Abstract
Phylogenetic analysis of a Coxsackievirus A16 (CA16) sequence from Shenzhen, China, and other Chinese and international CA16 sequences revealed a pattern of endemic cocirculation of strains of clusters B2a and B2b within subtype B2 viruses. Amino acid evolution and nucleotide variation in the VP1 region were slight for 5 years.
Coxsackievirus A16 (CA16), an RNA virus of the family Picornaviridae, is cocirculating with Human enterovirus 71 (HEV71) to cause hand, foot, and mouth disease (HFMD) worldwide (11). In recent years, numerous large outbreaks of HEV71-associated HFMD have been more severe than the disease caused by CA16 (5, 10, 11, 13), so the molecular characteristics of CA16 have not been fully described due to less attention being paid to this virus. However, fatal CA16 infection has also been described in children with HFMD-associated myocarditis (6) and in an adult with pneumonitis (8). Moreover, the cocirculation of HEV71 and CA16 during HFMD outbreaks has often been observed. Although CA16 is genetically close to HEV71, the levels of genetic diversity and molecular evolutions of CA16 and HEV71 are different. Therefore, it is necessary to determine the gene characteristics of CA16.
For this study, a total of 1,161 HFMD specimens were conserved by the Department of Microbiology at the Shenzhen Center for Disease Control and Prevention (CDC) from 2005 to 2009. A one-step, single-tube, real-time reverse transcription-PCR (RT-PCR) assay was used for rapid diagnosis of CA16. The method was standardized with selected primers and probes as previously described (9). A total of 69 strains (9, 18, 5, 21, and 16 strains conserved in 2005, 2006, 2007, 2008, and 2009, respectively) were randomly selected to conduct genetic analysis for identifying positive CA16 strains through real-time RT-PCR. The whole VP1 sequences of the selected CA16 strains were amplified by RT-PCR with the following in-house primers (3, 12): CA16-ZF, 5′-ATTGGTGCTCCCACTACAGC-3′, and CA16-ZR, 5′-GCAAGGTGCCGATTCACTACCCT.
Based on a homological analysis of nucleotides and amino acids from collected samples, the nucleotide and amino acid homologies were higher than 91.0 and 98.7%, respectively. However, neither a clear trend toward increased numbers of nonsynonymous substitutions nor major changes in evolution since 2005 were observed. Intrayear-diversity analysis revealed a consistent difference in pairwise comparisons of less than 7.34%. A certain number of nucleotide variations were observed between inter- and intrayear levels of diversity, although little change occurred in amino acid sequences in the above-described data.
The sequences of 69 CA16 strains were divided into three groups according to nucleotide alignment and phylogenetic analysis of VP1 sequences (data not shown). Thus, CA16-associated HFMD in Shenzhen, China, from 2005 to 2009 resulted from coincident circulation of three genetically distinct viruses.
