Abstract
PCR-based molecular assays have a central role in polyomavirus diagnostics. To assure optimal performance, target sequences should be regularly updated according to newly available sequences. The aim of this study was to review our in-house polyomavirus BK (BKV) and JC (JCV) real-time PCR assays. Database analysis revealed variations in the BKV target region which might affect the assay performance, while no significant changes were found in the JCV target region. We compared two degenerate versions of our BKV primers which accommodated at least 95% of all published genetic variants. Dilutions of cloned viral genomic DNA and probit analysis indicated an analytical sensitivity of the updated BKV assay of 4.15 copies/reaction and that of the JCV assay was 3.37 copies/reaction. The specificity was assessed by testing JCV- and BKV-positive samples that showed no cross-reactivity. The performance of the original and updated BKV assay was compared in 101 urine and 200 plasma samples submitted to our routine diagnostic laboratory revealed similar quantitative results. We conclude that our JCV and updated BKV real-time PCR assays are robust and detect rare variants possibly encountered in the clinical routine.
INTRODUCTION
PCR-based molecular assays have a central role in diagnosing and monitoring of polyomavirus diseases such as polyomavirus virus-associated nephropathy (PyVAN), commonly caused by polyomavirus BK (BKV), and PyV-associated progressive multifocal leukoencephalopathy (PML), caused by polyomavirus JC (JCV) (5). Since 35 to 90% of healthy adult are seropositive for BKV and JCV, serological assays are rarely used for diagnosing ongoing PyV-associated diseases (3, 9, 24). Instead, BKV DNA loads in urine and blood have become pivotal laboratory assays in the management of kidney transplants (11, 12, 17). Urine BKV PCR allows to rule out PyVAN with a high negative predictive value above 95%, whereas plasma BKV loads >4 log10 genome equivalents (GEq)/ml persisting for more than 3 weeks have a positive predictive value of 50 to 80% (8, 17, 20, 34). Accordingly, BKV loads are currently recommended for screening and monitoring kidney transplant patients (16, 20, 23).
Similarly, detection of JCV DNA in cerebrospinal fluid (CSF) by PCR is recommended as first line diagnostic test for patients with possible PML. The diagnostic sensitivity of the qualitative JCV DNA PCR in CSF ranged from 70% to >90%, and the specificity ranged from 80% to more than 99% (6, 18). In recent years, most laboratories developed quantitative real-time PCR assays, which also provide an estimate of JCV DNA level in the CSF (5).
Given the central role of the PCR assay for diagnosing virus-associated pathologies, quality control issues regarding DNA preparation and the characteristics of the PCR assay become essential in the clinical virology laboratory. To ensure optimal performance, the target sequences should be chosen carefully in conserved regions and be regularly evaluated against newly available sequences (15). Although this is almost imperative for RNA viruses such as influenza virus which are prone to higher error rates during genome replication, as well as reassortments, it should be noted that sequence variabilities may also occur in DNA viruses, including BKV and JCV. Here, sequence variations concern typically the noncoding control region, the capsid protein VP1 and less frequently the large T antigen (1, 4, 13, 14, 22, 25, 29, 31, 32). Moreover, six new PyVs have been detected in human specimens in recent years bearing hitherto-unknown genomic sequences that may affect the analytic specificity of established assays (7, 30, 33). As more PyV sequences become available, there is a need to reevaluate and potentially optimize existing real-time PCR assays for BKV and JCV.
MATERIALS AND METHODS
Oligonucleotides and reference plasmid.
The primers and probes of the original and updated assays are listed in Table 1. The reference plasmid pBKV2 contains the whole BKV genome (strain Dunlop) cloned into a pGEM vector (Promega, Madison, WI) (14). Our reference plasmid pJCV4 contains the whole JCV genome of strain MAD4 cloned in a pGEM vector (13).
Table 1.
