Abstract
In addition to the large virulence plasmid pO157, a novel 38-kb conjugative plasmid, pO157_Sal, was identified and sequenced from an Escherichia coli O157:H7 outbreak-associated Chinese isolate that shares high similarity with a plasmid in Salmonella enterica serovar Agona. The plasmid was found in 15 of 326 isolates, 12 of which were of the same pulsed-field gel electrophoresis type.
Escherichia coli O157:H7 is an internationally well known pathogen first identified as the cause of a multistate outbreak in the United States in 1982 (22). It belongs to the enterohemorrhagic E. coli (EHEC) pathovar which comprises the etiological agents of bloody diarrhea and hemolytic uremic syndrome (HUS). Farm animals serve as the main reservoir of the pathogen, which does not usually cause disease. However, many O157:H7 human infections and outbreaks have been reported worldwide (1, 8, 9, 13). In 1999, a large outbreak causing 177 deaths occurred in Xuzhou City and in neighboring Anhui Province in China. Pulsed-field gel electrophoresis (PFGE) found that human isolates from the Xuzhou outbreak belonged to the same pulse type. Isolates of identical pulse types were also isolated from several types of animals in the villages, including pigs, cattle, goats, and chickens, suggesting that the outbreak isolates have a single origin. Later sampling from a variety of sources identified a diverse range of pulse types of O157:H7 isolates (27).
The O157:H7 genome contains the locus of enterocyte effacement island, an essential virulence factor for enteropathogenicity (11). It also contains the Stx2 and/or Stx1 phages and the pO157 plasmid, which give O157:H7 its characteristic properties (16). pO157, a 92-kb F-like plasmid, is found almost invariably in all E. coli O157:H7 clinical isolates (18) and shares sequence similarities with plasmids present in other EHEC serotypes (15, 23). A number of virulence factors encoded by pO157, including an enterohemolysin (ehxA), a type II secretion system, a serine protease (espP), a catalase-peroxidase (katP), a potential adhesin (toxB), a Cl esterase inhibitor (stcE), and attaching and effacing gene-positive conserved fragments (ecf), were identified (2, 25). ecf encodes a functional LpxM homologue that catalyzes the addition of myristate to the lipid A moiety of lipopolysaccharide (LPS) (25). Earlier plasmid-profiling studies showed the variable presence in O157:H7 isolates of, in addition to pO157, several plasmids ranging from 2 to 6 kb (19, 21). Two small plasmids have been sequenced, including a 6-kb pCOLD157 plasmid which encodes colicin functions (10) and a cryptic 3.3-kb pOSak1 plasmid from the Sakai strain, whose genome has been sequenced (17). In this study, we report the sequence of a novel 38-kb conjugative plasmid from an O157:H7 isolate and its nearly unique presence in the outbreak isolates in China.
Strains, plasmid sequencing, and PCR screening.
The O157:H7 strains used in this study were isolated from feces of human patients and animals between 1988 and 2005 from Xuzhou City, Jiangsu Province (194 strains), China, and the neighboring Anhui (30 strains), Henan (54 strains), and Shandong (12 strains) Provinces. Twenty-six isolates from other parts of China (Yunnan [21 strains], Tianjin [5 strains]), the United States (2 strains), and Japan (8 strains) were also included, as well as three fully sequenced strains, EDL933, Sakai, and TW14359 (Table 1). Sequencing of the novel plasmid from isolate Xuzhou-21 was done using an ABI BigDye Terminator V3.1 cycle sequencing kit and an ABI 3730 automated DNA analyzer (Applied Biosystems). To screen for the presence of the plasmid in other isolates, four pairs of PCR primers were designed (see Table S1 in the supplemental material). PCR was performed in a reaction volume of 20 μl containing 1× PCR buffer, 200 μM (each) dATP, dCTP, dTTP, and dGTP, 0.5 μM (each) primers, 1 U of Taq DNA polymerase (TaKaRa, Dalian, China), and 50 ng of genomic DNA templates. Amplification utilized an initial denaturing step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 1 min. A final step of 72°C for 7 min was used for final extension. PCR products (5 μl) were visualized on ethidium bromide-stained 1.2% agarose gels by illumination with UV light.
Table 1.