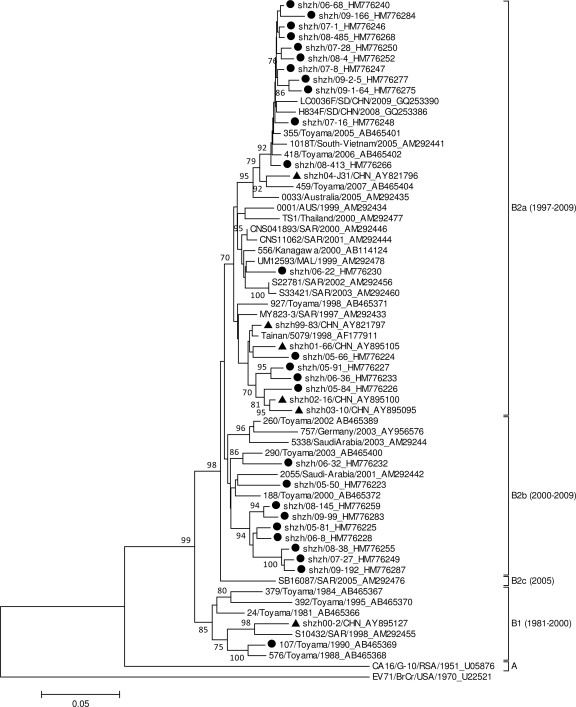
A few previous attempts have been made to classify CA16 strains on the basis of VP4 and VP1 sequence analysis as three different major clusters, called A, B, and C (2–4). However, CA16 strains were also reported to group into genotypes A and B (7, 12). In order to further determine the genotype of Shenzhen CA16 strains, a phylogenetic dendrogram was constructed with 25 CA16 strains collected from Shenzhen (five strains per year were randomly selected based on the cases of three different groups) from 2005 to 2009 and 37 genotype-identified CA16 sequences from other Chinese locations or other countries, as shown in Fig. 1. Genotypes were described in a way similar to that used for HEV71 (1), and a difference of at least 15% in VP1 genes was used to distinguish genotypes. The VP1 phylogenetic tree indicated that strain G-10 was different from other strains by 25.0% and designated the sole member of genogroup A. The genetic variation among nucleotide sequences of other CA16 strains except those of genogroup A was less than 13.8%. Therefore, all CA16 strains, including Shenzhen CA16 strains, were classified as members of gene group B. Viruses in genotype B could be chronologically divided into B1 and B2 subtypes in the phylogenetic tree, with bootstrap support of 99%, and subtype B2 could be further divided into clusters B2a, B2b, and B2c, with bootstrap support of 98%. The phylogenetic classification in our study was consistent with those of previous studies (12). A single difference between studies from other groups and our group was the definition of subtype B1.
Fig. 1.
Phylogenetic dendrogram constructed by using 891 nucleotides, the whole VP1 gene sequence of 25 Shenzhen CA16 strains, and sequences from 39 other Chinese and international CA16 strains. Details of each strain can be found in GenBank. The prototype HEV71 strain (BrCr) was used as an outgroup. Triangles and filled circles indicate CA16 strains derived from HFMD cases from 1999 to 2004 in a previous study and from 2005 to 2009 in this study in Shenzhen, China. Genotypes are shown at the right of the tree, and bootstrap values (percentage of 1,000 pseudoreplicates) are shown at the nodes of major clades. The scale at the bottom indicates a measurement of relative phylogenetic distance. SD, Shangdong Province, China; CHN, China; AUS, Australia; MAL, Malaysia.
The prototype G-10 strain of CA16, first identified in South Africa in 1951, appeared as the single genotype A strain. During a 60-year evolution, CA16 circulation experienced two periods. During the first stage, from 1981 to 2000, CA16 strains belonged to genotype B1. Since 1997, genotype B1 has gradually been replaced by genotype B2. In the past 10 years (the second stage), CA16 from clusters B2a and B2b became the predominant virus circulating in mainland China and in neighboring countries and regions, including Taiwan, Japan, Vietnam, Malaysia, and Thailand. During the same period, only one representative strain of cluster B2c (SB16078/SAR/2005, where “SAR” denotes Sarawak, Malaysia) was isolated from Malaysia. Shenzhen strains detected in this study belonged to clusters B2a and B2b. Thus, recent genotypes of CA16 in Shenzhen were similar to those detected in other Chinese provinces and countries. However, Shenzhen CA16 strains probably originated from earlier CA16 strains and do not represent the source of genotype B2. Therefore, the further investigation of an earlier origin for genotype B2 strains in China was deserved.
Average diversities of whole VP1 genes among CA16 strains of genotypes A, B1, B2a, B2b, and B2c were compared (data not shown). An approximately 26% difference was observed between strains of prototype A and other subtypes, namely, B1 to B2c; a >10% variation was observed between B1 and B2a to B2c, and a <7.4% difference was observed among the B2a, B2b, and B2c groups. Moreover, all CA16 strains revealed a 43.0 to 49.1% difference from the HEV71 prototype strain BrCr. These studies provided strong evidence for subtype classification of CA16 strains. Genotype B in our study was consistent with genotype B and C in previous studies (2–4), so B and C clusters should be combined into one genotype.