Primers and probes
| Primer or probe | Sequence (5′–3′)a | Assay |
|---|---|---|
| BK-Forward | AGCAGGCAAGGGTTCTATTACTAAAT | Original BKV PCR |
| BK-Reverse | GAAGCAACAGCAGATTCTCAACA | Original BKV PCR |
| BK-Probe | 5′FAM-AAGACCCTAAAGACTTTCCCTCTGATCTACACCAGTTT-TAMRA | Original BKV PCR |
| BK-Deg-Forward | AGCAGGCAAGRGTTCTATTACTAAAT | BKV-2.0 PCR |
| BK-Deg2-Forward | AGCAGGCAAGDGTTCTATTACTAAAT | BKV-2.1 PCR |
| BK-Deg-Reverse | GARGCAACAGCAGATTCYCAACA | BKV-2.0 and -2.1 PCR |
| BK-Deg-Probe | 5′FAM-AAGACCCTAAAGACTTTCCYTCTGATCTACACCAGTTT-TAMRA | BKV-2.0 and -2.1 PCR |
| JC-Forward | CTAAACACAGCTTGACTGAGGAATG | Original JCV PCR |
| JC-Reverse | CATTTAATGAGAAGTGGGATGAAGAC | Original JCV PCR |
| JC-Probe | 5′FAM-TAGAGTGTTGGGATCCTGTGTTTTCATCATCACT-TAMRA | Original JCV PCR |
Degenerated positions in the updated BKV oligonucleotides are indicated in boldface.
BLAST search.
The target sequences of our BKV and JCV real-time PCR assays were used as query for a homology search using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/blast.cgi). To avoid shortening of the sequence ends by the algorithm, 10 additional nucleotides were added to the query sequences upstream and downstream of the 5′ and 3′ ends, respectively. The default BLAST parameters were used (expect threshold, 10; word size, 28; scoring matrix, PAM30; match/mismatch scores, 1/−2; gap cost, linear), but the maximal number of target sequences was set to 20,000). To facilitate the analysis of the data, the layout of the search results was set to “query anchored with dots for identities.” The primers and probe binding sites in the resulting raw alignment were highlighted in a text processing software and analyzed individually. Identical sequences were summarized as one sequence variant in the edited alignment. The number of sequences constituting a particular variant was counted, and the percentage of the total number of sequences was calculated. Nucleotide polymorphisms present in at least 10% of the total number of the sequences were taken into account as degenerated positions in the updated primers. The adapted forward primer for the BK-PCR was tested in two variants: BK-Deg-Forward included a “R” (A or G) at position 11, whereas BK-Deg2-Forward had a “D” (A, T, or G) at this position. The JCV oligonucleotides were found to match 98, 95, and 91% of the available National Center for Biotechnology Information (NCBI) sequences for the forward primer, the reverse primer, and the probe, respectively, and were therefore considered not to require adaptations. The BLAST search were originally performed in May 2008 and repeated in January 2010.
Clinical specimens.
We prospectively collected 101 plasma and 100 urine samples of 131 patients sent to our laboratory for BKV diagnostics (89 men [median age, 54 years; interquartile range {IQR}, 40 to 62 years], 38 women [median age, 50 years; IQR, 35 to 61 years], and 4 patients of unknown gender). These samples were analyzed with the original and the new BKV real-time PCR assay (BKV-2.0 PCR). In addition, for the evaluation of the updated BKV assay using the BK-Deg2-Forward primer (BK-Deg2.1 PCR), we retrospectively analyzed 100 plasma samples of 77 patients (48 men [median age, 54 years; IQR, 36 to 58 years] and 29 women [median age, 48 years; IQR, 41 to 58 years]).
The JCV assay was evaluated on clinical samples of a cohort of 103 renal transplant recipients shedding decoy cells (8), as well as in 400 healthy blood donors (10).
DNA extraction.
DNA extraction was performed with a Corbett Xtractor (Qiagen, Hombrechtikon, Switzerland) and a VX DNA/RNA extraction kit (Qiagen). The input volume was 200 μl; the elution volume was 100 μl.
PCR conditions for the BKV and JCV assays.
