Abstract
For influenza viruses, pyrosequencing has been successfully applied to the high-throughput detection of resistance markers in genes encoding the drug-targeted M2 protein and neuraminidase. In this study, we expanded the utility of this assay to the detection of multiple receptor binding variants of the hemagglutinin protein of influenza viruses directly in clinical specimens. Specifically, a customized pyrosequencing protocol that permits detection of virus variants with the D, G, N, or E amino acid at position 222 in the hemagglutinin of the 2009 pandemic influenza A (H1N1) virus was developed. This customized pyrosequencing protocol was applied to the analysis of 241 clinical specimens. The use of the optimized nucleotide dispensation order allowed detection of mixtures of variants in 10 samples (4.1%) which the standard cyclic nucleotide dispensation protocol failed to detect. The optimized pyrosequencing protocol is expected to provide a more accurate tool in the analysis of virus variant composition.
INTRODUCTION
The influenza virus is an enveloped single-stranded negative-sense RNA virus that is an important respiratory pathogen which causes annual epidemics and occasional pandemics (27). As a result of a high mutation rate of viral RNA polymerase (4, 19), the influenza virus exists as a quasispecies, a complex mixture of closely related genomes. When selective pressure is applied, variants carrying favorable changes gain growth advantage and establish dominance (16). Influenza virus hemagglutinin (HA) is responsible for binding to cellular receptors and for membrane fusion during the virus entry into the host cell (23). Mutations in HA can allow the virus to adapt to a new host and evade the human host immune system (25) and can be responsible for altered receptor specificity, virulence, and other traits. Therefore, the ability to detect HA variants, even when they are present at a low frequency within the virus population, is desirable.
Currently, there are several assays that allow the monitoring of mutations at specific nucleotides. Sanger sequencing is considered the “gold standard” for analysis of virus genome variations (26). However, the interpretation of sequencing chromatograms can be difficult for mixed-genome variants containing insertions, deletions, or multiple nucleotide substitutions (23). Sanger sequencing (31), high-resolution melting curve (HRM) analysis (14), real-time quantitative reverse transcriptase PCR (qRT-PCR) (2), rolling cycle amplification (RCA) (28), single-strand confirmation polymorphism (SSCP) (10), resequencing array (29), and, recently, pyrosequencing (3, 7, 18, 20) have been employed for the analysis of specific mutations in genomes of different viruses.
Pyrosequencing technology is based on luminometric detection of the light signal produced by luciferase (21). The reliability and flexibility in assay development have made pyrosequencing a widely used method for various screening and diagnostic applications (13). Pyrosequencing platforms have been useful for high-throughput sample screening for molecular markers associated with drug resistance or other traits (3, 7, 12). Recently, pyrosequencing was used for the timely detection of resistance to adamantanes and oseltamivir during the 2009 influenza A virus (H1N1) pandemic (pH1N1) (6).
Although pyrosequencing is efficient and well suited for the analysis of short sequences, the technology is still fairly new and there are challenges for certain applications. For example, it was shown for the KRAS oncogene that the presence of a mixture containing more than two major genomic variants can complicate pyrogram interpretation (26). Such mixtures can also cause inaccurate identification of single nucleotide polymorphisms (SNPs).
Identification of molecular markers of virulence in influenza viruses remains a challenging task. The emergence and rapid global spread of a novel pH1N1 influenza virus in 2009 highlighted the need for high-throughput analysis to detect new variants of this rapidly evolving pathogen. Overall, the pH1N1 virus caused mild illness in most cases; however, numerous severe and fatal cases were also reported (30). Analysis of the HA gene of the 2009 pH1N1 virus revealed several mutations (Table 1) at the aspartic acid (D) residue at position 222 (225 in H3 numbering). An association of clinically severe cases with the D222G substitution (GAT → GGT) in the HA1 subunit was reported (11), although no such association was found in other studies (1). Also, a considerable frequency of N222 variants (GAT → AAT) was found among severe and fatal cases (11, 17). Noteworthy, changes at D222 have been associated with adaptation of a seasonal, nonpandemic human H1N1 influenza A virus to a new environment (e.g., adaptation to mice and embryonated chicken eggs) because this amino acid is a part of the receptor binding site (8, 9, 24). To further investigate the potential role of amino acid substitutions at position 222, it was highly desirable to develop an assay which would allow accurate, high-throughput detection of multiple variants within a clinical specimen. In this report, we describe the use of a customized dispensation order of nucleotides for pyrosequencing that enables the accurate detection of HA variants in pH1N1 viruses.