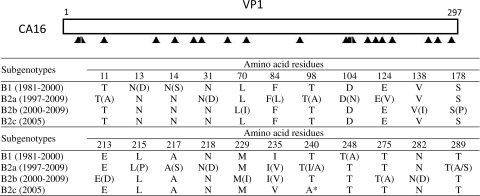
The variations in amino acid sequences in the VP1 region of group B genes were scattered throughout the entire region, as shown in Fig. 2. From the horizontal view, a total of 22 different amino acid residues were observed in all subtypes, and 92.6% of predicted VP1 amino acid residues were unchanged. The maximum variation number was only 12 among the same cluster (B2a), and most changes were not accumulated. Longitudinally, only one mutation of A or T at residue 240 was observed between B2c and other subtypes. Thus, amino acid evolution in the VP1 region was small, even though higher activity was exhibited in the same subtypes than in different subtypes, which was consistent with the results of previous investigations (3).
Fig. 2.
Variations in amino acids in the VP1 region among different clusters of strains of genotype B. Filled triangles indicate amino acid residues. The asterisk indicates a change between amino acids in different subtypes. The abbreviations of amino acids in parentheses indicate the variations in amino acids among different strains in the same clusters.
Nucleotide sequence accession numbers.
Whole VP1 sequences for 69 CA16 strains have been submitted to the GenBank database with the accession numbers HM776219 to HM7762187.
Acknowledgments
This work was supported by the 211 Project in Guangdong Province, China.
We thank Chunli Wu at the Department of Microbiology, Shenzhen Center for Disease Control and Prevention, Shenzhen, China, for the assistance with the phylogenetic analysis.
The parents or guardians of study subjects were given written informed consent forms that had been approved by the Ethics Committee at the Shenzhen Center for Disease Control and Prevention.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Brown B. A., Oberste M. S., Alexander J. P., Jr., Kennett M. L., Pallansch M. A. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hosoya M., et al. 2007. Genetic diversity of coxsackievirus A16 associated with hand, foot, and mouth disease epidemics in Japan from 1983 to 2003. J. Clin. Microbiol. 45:112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwai M., et al. 2009. Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot, and mouth disease patients in Toyama, Japan between 1981 and 2007. Jpn. J. Infect. Dis. 62:254–259 [PubMed] [Google Scholar]
- 4. Li L., et al. 2005. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 43:3835–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mao L. X., et al. 2010. Epidemiology of hand, foot, and mouth disease and genotype characterization of Enterovirus 71 in Jiangsu, China. J. Clin. Virol. 49:100–104 [DOI] [PubMed] [Google Scholar]
- 6. McMinn P., Stratov I., Nagarajan L., Davis S. 2001. Neurological manifestations of enterovirus 71 infection in children during an outbreak of hand, foot, and mouth disease in Western Australia. Clin. Infect. Dis. 32:236–242 [DOI] [PubMed] [Google Scholar]
- 7. Perera D., et al. 2007. Molecular phylogeny of modern coxsackievirus A16. Arch. Virol. 152:1201–1208 [DOI] [PubMed] [Google Scholar]
- 8. Wang J. R., et al. 2002. Change of major genotype of enterovirus 71 in outbreaks of hand-foot-and-mouth disease in Taiwan between 1998 and 2000. J. Clin. Microbiol. 40:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao X. L., et al. 2009. Simultaneous detection of human enterovirus 71 and coxsackievirus A16 in clinical specimens by multiplex real-time PCR with an internal amplification control. Arch. Virol. 154:121–125 [DOI] [PubMed] [Google Scholar]
- 10. Yang Z. H., et al. 2005. Detection of enterovirus 71 and coxsackievirus A16 from children with hand, foot and mouth disease in Shanghai, 2002. Zhonghua Er Ke Za Zhi 43:648–652 [PubMed] [Google Scholar]
- 11. Zhang Y., et al. 2009. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 44:262–267 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., et al. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. J. Clin. Microbiol. 48:619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., et al. 2010. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang City of China. Virol. J. 7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]