Every clinical sample was tested in quadruplicate using 5-μl DNA extracts from plasma or whole blood. The 25-μl PCR test contained 300 nM concentration of each primer, 200 nM FAM-labeled probe (Table 1), and 12.5 μl of 2-fold-concentrated amplification master mix (Eurogentec, Seraing, Belgium) containing the polymerase, 5 mM MgCl2, deoxynucleoside triphosphate (including dUTP), and uracil-N-glycosylase. Quantification was performed by using a standard curve generated by three concentrations (102, 104, and 106 copies/PCR) of the reference plasmid. The slope of the standard curve (log copies/PCR plotted against the cycle threshold [CT] value) ranged from −3.21 to −3.73 (mean, 3.54). The temperature profile consisted of a preincubation step at 50°C for 2 min to allow for enzymatic decontamination of potential uracyl-containing amplicons, followed by 95°C for 10 min for activation of the polymerase and 45 cycles of 95°C for 15 s and 60°C for 60 s. One of four replicates was spiked with 1,000 copies of the reference plasmid to monitor for PCR inhibition. Samples were considered detectable if at least two replicates had CT values below 45. The PCR assays were performed on an ABI 7500 fast cycler (Applied Biosystems) using a fixed threshold of 0.1 for PCR analysis.
Sensitivity and specificity of the BKV and JCV assays.
The reference plasmids pBKV2 and pJCV4, respectively, were diluted in 2-fold steps with Tris-EDTAbuffer containing 5 ng of salmon sperm DNA/ml. The dilution ranged from 100 GEq per PCR down to 0.39 GEq per PCR test. Each concentration was tested in at least two runs of five replicates. The lower limit of detection was determined by probit analysis using the SPSS software package version 18.0. In addition, the sensitivity and specificity of the BKV-assay was tested by spiking 10,000 GEq of cloned JCV DNA per PCR test. In addition, the updated BKV assays were performed on 10 clinical samples containing JCV DNA.
Statistical analysis.
The agreement between the original assay and both versions of the new assay was calculated with the Bland-Altman algorithm using the GraphPad Prism software version 5.00 (2).
RESULTS
Analysis of the BLAST search for BKV PCR.
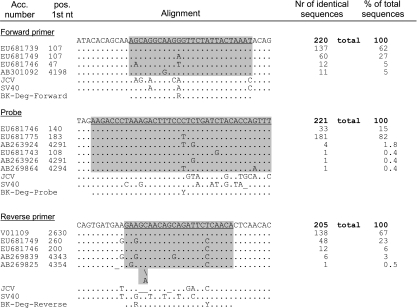
A BLAST search of the BKV target sequence (133 nucleotides [nt] in the LT gene) resulted in 221 hits (January 2010). The edited alignments for BKV are presented in Fig. 1. (The alignments for JCV [Fig. 2] are discussed below.) For the forward primer, 137 sequences (62%) were identical to the original primer-binding sequence. A single polymorphism at nt 11 (A instead of G) was present in 60 sequences (27%). At this position, a T was present in 5% of the sequence. This position was degenerated to an R (purine nucleotide, A or G) in the updated forward primer BK-Deg-Forward and to a D (A, T, or G) in BK-Deg2-Forward. The other polymorphisms accounting for <10% of the total sequences were not included in the updated primer. The reverse primer binding site was identical to the original primer sequence in 138 database entries (67%). However, a sequence variant containing two polymorphisms (G instead of A at position 3 and C instead of T at position 18) accounted for 23% of the sequences (48 database entries). These positions had degenerated to R and Y, respectively, in the updated reverse primer sequence (BK-Deg-Reverse). For the probe, 33 sequences (15%) were identical to the original primer binding sequence. At position 20, most sequences (181, 82%) contained a T instead of a C. This position was therefore degenerated to a Y (pyrimidine nucleotide, C or T) in the updated reverse probe BK-Deg-Probe (Fig. 1).
Fig. 1.
Alignment of the oligonucleotides of the BKV real-time PCR. The top sequence corresponds to the Dunlop prototype strain. Each line corresponds to a representative sequence for a variant. The accession number and the position of the first nucleotide in the database entry are indicated. The number of sequences constituting a variant and the percentage of the total number of sequences are shown in the two columns on the right. Nucleotides identical to the query sequence are indicated as a dot (.); substitutions are indicated by the corresponding nucleotide one-letter code. Gaps are indicated by an underscore; insertions are denoted by a backslash (\). The binding sites of the oligonucleotides are underlined in the query sequence and shaded gray in the alignment. The corresponding sequences of JCV and simian virus 40 (SV40) are shown for comparison. The sequences of the updated oligonucleotides BK-Deg-Forward, -Reverse, and -Probe are shown on the last line.