Table 1.
Virus variants at amino acid position 222 of pH1N1 hemagglutinin
Letters in boldface indicate the nucleotide change.
Wild type.
MATERIALS AND METHODS
Virus isolates and clinical specimens.
Seven pandemic A (H1N1) influenza virus isolates and 241 clinical specimens (throat swabs, nasal swabs and washes, nasopharyngeal swabs, and sputum specimens) submitted to the Centers for Disease Control and Prevention (CDC) between November 2009 and January 2010 as part of the U.S. virus surveillance activity were used for this study.
Primer design.
To detect mutations at the 222 position of the HA, three primers were designed with the use of PSQ Assay Design software, version 1.0.6 (Qiagen, Valencia, CA). The primers SW-HA-F696 (5′-CAAGAAGTTCAAGCCGGAAATAGC-3′, forward primer) and SW-HA-R799b (biotin-5′-ATTGCGAATGCATATCTCGGTAC-3′, reverse biotinylated primer) were used to amplify DNA fragments by RT-PCR. The primer SW-HA-F715 (5′-CAATAAGACCCAAAGTGAGG-3′, sequencing primer) was applied for detecting the target region.
RNA extraction and RT-PCR.
Viral RNA was extracted from 100 μl of sample using either a MagNA Pure Compact or MagNA Pure LC 2.0 nucleic acid isolation system (Roche Diagnostics, Basel, Switzerland). RNA was eluted in 50 μl and stored at −30°C. A SuperScript III one-step RT-PCR system with Platinum Taq High Fidelity enzyme (Invitrogen, Carlsbad, CA) was used for cDNA synthesis and amplification as described by the manufacturer. Briefly, the reaction was initiated at 50°C for 30 min. Then, Taq DNA polymerase was heat activated at 94°C for 2 min. This was followed by 45 cycles at 94°C for 15 s, 50°C for 30 s, and 68°C for 1 min. The final elongation time was 5 min. Primers SW-HA-F696 and SW-HA-R799-b were used for RT-PCR at a final concentration of 0.4 μM. The amplified RT-PCR products were visualized by electrophoresis on 2% agarose E-gels (Invitrogen, Carlsbad, CA).
Sequencing by dideoxy chain termination method (Sanger sequencing).
Primers designed for the RT-PCR step of pyrosequencing (SW-HA-F696 and nonbiotinylated SW-HA-R799) were used to amplify the cDNA fragment for Sanger sequencing. Amplified RT-PCR products were purified using ExoSAP-IT reagent (USB, Cleveland, OH). Sequence template was synthesized with an ABI Prism BigDye Terminator kit (Applied Biosystems, Foster City, CA). Products of sequencing reactions were treated with XTerminator solution (Applied Biosystems, Foster City, CA), according to the manufacturer's protocol. Sequences generated in an ABI Prism 3730 genetic analyzer (Applied Biosystems, Foster City, CA) were analyzed using Lasergene software, version 7.0 (DNAStar, Madison, WI).
Pyrosequencing.
Pyrosequencing reactions were performed on a PyroMark Q96 ID instrument as described by the manufacturer (Qiagen, Valencia, CA). The SW-HA-F715 sequencing primer was used for all reactions at a final concentration of 0.45 μM. Both the sequence analysis (SQA) and the SNP modes of the PyroMark Q96 ID instrument were utilized for analysis of substitutions at the 222 position in the HA. Two different nucleotide dispensation orders were used for pyrosequencing in SQA mode: the cyclic dispensation order (GATC)6 and the customized dispensation order ATGTAT(CAGT)6 (see Results). SNP analysis was conducted to determine the percentage of each variant within a mixture containing two virus variants. These percentages were calculated by the PyroMark ID software. The dispensation order for SNP analysis was generated by PyroMark ID software on the basis of the target sequence and the site for SNP analysis. For a mixture containing GAT and AAT variants, the generated dispensation was CGAGTCAGA.
