Since a fluoroquinolone-modifying enzyme gene, aac(6′)-Ib-cr, was first reported in 2006, it has rapidly spread among Enterobacteriaceae clinical isolates worldwide (7). AAC(6′)-Ib-cr differs from AAC(6′)-Ib by two amino acids, Trp102Arg and Asp179Tyr, and these substitutions allow it to reduce the antibacterial activities of norfloxacin and ciprofloxacin through acetylation of their piperazinyl substituent (6). Detection of aac(6′)-Ib-cr has so far depended mainly on genotyping, PCR, and sequencing (2). Recently, simultaneous high-resolution melting analysis and pyrosequencing were developed for detection (1, 3). However, these methods are costly and need specialized equipment. Therefore, the availability is limited to highly advanced institutions, such as research laboratories and university hospitals. In the present study, we developed a cost-effective and practical disk-based method to screen for AAC(6′)-Ib-cr producers.
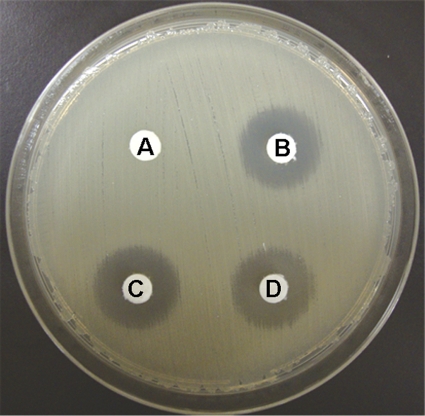
The Escherichia coli clinical isolates CR1 [aac(6′)-Ib-cr positive], N64 [aac(6′)-Ib positive], and BN41 [aac(6′)-Ib-cr and aac(6′)-Ib negative] were grown in LB broth containing norfloxacin (8 μg/ml), with shaking for 18 h at 35°C. The broth containing the same concentration of norfloxacin as the other tubes, but lacking any bacteria, was used as the control in this study. Ten microliters of each culture medium was applied on the blank disk set on a Mueller-Hinton agar plate inoculated with E. coli ATCC 25922 and incubated for 18 h at 35°C. The result is shown in Fig. 1. When the control medium was applied, an 18-millimeter growth-inhibitory zone (corresponding to 80 ng norfloxacin per disk) was observed. The significant decrease of a growth-inhibitory zone was observed when the culture medium of the aac(6′)-Ib-cr-positive E. coli strain CR1 was applied, while neither of the aac(6′)-Ib-cr-negative strains, N64 and BN41, showed a decrease in zone diameter. The decrease in zone diameter is an indicator of AAC(6′)-Ib-cr production and would be attributed to the inactivation of norfloxacin in the culture medium by the AAC(6′)-Ib-cr enzyme produced during growth (4).
Fig. 1.
Result of disk-based detection. (A) E. coli strain CR1 [aac(6′)-Ib-cr (+)]; (B) E. coli strain N64 [aac(6′)-Ib (+)]; (C) E. coli strain BN41 (negative for both genes); (D) control medium.
In this study, a total of 89 E. coli clinical isolates from our own strain collection, which were obtained from 49 facilities throughout Japan between 2002 and 2010, were subjected to the developed disk-based method. The MICs of norfloxacin for these strains were ≥16 μg/ml, and they showed visible growth in liquid broth containing 8 μg/ml of norfloxacin. The presence and absence of the aac(6′)-Ib-cr and aac(6′)-Ib genes in these isolates were preliminarily determined by PCR and nucleotide sequencing (5) but were not examined for the presence of other fluoroquinolone resistance mechanisms, such as qnr and mutations in DNA gyrase and topoisomerase IV, in the present study. As a result, all of the 19 aac(6′)-Ib-cr-positive strains showed a decrease in zone diameter of >10 mm, while all of the 70 aac(6′)-Ib-cr-negative strains, including 10 aac(6′)-Ib-positive ones, showed a growth-inhibitory zone that was the same size as that of the control medium. These results indicated that the new disk-based method developed here might have a specificity and sensitivity equivalent to those of genotyping using PCR and nucleotide sequencing. Therefore, the method seems to be effective for screening of AAC(6′)-Ib-cr producers, although application of the method is limited to bacterial strains that can grow in medium including norfloxacin.
We applied the method to aac(6′)-Ib-cr-positive clinical isolates of Klebsiella pneumoniae (n = 3), Serratia marcescens (n = 1), and Citrobacter freundii (n = 1), and we confirmed the >10-mm reduction in zone diameter seen in aac(6′)-Ib-cr-positive E. coli. This result indicates the possibility that the method could be applied not only to E. coli but also to the other bacterial strains belonging to the family Enterobacteriaceae. Extension of this method to members of the Enterobacteriaceae family other than E. coli would, however, require substantial additional experimentation aimed at demonstrating that the method is more broadly applicable to these other species.
We conclude that our method is simple and highly sensitive and specific for identification of AAC(6′)-Ib-cr producers and that it would be of great assistance in screening for AAC(6′)-Ib-cr producers and their epidemiology in clinical microbiology laboratories.
Acknowledgments
This study was supported by the Ministry of Health, Labor, and Welfare of Japan (grant H21-Shinkou-Ippan-008).
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Bell J. M., Turnidge J. D., Andersson P. 2010. aac(6′)-Ib-cr genotyping by simultaneous high-resolution melting analyses of an unlabeled probe and full-length amplicon. Antimicrob. Agents Chemother. 54:1378–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cano M. E., et al. 2009. Detection of plasmid-mediated quinolone resistance genes in clinical isolates of Enterobacter spp. in Spain. J. Clin. Microbiol. 47:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guillard T., et al. 2010. Rapid detection of aac(6′)-Ib-cr quinolone resistance gene by pyrosequencing. J. Clin. Microbiol. 48:286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jung C. M., Heinze T. M., Strakosha R., Elkins C. A., Sutherland J. B. 2009. Acetylation of fluoroquinolone antimicrobial agents by an Escherichia coli strain isolated from a municipal wastewater treatment plant. J. Appl. Microbiol. 106:564–571 [DOI] [PubMed] [Google Scholar]
- 5. Park C. H., Robicsek A., Jacoby G. A., Sahm D., Hooper D. C. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robicsek A., et al. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 7. Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]