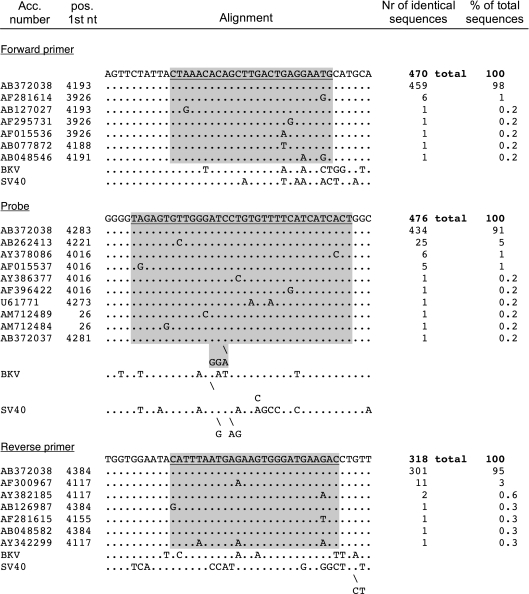
Fig. 2.
Alignment of the oligonucleotides of the JCV real-time PCR. The top sequence corresponds to the MAD-1 prototype strain. Each line corresponds to a representative sequence for a variant. The accession number and the position of the first nucleotide in the database entry are indicated. The number of sequences constituting a variant and the percentage of the total number of sequences are shown in the two columns on the right. Nucleotides identical to the query sequence are indicated as a dot (.); substitutions are indicated by the corresponding nucleotide one-letter code. Gaps are indicated by an underscore; insertions are indicated by a backslash (\). The binding sites of the oligonucleotides are underlined in the query sequence and shaded gray in the alignment. The corresponding sequences of BKV and SV40 are shown for comparison.
Sensitivity and specificity of the new BKV-2 assays.
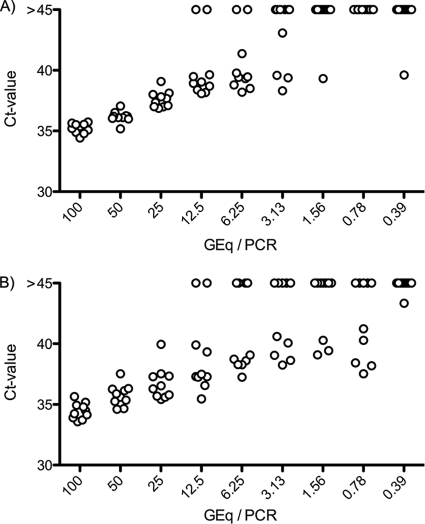
The sensitivity of the BKV-2.0 PCR assay was evaluated by using limiting dilution of a cloned BKV genome (plasmid pBKV2). The lower limit of detection was determined by probit analysis to be 4.15 GEq/PCR (Fig. 3). After correction for the dilution factor, this is equivalent to a lower limit of detection of 415 GEq/ml for our routine blood or urine samples. This value was identical to the original assay (data not shown). The sensitivity of the BKV-2.1 PCR was identical. The same limit of detection was obtained in the presence of 1 × 10E4 JCV GEq/PCR, excluding underquantification in the presence of competing JCV DNA (data not shown).
Fig. 3.
Sensitivity test of the PCR assays. (A) Updated BKV-2.0 assay. The plasmid pBKV2 harboring the whole BKV genome was diluted in 2-fold steps to a theoretical concentration of 0.39 copies per PCR. The lower limit of detection calculated by probit analysis was 4.15 GEq/PCR. (B) JCV assay. The plasmid pJCV4 harboring the whole JCV genome was similarly diluted in 2-fold steps. The calculated lower limit of detection was 3.37 GEq/PCR. The dots indicate the obtained CT values for the individual replicates.
Performance of the BKV-2.0 PCR assay compared to the original assay.