Population cloning.
To confirm the presence of virus variants within mixtures, cloning of RT-PCR products amplified with primers SW-HA-F696 and nonbiotinylated SW-HA-R799 was performed using a TOPO TA cloning kit as described by the manufacturer (Invitrogen, Carlsbad, CA). One hundred individual bacterial colonies containing recombinant plasmids carrying an individual RT-PCR fragment were randomly picked and analyzed by pyrosequencing of the PCR product amplified with primer SW-HA-F696 and biotinylated primer SW-HA-R799b. Pyrosequencing analysis was performed as described above.
Statistical analysis.
t-test analysis was used to determine whether there was a statistically significant difference between the percentage calculated for virus variants in a mixture tested with the customized nucleotide dispensation order in SQA mode or in SNP mode. Statistical significance was set at a P value of <0.05. All probabilities were two-tailed.
RESULTS
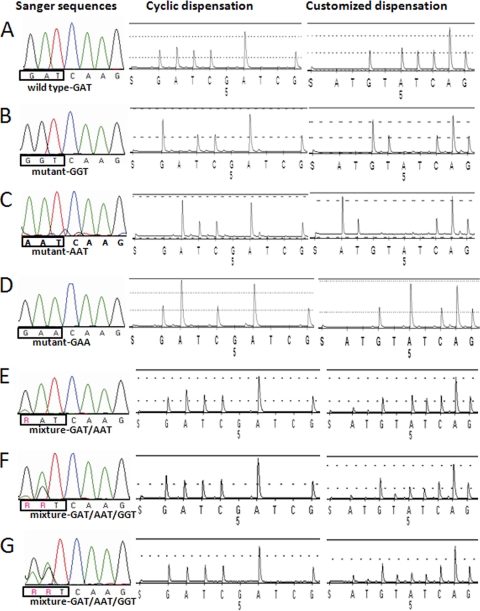
Pandemic H1N1 influenza viruses with changes at each nucleotide of the triplet for amino acid 222 of the HA protein were detected by Sanger sequencing. Specifically, a single nucleotide substitution at the first (GAT → AAT), the second (GAT → GGT), or the third (GAT → GAA) position (Fig. 1A to D) was seen. In addition, sequencing showed that some viruses contained a mixture of variants (Fig. 1E to G). For example, the chromatogram in Fig. 1E showed a polymorphism for the first nucleotide (G/A), which suggests the presence of two variants: GAT and AAT. Another virus had two polymorphisms, at the first and second positions (Fig. 1F and G), which might reflect the presence of up to four variants: GAT, AAT, GGT, and AGT, encoding aspartic acid (D), asparagine (N), glycine (G), and serine (S), respectively. However, the actual variants cannot be reliably ascertained on the basis of Sanger sequencing. Therefore, isolates (n = 7) with an individual GAT, AAT, GGT, or GAA variant as well as mixtures were selected to be analyzed using the pyrosequencing assay.
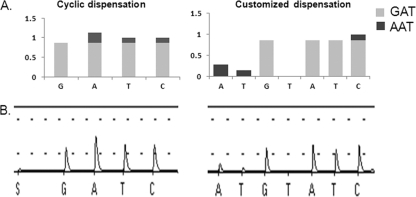
Fig. 1.
Analysis of virus isolates containing an individual virus variant (A to D) or a mixture of virus variants (E to G) at position 222 of hemagglutinin protein. Sanger sequencing chromatograms are shown in the first column (A nucleotides, green; C nucleotides, blue; T nucleotides, red; G nucleotide, black; R, G or A); pyrosequencing cyclic nucleotide dispensation (GATC)6 pyrograms are shown in the second column; pyrosequencing customized nucleotide dispensation ATGTAT(CAGT)6 pyrograms are shown in the third column. (A) Wild-type GAT; (B) mutant GGT; (C) mutant AAT; (D) mutant GAA; (E) mixture of AAT and GAT; (F and G) mixture of AAT, GGT, and GAT.
Analysis of virus isolates using the pyrosequencing assay (cyclic nucleotide dispensation).
