubMed] [Google Scholar]
- 16. Maubon D., et al. 2010. Therapeutic impact and diagnostic performance of multiplex PCR in patients with malignancies and suspected sepsis. J. Infect. 61:335–342 [DOI] [PubMed] [Google Scholar]
- 17. Mussap M., et al. 2007. New diagnostic tools for neonatal sepsis: the role of a real-time polymerase chain reaction for the early detection and identification of bacterial and fungal species in blood samples. J. Chemother. 19:31–34 [DOI] [PubMed] [Google Scholar]
- 18. Peters R. P., van Agtmael M. A., Danner S. A., Savelkoul P. H., Vandenbroucke-Grauls C. M. 2004. New developments in the diagnosis of bloodstream infections. Lancet Infect. Dis. 4:751–760 [DOI] [PubMed] [Google Scholar]
- 19. Reier-Nilsen T., Farstad T., Nakstad B., Lauvrak V., Steinbakk M. 2009. Comparison of broad range 16S rDNA PCR and conventional blood culture for diagnosis of sepsis in the newborn: a case control study. BMC Pediatr. 19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rothman R. E., et al. 2002. Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J. Infect. Dis. 186:1677–1681 [DOI] [PubMed] [Google Scholar]
- 21. Tissari P., et al. 2010. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet 375:224–230 [DOI] [PubMed] [Google Scholar]
- 22. Tsalik E. L., et al. 2010. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. J. Clin. Microbiol. 48:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valles J., Rello J., Ochagavia A., Garnacho J., Alcala M. A. 2003. Community-acquired bloodstream infection in critically ill adult patients: impact of shock and inappropriate antibiotic therapy on survival. Chest 123:1615–1624 [DOI] [PubMed] [Google Scholar]
- 24. Varani S., et al. 2009. Diagnosis of bloodstream infections in immunocompromised patients by real-time PCR. J. Infect. 58:346–351 [DOI] [PubMed] [Google Scholar]
- 25. Vincent J. L., et al. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329 [DOI] [PubMed] [Google Scholar]
- 26. Wallet F., et al. 2010. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin. Microbiol. Infect. 16:774–779 [DOI] [PubMed] [Google Scholar]
- 27. Weinstein M. P., et al. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 24:584–602 [DOI] [PubMed] [Google Scholar]
- 28. Westh H., et al. 2009. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect. 5:544–551 [DOI] [PubMed] [Google Scholar]
- 29. Yanagihara K., et al. 2010. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit. Care 14:R159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zaragoza R., et al. 2003. The influence of inadequate empirical antimicrobial treatment on patients with bloodstream infections in an intensive care unit. Clin. Microbiol. Infect. 9:412–418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.