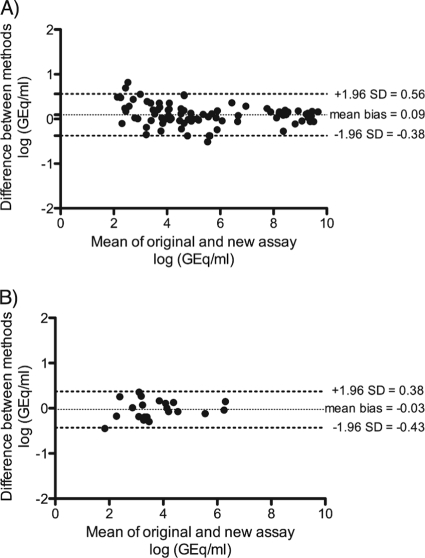
We prospectively collected 101 plasma and 100 urine samples of 131 patients sent to our laboratory for BKV diagnostics. Most of the patients were undergoing routine BKV testing following kidney transplantation. These samples were analyzed with the original and the BKV-2 real-time PCR assay. Both assays were positive for 66 (66%) and negative in 33 (33%) urine samples. There was only one discrepant result for a sample that tested positive with the original assay and negative with the updated assay. This sample had a quantitative value of 508 copies/ml near the limit of detection of the assay and therefore was more subject to strong variations of quantification. Overall, the sensitivity of the updated assay was 99% and the specificity was 98% for urine when the original assay was used as a reference. Most of the plasma samples (80 of 101 [80%]) were negative by both assays, as expected since BKV DNAemia is rarer than BKV DNAuria. Both assays were positive for 17 samples. Four samples were discrepant. Two were positive with the new assay and negative with the original assay (124 and 321 copies/ml). Two were positive with the original assay and negative with the updated assay (105 and 134 copies/ml). All four discrepant samples had quantitative values below the calculated limit of detection of the assays. Overall, the sensitivity of the updated assay was 89% and the specificity 98% for plasma when the original assay was used as a reference. The quantitative agreement of the original and the new assay was excellent, with a Bland-Altman bias of 0.09 and a confidence interval of 0.56 to −0.38 (Fig. 4A).
Fig. 4.
Agreement of the quantification the original with the new BKV assays. (A) Bland-Altman plot of the original assay compared to the new assay using the BK-Deg-Forward primer (BKV-2.0 PCR). A total of 83 positive samples was analyzed (66 urine and 17 plasma samples). The mean bias was 0.09, and the 95% limit of agreement was between −0.38 and 0.56. (B) Bland-Altman plot of the original assay compared to the new assay using the BK-Deg2-Forward primer (BKV-2.1 PCR). A total of 21 positive samples were analyzed (all plasmas). The mean bias was −0.03, and the 95% limit of agreement was between −0.43 and 0.38.
Performance of BKV-2.1 PCR compared to the original assay.
For the evaluation of the updated BKV assay using the BK-Deg2-Forward primer, we collected prospectively 100 plasma samples of 77 patients sent to our laboratory for BKV diagnostics. Most of the patients were undergoing routine BKV check after kidney transplantation. These samples were analyzed with the original and the second version of the new BKV-2 PCR assay.
In this panel, most of the plasma samples (75 of 100, 75%) were negative according to both assays. Both assays were positive for 21 samples. Four samples were discrepant. One was positive with the updated assay and negative with the original assay (158 copies/ml), and three were positive with the original assay and negative with the updated assay (105, 208, and 240 copies/ml). All four discrepant results had quantitative values below the limit of detection of the assays. The sensitivity of the updated assay with the BK-Deg2-Forward primer was 95%, and the specificity was 97% when the original assay was used as a reference. The agreement between the BKV-2 PCR assay and the original assay calculated with the Bland-Altman algorithm was excellent (bias, −0.03; confidence interval, 0.38 to −0.43 [Fig. 4B]).
Analysis of the BLAST search for JCV PCR.
The BLAST search of the JCV target sequence (172 nt in the LT gene) resulted in 476 hits, i.e., more than twice the number of what was found for BKV. The edited alignment for JCV is shown in Fig. 2. Since no sequence variant accounted for >5% of the total sequences, the assay did not require any modification.
Performance of the JCV assay.
The sensitivity of the JCV PCR assay was evaluated using limiting dilution of a cloned BKV genome (plasmid pJCV4). The lower limit of detection was determined by probit analysis to be 3.37 GEq/PCR (Fig. 4B). After correction for the dilution factor used in blood and urine, this is equivalent to a lower limit of detection of 337 GEq/ml for these clinical samples.
The clinical performance of the JCV assay was demonstrated in a previously published study in patients at high-risk for JCV replication (8), as well as in low-risk healthy blood donors (10). In the former study, 27.2% of the urine samples from renal transplant recipients containing decoy cells, a result indicative of PyVAN, were positive for JCV DNA with our assay. In the patients with biopsy-proven PyVAN, 21.4% were positive with JCV DNA. In the latter study, 400 blood donors were screened for JCV DNA in urine and blood samples. Low-level JCV shedding was detected in 19% of the urine samples (10).