Characteristics of E. coli O157:H7 isolates used in this study
| Strain(s) | Location of isolation | Presence of pO157_Sal | Source | Toxin(s)a | PFGE typeb | STc | Yr of isolation |
|---|---|---|---|---|---|---|---|
| Xuzhou-21 | Xuzhou | + | Human | 1 and 2 | 0001 | 96 | 1999 |
| Xuzhou-65 | Xuzhou | + | Human | 1 and 2 | 0001 | 96 | 1999 |
| Xuzhou-143 | Xuzhou | + | Human | 1 and 2 | 0001 | 96 | 1999 |
| Xuzhou-223 | Xuzhou | + | Human | 1 and 2 | 0001 | 96 | 1999 |
| Xuzhou-284 | Xuzhou | + | Human | 1 and 2 | 0001 | 96 | 1999 |
| 99A024 | Xuzhou | + | Chicken | 1 and 2 | 0001 | 96 | 1999 |
| 99A032 | Xuzhou | + | Pig | 1 and 2 | 0001 | 96 | 1999 |
| 99A038 | Xuzhou | + | Chicken | 1 and 2 | 0001 | 98 | 1999 |
| 2063 | Xuzhou | + | Chicken | 1 and 2 | 0175 | NDe | 1999 |
| X114 | Xuzhou | + | Chicken | 1 and 2 | 0115 | 23 | 1999 |
| X119 | Xuzhou | + | Goat | 1 and 2 | 0001 | 96 | 1999 |
| X120 | Xuzhou | + | Bovine | 1 and 2 | 0001 | 96 | 1999 |
| F828 | Xuzhou | + | Pig | 1 and 2 | 0173 | ND | 1999 |
| F846 | Xuzhou | + | Fly | 1 and 2 | 0001 | ND | 1999 |
| F971 | Xuzhou | + | Goat | 1 and 2 | 0001 | ND | 1999 |
| EDL933 | United States | − | Human | 1 and 2 | ND | 23 | 1982 |
| Sakai | Japan | − | Human | 1 and 2 | ND | 23 | 1996 |
| TW14359 | United States | − | Human | 2 | ND | 100 | 2006 |
| Others (311 strains)d | I | − | II | 1 and 2 or 2 | III | IV | 1988–2005 |
1 and 2 refer to stx1 and stx2, respectively.
Pulsed-field gel electrophoresis (PFGE) XbaI pattern (see Fig. S2 in the supplemental material).
ST, sequence type based on multilocus sequence typing of 15 housekeeping genes.
I, Jiangsu Province, Anhui Province, Shandong Province, Yunnan Province, Henan Province, and Tianjin City, China; II, human, bovine, goat, pig, chicken, and vegetable; III, 137 PFGE types in total; IV, 5 sequence types in total.
ND, not determined.
Identification and sequencing of a novel conjugative plasmid in Xuzhou outbreak O157:H7 isolates.
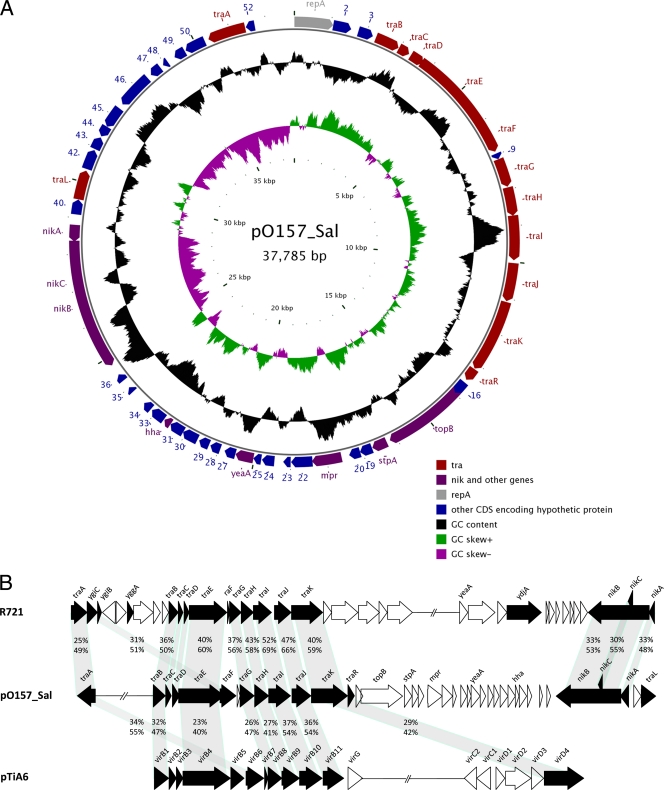
Plasmid analysis of an outbreak isolate (Xuzhou-21) from Xuzhou, China (unpublished data), revealed that the isolate contains a large plasmid in addition to the pO157 plasmid. We sequenced the plasmid from Xuzhou-21 and called it pO157_Sal, as the plasmid is closely related to an uncharacterized plasmid in Salmonella enterica serovar Agona strain SL483 (GenBank accession number CP001137). The circular plasmid is 37,785 bp. Its G+C content is 40.55%, which is lower than that of the chromosome of O157:H7. Fifty-two open reading frames (ORFs) encoding a product of at least 50 amino acids in length were identified (Fig. 1A and see Table S2 in the supplemental material). There are no known virulence-associated genes on the plasmid. However, 30 ORFs, some of which