First, the viruses were analyzed in the pyrosequencing assay using a standard cyclic dispensation order, (GATC)6, in SQA mode. With this approach, the single variants were readily identified (Fig. 1A to D, cyclic dispensation). However, results for the samples containing mixtures of variants were not in agreement with the sequences determined by the Sanger method. For example, a mixture of two variants, GAT and AAT, was interpreted as a single wild-type variant, GAT, by the PyroMark ID software analysis (Fig. 1E). Visual inspection of the corresponding pyrogram (Fig. 1E) suggested the presence of the additional variant (AAT), on the basis of the increased height of the first A peak in relation to the first G and T peaks.
In another instance, Sanger sequencing indicated the presence of GAT and GGT variants, while the PyroMark ID software still identified only the wild-type variant, GAT (Fig. 1F). Contrarily, the increased height of the first G peak in comparison to the height of the first A peak in the pyrogram suggested that in addition to the wild type, the variant GGT was present. However, in another sample (Fig. 1G), even close inspection of pyrograms did not provide any evidence for the presence of additional variants detected using the Sanger method.
Target-specific customization of nucleotide dispensation.
Since pyrosequencing using the standard cyclic nucleotide dispensation was unable to accurately determine the variants in the mixtures, a customized dispensation order was designed with the intention to improve the resolution of pyrograms. Because all three variants of interest (GAT, GGT, and AAT) share a T in the third position, we proposed a new customized order: ATGTAT(CAGT)6. Consequently, when the first A nucleotide was dispensed, both A nucleotides for an AAT variant were incorporated. The subsequently dispensed T would then be incorporated as the T of an AAT variant. Therefore, the peak height for the A nucleotide would be twice that of the T peak and the T peak would reflect the total portion of the AAT virus variant in the mixture. The next dispensation of a G would be incorporated as the first nucleotide of a GAT variant or the first and second nucleotides of a GGT variant. The presence of the T peak for the T nucleotide dispensed after the G would confirm the presence of a GGT variant. In this case, the height of the T peak was used to calculate the final proportion of the GGT variant. The following A nucleotide dispensation would extend a GAT variant, and the final T dispensed prior to the cyclic (CAGT)6 portion of the dispensation order would indicate the last peak for a GAT variant. This T could also be used to analyze the proportion of a GAT variant present in a mixture. Thus, the proposed order of dispensation allowed separation of each of the three variants on the basis of a location of the T nucleotide on a pyrogram.
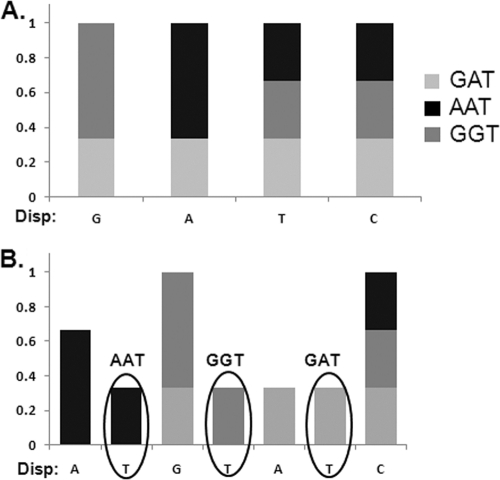
For a theoretical sample containing equal amounts of three variants (AAT, GGT, and GAT), a T nucleotide at the second, fourth, or sixth position would indicate the presence of all three variants, respectively (Fig. 2B). Using the standard cyclic order of nucleotide dispensation, this theoretical sample would be incorrectly identified to have the GAT variant solely present in the sample, as depicted in Fig. 2A.
Fig. 2.
Histogram comparison of two theoretical pyrograms for cyclic (GATC)6 (A) and customized ATGTAT(CAGT)6 (B) nucleotide dispensation orders. The x axis shows the nucleotide dispensed. The y axis represents the proportion of each nucleotide relative to the variant present. Three virus variants (GAT, GGT, and AAT) are present in equal proportions. Black ovals on histogram B show the signal for the dispensation of the T nucleotide. The first T dispensation is for the AAT variant, the second T for the GGT variant, and the third T for the GAT variant. The level of signals for T nucleotides can be used for quantification of all three virus variants.