DISCUSSION
In the last decade, the wide-spread use of PCR in virus diagnostics and the increasing availability of sequencing facilities have resulted in an increase of the number of viral sequences in public databases. The number of sequences of influenza viruses available in the NCBI database, for example, has grown from less than 5,000 in 1997 to more than 100,000 in 2010 (http://www.ncbi.nlm.nih.gov/genomes/flu/growth.html). Similarly, the number of available BKV and JCV DNA sequences has also increased, and many molecular assays for detection and quantification have been published since the development of our PCR assays more than 10 years ago (17, 19, 26, 27). Even though PyVs are DNA viruses depending on the host cell DNA polymerase, expression of the large T-antigen has been associated with genetic instability and under conditions of high-level replication, variants in the PyV genome appear to emerge with changing majority species. This concerns particularly the viral noncoding control region of BKV and JCV, which we and others have recently reexamined (1, 13, 14, 28, 29). Similarly, changes were also noted in the PyV capsid regions (1, 31), which may negatively affect the detection and quantification of these polyomavirus variants (25).
Recently, a systematic single-center study compared four published and three newly designed BKV load assays targeting different sequences of the large T antigen and the VP1 gene, among others, our original BKV PCR (assay T1 [21]). Marked variability was noted that was associated with polymorphisms particularly among the less common BKV subtype III and IV isolates. The overall performance of our assay here was robust, but some quantitative discrepancies were observed when a cloned Dunlop sequence was used as a quantification standard. We hypothesized that this was due to the oligonucleotides binding perfectly to the Dunlop standard but not to the extracted DNA, resulting in a suboptimal amplification of the clinical samples and therefore a lower quantification. Better results were obtained when a quantification standard derived from pooled urine samples was used, probably because this standard had a mismatch pattern similar to that of the tested clinical samples. In the same study, suboptimal performances were predicted with subtypes III and IV due to multiple mismatch in the oligonucleotide binding sites (21). It was therefore necessary to reevaluate our assays and to take into account sequences newly added to databases. In their study, Hoffman et al. (21) solved the problem of suboptimal quantification of some subtypes by designing an assay targeting two regions. We used an alternative approach by including degenerated position in the primers and probe, taking into account the most frequent sequence variants and therefore allowing improved detection and quantification of rare BKV subtypes. The two versions of the updated assay performed equally well compared to the original assay on a quantitative level in an evaluation that included 101 urine and 200 plasma samples. The few discrepant results were below the calculated limit of detection since the probit analysis indicates a threshold viral load detected in 100% of the cases. Below this level, positive signals can be picked up, especially if multiple replicas are performed, e.g., in triplicates as in our routine.
We observed no striking difference between the original and our new BKV-2 PCR assays. Possibly, BKV variants harboring mismatches in the oligonucleotide binding sites were not represented in the tested panels. In a recent study in collaboration with the local blood donation center, we showed that BKV subtype I is the most frequently detected variant in the healthy population in the region of Basel (10). Since the sequence of this subtype almost perfectly matches the target sequence of the original assay, the improvement due to the updated primers might not be critical for subtype I. However, rarer subtypes, such as subtype III, might be better quantified with the updated assay. Since most of the samples were collected at the local hospital, it is likely that the dominant BKV subtype might be overrepresented in this panel. Since the updated assays performed as well as the original assay with regard to sensitivity and specificity, the substitution of the assays could be done without any drawback for routine diagnostics. Of note is the fact that the calculated limit of detection of our assay (415 GEq/ml) is well below the BK viral load of 10,000 GEq/ml that defines presumptive PyVAN (16). Bland-Altman analysis indicates that the quantitative differences are minor and likely without impact for the clinical interpretation.