may code for virulence genes, have been identified as encoding hypothetical proteins. The average identity of the 48 ORFs shared by pO157_Sal and the S. enterica serovar Agona plasmid is 94%, ranging from 46% to 100%, with the majority of the ORFs (42) being over 90% identical. The two plasmids are colinear. Four pO157_Sal ORFs are unique. These two plasmids clearly share the same origin but have also diverged considerably.
Fig. 1.
Plasmid map and comparison with other conjugative plasmids. (A) Representational circular map of pO157_Sal. The inside circle is the G+C skew. The middle circle is the G+C content. The outside circle is the genetic organization of ORFs within the plasmid. The direction of transcription of each ORF is indicated. Genes are color coded as indicated. (B) Comparison of the tra regions of pO157_sal, R721, and pTiA6. Sequences for other plasmids were obtained from GenBank (accession numbers NC_002525 and NC_002377 for R721 and pTiA6, respectively). Filled arrows and arrowheads are T4SS-related genes. The percentages of amino acid identity (above) and similarity (below) are shown between the homologous genes.
The plasmid has a full set of genes for the type 4 secretion system (T4SS). Figure 1B shows the comparison of the tra (transfer) genes of the T4SS with the E. coli conjugative IncI2 plasmid R721 (12) and Agrobacterium tumefaciens plant tumor-inducing Ti plasmid pTiA6 (28). The tra genes resemble, in an approximately colinear fashion, members of the tra gene cluster of R721. In comparison to the prototype T4SS of the Agrobacterium pTi plasmid, traA, traB-traF, traG-traJ, and traK encode homologues of virB6, virB1-virB5, virB8-virB11, and virD4, respectively. However, no virB7 homologue was found in pO157_Sal. The proteins of the T4SS machinery can be grouped according to their functions and/or cellular locations (4, 24). The first group comprises three cytoplasm- or inner membrane-associated ATPases, namely, TraE, TraJ, and TraK; the second group comprises proteins forming core complexes in the periplasm and/or membrane, including TraA, TraG, TraH, and TraI; and the third group comprises TraC and TraF, components of the pilus. TraD is also a pilus-associated protein. TraB is a lytic transglycosylase that degrades the peptidoglycan cell wall at the site of T4SS assembly. Phylogenetic analysis of TraE showed that the pO157_Sal T4SS belongs to cluster 4, as defined by Frank et al. (6) (see Fig. S1 in the supplemental material), together with the Erwinia tasmaniensis plasmid pET35, Haemophilus influenzae plasmid pF3028 (14), Aeromonas culicicola plasmid pAc3249A, Vibrio harveyi plasmid pVIBHAR, and E. coli plasmid R721 (12). Most of these plasmids were reported from genome sequencing and are uncharacterized.