Since the respective peak heights in a pyrogram were proportional to the amount of incorporated nucleotide, the customized dispensation proposed here allowed the quantification of each of the three variants in a mixture based on the analysis of T peak heights. The following algorithm was derived to calculate the proportion of each variant: (Tn − TBkg)/(T2 + T4 + T6 − 3 × TBkg) × 100% = SNPn, where Tn (n = 2, 4, or 6) is the peak height of the T nucleotide dispensed at positions 2, 4, and 6 and Bkg is the average background of T calculated from the peak heights of the T nucleotides dispensed at positions 10, 14, and 18 where no T residues exist in the sequence.
Comparison of customized nucleotide dispensation order, SNP analysis, and cloning analysis.
The next step was to compare the results using customized nucleotide dispensation order, SNP analysis, and cloning analysis for the same specimen. A virus isolate which contained two virus variants, GAT and AAT, was used. The pyrogram generated with the customized nucleotide dispensation order clearly showed the presence of both variants (Fig. 3).
Fig. 3.
Detection of mixture of GAT and AAT virus variants at amino acid position 222 of hemagglutinin. (A) The theoretical histogram for customized nucleotides was made on the basis of the variant proportion detected for virus mixtures GAT and AAT by cloning analysis, which were present at 84% and 16%, respectively. (B) Pyrograms generated by PyroMark Q96 ID software. The x axis shows the nucleotide dispensed. The y axis represents the proportion of each nucleotide relative to the variant present.
Using the customized nucleotide dispensation, the proportion of the sample with the GAT and AAT variants was calculated using the peak height data described in the algorithm (Table 2) and was found to contain 86.8% ± 0.2% of the GAT variant and 13.2% ± 0.2% of the AAT variant. The analysis of this specimen in SNP mode showed that GAT and AAT variants were present in proportions of 87.7% ± 0.1% and 12.3% ± 0.1%, respectively. The similarity of the data obtained by the two approaches was statistically significant (P < 0.02) (Table 2).
Table 2.
Comparison of virus variant proportions in clinical specimens containing two virus variants, GAT and AAT
| aaa at position 222 (codon) | % virus variants ± SD |
No. of colonies screenedd | |
|---|---|---|---|
| SQAb | SNPc | ||
| D (GAT) | 86.8 ± 0.2 | 87.7 ± 0.1 | 84 |
| N (AAT) | 13.2 ± 0.2 | 12.3 ± 0.1 | 16 |
aa, amino acid.
Percentage of GAT and AAT virus variants calculated using T peak height for the second and sixth dispensed nucleotides dispensation [ATGTAT (CAGT)6]. T peak background was subtracted from T peak height.
SNP analysis determined by PyroMark Q96 ID software. Dispensation order was generated automatically for the sequence G/AATCAG.
One hundred individual colonies were analyzed for HA222.
For the same specimen, DNA cloning showed the proportions of GAT and AAT variants to be 84% and 16%, respectively (Table 2). Thus, the customized nucleotide dispensation order for pyrosequencing could accurately identify the proportions of the virus variants in mixtures.
High-throughput analysis of clinical specimens using pyrosequencing.
To assess the ability of the customized pyrosequencing assay to detect variants at position 222, 241 clinical specimens positive for pH1N1 virus were analyzed. Among those tested, GAT (n = 225), GGT (n = 3), AAT (n = 2), and GAA (n = 1) were individual variants (Table 3). In addition, 10 specimens (4.1%) contained more than a single variant (Table 3). These mixtures would not have been identified using the cyclic dispensation order.
Table 3.
Comparison of the number of HA variants detected between cyclic and customized nucleotide dispensationa
| Virus variant(s) present in specimen | aa at position 222 | No. of variants |
|
|---|---|---|---|
| Cyclic nucleotide dispensation | Customized nucleotide dispensation | ||
| GAT | D | 235 | 225 |
| GGT | G | 3 | 3 |
| AAT | N | 2 | 2 |
| GAA | E | 1 | 1 |
| GAT and GGT | D/G | NA | 1 |
| GAT and AAT | D/N | NA | 3 |
| GGT and AAT | G/N | NA | 1 |
| GAT, GGT, and AAT | D/G/N | NA | 5 |
aa, amino acid; NA, not applicable.