Our JCV assay appeared to target a conserved region of the genome, with only a few new variants and likely little effect on the performance. The JCV assay was extensively evaluated on clinical samples from high-risk patients such as kidney transplant recipients with high-level viruria and decoy cell shedding and histologically confirmed BKV- or JCV-PyVAN (8), as well as in a low-risk population of blood donors (10). Therefore, no update of primers and probe appeared to be required. However, given the central role of PCR for the diagnostics of JCV PML, periodic reevaluation of the sequence should be performed. Recent recommendations for the diagnosis of PyV-associated PML state that the level of detection for JCV DNA should be in the range of 50 GEq/ml of CSF. However, while some research groups report to reach this limit, most publications fail to demonstrate rigorous data to support this claim. Since a sensitivity of 1 GEq per PCR is theoretically possible, its formal demonstration requires a statistical analysis because of the stochastic distribution at this low copy number. Our probit analysis provides data indicating a sensitivity of 3.37 GEq per PCR test, which is very close to the theoretical limit. For our JCV assay, this diagnostic milestone of 50 copies per ml could therefore only be reached with a 14-fold concentration of the starting material, requiring at least a volume of 1.4 ml of CSF for 100 μl of eluted nucleic acids. Clinicians and laboratory experts need to be aware of this factors, since CSF is frequently a limiting factor in clinical diagnostics, especially when the list of differential diagnosis is broad. Moreover, robust quantitative extraction procedures not enriching inhibitory substances must be established to avoid a negative impact on the sensitivity of the PCR assays.
One limitation of the present study is the fact that we based the choice of degenerated positions on the frequency of the polymorphisms in the sequences available in the NCBI database. This might not reflect the epidemiological frequency of the individual variants, so that some polymorphisms might be overrepresented in our analysis. We observed that polymorphisms of the target sequence of our assay can be found in 32% of the sequences at position 11 of the forward primer, in 23% of the sequences at positions 3 and 18 of the reverse primer, and in 82% of the sequences at position 20 of the probe. By making use of degenerated oligonucleotides, we took into account these polymorphisms and showed that the updated assays were as efficient as the original PCR, with no apparent drawback for the sensitivity and might improve the detection of rare variants. We have therefore switched to this updated BKV-2.1 PCR assay in the clinical routine.
ACKNOWLEDGMENTS
We thank Simone Edelmann and the team of the Molecular Diagnostics Laboratory at the Institute for Medical Microbiology of the University of Basel for their excellent technical support.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Baksh F. K., et al. 2001. Molecular genotyping of BK and JC viruses in human polyomavirus-associated interstitial nephritis after renal transplantation. Am. J. Kidney Dis. 38:354–365 [DOI] [PubMed] [Google Scholar]
- 2. Bland J. M., Altman D. G. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307–310 [PubMed] [Google Scholar]
- 3. Bodaghi S., et al. 2009. Antibody responses to recombinant polyomavirus BK large T and VP1 proteins in pediatric kidney transplant patients. J. Clin. Microbiol. 47:2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., et al. 2004. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J. Gen. Virol. 85:2651–2663 [DOI] [PubMed] [Google Scholar]
- 5. Cinque P., Dumoulin A., Hirsch H. H. 2009. Diagnosis of polyomavirus infection, replication and disease, p. 401–424 In Jerome K. (ed.), Laboratory diagnosis of viral infections. Informa Healthcare, New York, NY [Google Scholar]
- 6. Cinque P., Scarpellini P., Vago L., Linde A., Lazzarin A. 1997. Diagnosis of central nervous system complications in HIV-infected patients: cerebrospinal fluid analysis by the polymerase chain reaction. AIDS 11:1–17 [DOI] [PubMed] [Google Scholar]
- 7. Dalianis T., Ramqvist T., Andreasson K., Kean J. M., Garcea R. L. 2009. KI, WU, and Merkel cell polyomaviruses: a new era for human polyomavirus research. Semin. Cancer Biol. 19:270–275 [DOI] [PubMed] [Google Scholar]
- 8. Drachenberg C. B., et al. 2007. Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: a prospective evaluation. Transplantation 84:323–330 [DOI] [PubMed] [Google Scholar]