The T4SS can function as a conjugation machine in conjugative plasmids and as an effector translocator in bacterial pathogens (3). The T4SS in pO157_Sal is most likely involved in conjugation, as no other known effectors on the plasmid were found. pO157_Sal also contains other genes typical of a conjugative plasmid, including the genes nikB, nikC, and nikA for relaxosome formation (Fig. 1B). The plasmid is likely to belong to the IncI family of plasmids based on the sequence homology of relaxase (7). Sequence analysis showed six direct repeats (consensus sequence, 5′-GCAAACA-3′) upstream of nikA, a possible site of origin of transfer (oriT). Therefore, this new O157:H7 plasmid is likely to be a conjugative plasmid.
Conjugal transfer of pO157_Sal.
To demonstrate whether pO157_Sal is a conjugative plasmid, the kanamycin resistance (Kmr) gene was introduced into pO157_Sal using a one-step gene inactivation method as described by Datsenko and Wanner (5) and was inserted between pO157_Sal_36 and the nikB gene without disrupting any ORF. Plasmid mobilization was monitored by using filter paper mating as described by Yoshida et al. (26). E. coli O157:H7 Xuzhou-21, containing pO157_Sal (Kmr), was used as the donor and E. coli TB1 (streptomycin resistance) as the recipient. The donor and recipient were grown on Luria broth (LB) medium to an optical density at 600 nm (OD600) of 0.5, mixed equally, and then incubated on filter paper for 4 h. The filter paper mixture was then resuspended in LB medium, and dilutions were plated on LB agar containing streptomycin and kanamycin to select for transconjugants. Mobilization efficiency was calculated as the number of transconjugant colonies divided by the number of donor colonies. We found that pO157_Sal was mobilized with an average efficiency of 4 × 10−5. This result shows that pO157_Sal is a conjugative plasmid.
Distribution of pO157_Sal.
We used PCR testing of four pO157_Sal genes, traA, traE, mpr, and traL, to determine how many O157:H7 isolates carried the pO157_Sal plasmid. We screened a total of 326 isolates, including 314 isolates from a variety of sources from China and 12 isolates from other countries (Table 1). Fifteen isolates were found to be positive for all four genes. We also tested four other E. coli isolates, but none was found to be positive for the plasmid. All 15 pO157_Sal-positive isolates, including the five human isolates from the 1999 Xuzhou outbreak (27), were isolated in Xuzhou, China. These isolates were typed by PFGE previously (20). Twelve of the pO157_Sal-positive isolates were pulse type 0001, and all pulse type 0001 isolates were positive for the plasmid. Another three pO157_Sal-positive isolates (X114, F828, 2063) belonged to pulse types 0115, 0173, and 0175, respectively, all of which have only one isolate. Pulse types 0115 and 0173 are closely related to pulse type 0001, differing by two bands. However, pulse type 0175 is dissimilar to pulse type 0001, with five bands different (see Fig. S2 in the supplemental material). Therefore, these three pO157_Sal-positive isolates may not have the same origin as the other pO157_Sal-positive isolates.
Conclusions.
We have identified and sequenced a novel conjugative plasmid from O157:H7. The pO157_Sal plasmid belongs to the IncI family of conjugative plasmids. pO157_Sal contains 52 ORFs with 13 tra genes, a complete set for the T4SS. PCR screening of 326 O157:H7 isolates found that the majority of the pO157_Sal-positive isolates, which were isolated from humans and several other sources, belong to pulse type 0001. All human isolates from the 1999 Xuzhou outbreak, which resulted in high fatality from HUS in China, were shown to carry the pO157_Sal plasmid. Further studies will be conducted to determine the role of pO157_Sal in virulence.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this study is CP001927.