DISCUSSION
In this study we developed a sensitive assay, utilizing the quantitative nature of pyrosequencing technology, for the detection of individual and mixed variants at position 222 in HA of the pH1N1 2009 influenza virus. Pyrosequencing technology measures a light signal produced after a nucleotide is incorporated into the sequence by the polymerase (21). When a nucleotide is dispensed but not incorporated, a gap is formed in the pyrogram (Fig. 1). We demonstrated that the SQA protocol with a customized nucleotide dispensation order, designed to determine virus variants and their proportions, was more informative than the traditional Sanger sequencing method as well as the pyrosequencing with the cyclic nucleotide dispensation order. Although the SNP pyrosequencing assay is a powerful tool for polymorphism quantification, it cannot be applied to unknown nucleotide polymorphisms. Furthermore, the PyroMark ID software does not permit the SNP analysis of identical nucleotide polymorphisms present at two or more consecutive positions. The advantage of the customized dispensation order was seen when mixtures that would be misinterpreted on the basis of pyrosequencing with the cyclic nucleotide dispensation order were clearly identified in the clinical specimens. No AGT (S) variants were detected in this study utilizing either dispensation order, although it was a possible variant if only Sanger sequencing is used.
The quantitative nature of pyrosequencing allowed the calculation of virus variant proportions with the use of the peak heights within a pyrogram generated using the customized dispensation order. The results from three different approaches, SNP analysis, cloning, and SQA analysis, were in good agreement and supported the use of the formula (Tn − TBkg)/(T2 + T4 + T6 − 3 × TBkg) × 100% = SNPn for the calculation of the virus variant proportions based on peak heights on SQA programs.
The customized nucleotide dispensation order approach has already proven useful in detecting the emergence of variants in other viruses, e.g., in analyzing the transition of wild-type to lamivudine-resistant variants in hepatitis B viruses (15). In this study, we were able to further expand the application of a customized nucleotide dispensation order to the quantitation of each of the virus variants in a mixture.
Pyrosequencing technology has been actively employed for the analysis of the influenza virus genome and has proven to be a robust platform for screening mutations associated with drug resistance in M2 and NA genes (5). The new approach extends the usefulness of pyrosequencing in the detection of multiple virus genomic variants for position 222 in HA. In addition, it was shown to be more informative than Sanger sequencing for quantitative analysis of a polymorphic site. It allows the circumvention of the time-consuming and costly cloning of RT-PCR products from specimens containing virus variants. This approach can also be useful in identifying the presence of multiple variants within influenza viruses and other pathogens. In the future, this assay could be used to rapidly screen for mutations at position 222 of HA or customized to screen for other mutations.
ACKNOWLEDGMENTS
We thank our colleagues from Sequencing Activity, Influenza Division, CDC, for their valuable technical assistance on the project. We also thank Alicia Fry and Marie-Joelle Miron for their useful discussions and contributions.