- 9. Egli A., Dumoulin A., Köhli S., Hirsch H. H. 2009. Polyomavirus BK after kidney transplantation: role of molecular and immunological markers. Trends Transplant. 3:85–102 [Google Scholar]
- 10. Egli A., et al. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J. Infect. Dis. 199:837–846 [DOI] [PubMed] [Google Scholar]
- 11. Egli A., et al. 2007. Cytomegalovirus and polyomavirus BK posttransplant. Nephrol. Dial. Transplant. 22(Suppl. 8):viii72–viii82 [DOI] [PubMed] [Google Scholar]
- 12. Funk G. A., Gosert R., Comoli P., Ginevri F., Hirsch H. H. 2008. Polyomavirus BK replication dynamics in vivo and in silico to predict cytopathology and viral clearance in kidney transplants. Am. J. Transplant. 8:2368–2377 [DOI] [PubMed] [Google Scholar]
- 13. Gosert R., Kardas P., Major E. O., Hirsch H. H. 2010. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. J. Virol. 84:10448–10456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gosert R., et al. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 205:841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunson R. N., Collins T. C., Carman W. F. 2006. Practical experience of high throughput real-time PCR in the routine diagnostic virology setting. J. Clin. Virol. 35:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirsch H. H., et al. 2005. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation 79:1277–1286 [DOI] [PubMed] [Google Scholar]
- 17. Hirsch H. H., et al. 2002. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 347:488–496 [DOI] [PubMed] [Google Scholar]
- 18. Hirsch H. H., et al. 1998. HIV-1-infected patients with focal neurologic signs: diagnostic role of PCR for Toxoplasma gondii, Epstein-Barr virus, and JC virus. Clin. Microbiol. Infect. 4:577–584 [DOI] [PubMed] [Google Scholar]
- 19. Hirsch H. H., Mohaupt M., Klimkait T. 2001. Prospective monitoring of BK virus load after discontinuing sirolimus treatment in a renal transplant patient with BK virus nephropathy. J. Infect. Dis. 184:1494–1496 [DOI] [PubMed] [Google Scholar]
- 20. Hirsch H. H., Randhawa P. 2009. BK virus in solid organ transplant recipients. Am. J. Transplant. 9(Suppl. 4):S136–S146 [DOI] [PubMed] [Google Scholar]
- 21. Hoffman N. G., Cook L., Atienza E. E., Limaye A. P., Jerome K. R. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J. Clin. Microbiol. 46:2671–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin L., Gibson P. E., Knowles W. A., Clewley J. P. 1993. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J. Med. Virol. 39:50–56 [DOI] [PubMed] [Google Scholar]
- 23. Kasiske B. L., et al. 2009. KDIGO clinical practice guideline for the care of kidney transplant recipients. Am. J. Transplant 9(Suppl. 3):S1–S155 [DOI] [PubMed] [Google Scholar]
- 24. Khanna N., et al. 2009. JC virus-specific immune responses in human immunodeficiency virus type 1 patients with progressive multifocal leukoencephalopathy. J. Virol. 83:4404–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landry M. L., Eid T., Bannykh S., Major E. 2008. False-negative PCR despite high levels of JC virus DNA in spinal fluid: implications for diagnostic testing. J. Clin. Virol. 43:247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nickeleit V., et al. 1999. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J. Am. Soc. Nephrol. 10:1080–1089 [DOI] [PubMed] [Google Scholar]
- 27. Nickeleit V., et al. 2000. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N. Engl. J. Med. 342:1309–1315 [DOI] [PubMed] [Google Scholar]
- 28. Olsen G. H., et al. 2006. Genetic variability in BK Virus regulatory regions in urine and kidney biopsies from renal-transplant patients. J. Med. Virol. 78:384–393 [DOI] [PubMed] [Google Scholar]
- 29. Olsen G. H., Hirsch H. H., Rinaldo C. H. 2009. Functional analysis of polyomavirus BK noncoding control region quasispecies from kidney transplant recipients. J. Med. Virol. 81:1959–1967 [DOI] [PubMed] [Google Scholar]
- 30. Schowalter R. M., Pastrana D. V., Pumphrey K. A., Moyer A. L., Buck C. B. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sunyaev S. R., Lugovskoy A., Simon K., Gorelik L. 2009. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML). PLoS Genet. 5:e1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tremolada S., et al. 2010. Polymorphisms of the BK virus subtypes and their influence on viral in vitro growth efficiency. Virus Res. 149:190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Meijden E., et al. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromised patient. PLoS Pathog. 6:e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Viscount H. B., et al. 2007. Polyomavirus polymerase chain reaction as a surrogate marker of polyomavirus-associated nephropathy. Transplantation 84:340–345 [DOI] [PubMed] [Google Scholar]