Supplementary Material
Acknowledgments
This work was supported by grants (2008ZX10004-001, 2008ZX10004-009, and 2009ZX10004-101) from the National Key Programs for Infectious Diseases of China.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Bell B. P., et al. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 272:1349–1353 [PubMed] [Google Scholar]
- 2. Brunder W., Schmidt H., Karch H. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305–3315 [DOI] [PubMed] [Google Scholar]
- 3. Cascales E., Christie P. J. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christie P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frank A. C., Alsmark C. M., Thollesson M., Andersson S. G. 2005. Functional divergence and horizontal transfer of type IV secretion systems. Mol. Biol. Evol. 22:1325–1336 [DOI] [PubMed] [Google Scholar]
- 7. Garcillan-Barcia M. P., Francia M. V., de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 8. Grant J., et al. 2008. Spinach-associated Escherichia coli O157:H7 outbreak, Utah and New Mexico, 2006. Emerg. Infect. Dis. 14:1633–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hart J., Smith G. 2009. Verocytotoxin-producing Escherichia coli O157 outbreak in Wrexham, North Wales, July 2009. Euro Surveill. 14(32):pii=19300 [PubMed] [Google Scholar]
- 10. Hofinger C., Karch H., Schmidt H. 1998. Structure and function of plasmid pColD157 of enterohemorrhagic Escherichia coli O157 and its distribution among strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 36:24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaper J. B. 1998. The locus of enterocyte effacement pathogenicity island of Shiga toxin-producing Escherichia coli O157:H7 and other attaching and effacing E. coli. Jpn. J. Med. Sci. Biol. 51(Suppl.):S101–S107 [DOI] [PubMed] [Google Scholar]
- 12. Kim S. R., Komano T. 1992. Nucleotide sequence of the R721 shufflon. J. Bacteriol. 174:7053–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King L. A., et al. 2009. Community-wide outbreak of Escherichia coli O157:H7 associated with consumption of frozen beef burgers. Epidemiol. Infect. 137:889–896 [DOI] [PubMed] [Google Scholar]
- 14. Kroll J. S., Farrant J. L., Tyler S., Coulthart M. B., Langford P. R. 2002. Characterisation and genetic organisation of a 24-MDa plasmid from the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Plasmid 48:38–48 [DOI] [PubMed] [Google Scholar]
- 15. Levine M. M., et al. 1987. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 156:175–182 [DOI] [PubMed] [Google Scholar]
- 16. Lim J. Y., Yoon J., Hovde C. J. 2010. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20:5–14 [PMC free article] [PubMed] [Google Scholar]
- 17. Makino K., et al. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1–9 [DOI] [PubMed] [Google Scholar]
- 18. Nataro J. P., Kaper J. B. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostroff S. M., et al. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994–998 [DOI] [PubMed] [Google Scholar]
- 20. Pang B., Zheng J. H. H., Sun H., Zhao A., Xu J. 2002. Molecular typing of Shiga-toxin producing Escherichia coli O157:H7 isolated in China with pulsed field gel electrophoresis. Zhonghua Liu Xing Bing Xue Za Zhi 23:123–126 [PubMed] [Google Scholar]
- 21. Paros M., Tarr P. I., Kim H., Besser T. E., Hancock D. D. 1993. A comparison of human and bovine Escherichia coli O157:H7 isolates by toxin genotype, plasmid profile, and bacteriophage lambda-restriction fragment length polymorphism profile. J. Infect. Dis. 168:1300–1303 [DOI] [PubMed] [Google Scholar]
- 22. Riley L. W., et al. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681–685 [DOI] [PubMed] [Google Scholar]
- 23. Schmidt H., Karch H., Beutin L. 1994. The large-sized plasmids of enterohemorrhagic Escherichia coli O157 strains encode hemolysins which are presumably members of the E. coli alpha-hemolysin family. FEMS Microbiol. Lett. 117:189–196 [DOI] [PubMed] [Google Scholar]
- 24. Yeo H. J., Waksman G. 2004. Unveiling molecular scaffolds of the type IV secretion system. J. Bacteriol. 186:1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon J. W., Lim J. Y., Park Y. H., Hovde C. J. 2005. Involvement of the Escherichia coli O157:H7(pO157) ecf operon and lipid A myristoyl transferase activity in bacterial survival in the bovine gastrointestinal tract and bacterial persistence in farm water troughs. Infect. Immun. 73:2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshida T., Takahashi I., Tubahara H., Sasakawa C., Yoshikawa M. 1984. Significance of filter mating in integrative incompatibility test for plasmid classification. Microbiol. Immunol. 28:63–73 [DOI] [PubMed] [Google Scholar]
- 27. Zheng H., et al. 2005. stx2vha is the dominant genotype of Shiga toxin-producing Escherichia coli O157:H7 isolated from patients and domestic animals in three regions of China. Microbiol. Immunol. 49:1019–1026 [DOI] [PubMed] [Google Scholar]
- 28. Zhu J., et al. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.