The findings and conclusions of this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Anton A., et al. 2010. D225G mutation in the hemagglutinin protein found in 3 severe cases of 2009 pandemic influenza A (H1N1) in Spain. Diagn. Microbiol. Infect. Dis. 67:207–208 [DOI] [PubMed] [Google Scholar]
- 2. Bolotin S., et al. 2009. Development of a novel real-time reverse-transcriptase PCR method for the detection of H275Y positive influenza A H1N1 isolates. J. Virol. Methods 158:190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bright R. A., et al. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175–1181 [DOI] [PubMed] [Google Scholar]
- 4. de la Torre J. C., Holland J. J. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 64:6278–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deyde V. M., Gubareva L. V. 2009. Influenza genome analysis using pyrosequencing method: current applications for a moving target. Expert Rev. Mol. Diagn. 9:493–509 [DOI] [PubMed] [Google Scholar]
- 6. Deyde V. M., et al. 2010. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob. Agents Chemother. 54:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duwe S., Schweiger B. 2008. A new and rapid genotypic assay for the detection of neuraminidase inhibitor resistant influenza A viruses of subtype H1N1, H3N2, and H5N1. J. Virol. Methods 153:134–141 [DOI] [PubMed] [Google Scholar]
- 8. Gambaryan A. S., Robertson J. S., Matrosovich M. N. 1999. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology 258:232–239 [DOI] [PubMed] [Google Scholar]
- 9. Gubareva L. V., Kaiser L., Matrosovich M. N., Soo Hoo Y., Hayden F. G. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523–531 [DOI] [PubMed] [Google Scholar]
- 10. Kalvatchev Z., Walder R., Garzaro D., Barrios M. 2000. Detection of genetic diversity among bovine immunodeficiency virus population by single-strand conformation polymorphism analysis. Viral Immunol. 13:373–381 [DOI] [PubMed] [Google Scholar]
- 11. Kilander A., Rykkvin R., Dudman S. G., Hungnes O. 2010. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill. 15(9):pii=19546 [DOI] [PubMed] [Google Scholar]
- 12. Lackenby A., Democratis J., Siqueira M. M., Zambon M. C. 2008. Rapid quantitation of neuraminidase inhibitor drug resistance in influenza virus quasispecies. Antivir. Ther. 13:809–820 [PubMed] [Google Scholar]
- 13. Langaee T., Ronaghi M. 2005. Genetic variation analyses by pyrosequencing. Mutat. Res. 573:96–102 [DOI] [PubMed] [Google Scholar]
- 14. Lin J. H., et al. 2008. Rapid differentiation of influenza A virus subtypes and genetic screening for virus variants by high-resolution melting analysis. J. Clin. Microbiol. 46:1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindstrom A., Odeberg J., Albert J. 2004. Pyrosequencing for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 42:4788–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mas A., Lopez-Galindez C., Cacho I., Gomez J., Martinez M. A. 2010. Unfinished stories on viral quasispecies and Darwinian views of evolution. J. Mol. Biol. 397:865–877 [DOI] [PubMed] [Google Scholar]
- 17. Miller R. R., MacLean A. R., Gunson R. N., Carman W. F. 2010. Occurrence of haemagglutinin mutation D222G in pandemic influenza A(H1N1) infected patients in the West of Scotland, United Kingdom, 2009-10. Euro Surveill. 15(16):19498=pii [PubMed] [Google Scholar]
- 18. O'Meara D., et al. 2001. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J. Clin. Microbiol. 39:464–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parvin J. D., Moscona A., Pan W. T., Leider J. M., Palese P. 1986. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J. Virol. 59:377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pourmand N., et al. 2006. Rapid and highly informative diagnostic assay for H5N1 influenza viruses. PLoS One 1:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ronaghi M., Karamohamed S., Pettersson B., Uhlen M., Nyren P. 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242:84–89 [DOI] [PubMed] [Google Scholar]
- 22. Reference deleted.
- 23. Skehel J. J., Wiley D. C. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 24. Smee D. F., et al. 2007. Influenza A (H1N1) virus resistance to cyanovirin-N arises naturally during adaptation to mice and by passage in cell culture in the presence of the inhibitor. Antivir. Chem. Chemother. 18:317–327 [DOI] [PubMed] [Google Scholar]
- 25. Stewart P. L., Nemerow G. R. 1997. Recent structural solutions for antibody neutralization of viruses. Trends Microbiol. 5:229–233 [DOI] [PubMed] [Google Scholar]
- 26. Tsiatis A. C., et al. 2010. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J. Mol. Diagn. 12:425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viboud C., et al. 2006. Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine 24:6701–6707 [DOI] [PubMed] [Google Scholar]
- 28. Wang B., et al. Detection of the rapid emergence of the H275Y mutation associated with oseltamivir resistance in severe pandemic influenza virus A/H1N1 09 infections. Antiviral Res. 87:16–s21 [DOI] [PubMed] [Google Scholar]
- 29. Wong C. W., et al. 2004. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 14:398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 362:1708–1719 [DOI] [PubMed] [Google Scholar]
- 31. Yang Z. J., Tu M. Z., Liu J., Wang X. L., Jin H. Z. 2006. Comparison of amplicon-sequencing, pyrosequencing and real-time PCR for detection of YMDD mutants in patients with chronic hepatitis B. World J. Gastroenterol. 12:7192–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]