Abstract
Trypanosoma brucei gambiense group 1 is the major causative agent of the Gambian human African trypanosomiasis (HAT). Accurate diagnosis of Gambian HAT is still challenged by lack of precise diagnostic methods, low and fluctuating parasitemia, and generally poor services in the areas of endemicity. In this study, we designed a rapid loop-mediated isothermal amplification (LAMP) test for T. b. gambiense based on the 3′ end of the T. b. gambiense-specific glycoprotein (TgsGP) gene. The test is specific and amplifies DNA from T. b. gambiense isolates and clinical samples at 62°C within 40 min using a normal water bath. The analytical sensitivity of the TgsGP LAMP was equivalent to 10 trypanosomes/ml using purified DNA and ∼1 trypanosome/ml using supernatant prepared from boiled blood, while those of classical PCR tests ranged from 10 to 103 trypanosomes/ml. There was 100% agreement in the detection of the LAMP product by real-time gel electrophoresis and the DNA-intercalating dye SYBR green I. The LAMP amplicons were unequivocally confirmed through sequencing and analysis of melting curves. The assay was able to amplify parasite DNA from native cerebrospinal fluid (CSF) and double-centrifuged supernatant prepared from boiled buffy coat and bone marrow aspirate. The robustness, superior sensitivity, and ability to inspect results visually through color change indicate the potential of TgsGP LAMP as a future point-of-care test.
INTRODUCTION
Trypanosoma brucei gambiense is the causative agent of the Gambian form of human African trypanosomiasis (HAT) in sub-Saharan Africa and is responsible for over 90% of all HAT cases (7). The Gambian HAT is endemic to rural areas of West and Central Africa, where deterioration of control activities, severe disruptions of health services, and population movements into high-risk areas have led to resurgence of the disease (47). T. b. gambiense infection is characterized by low parasitemia with no specific clinical symptoms (15), especially during the early stage, when the trypanosomes are confined to the hemolymphatic system. This has limited the use of standard diagnostic techniques, with an estimated 20 to 30% of patients being undiagnosed (42). The serological test—card agglutination test for trypanosomiasis (CATT)—for T. b. gambiense (24) is widely used; however, the test has varying sensitivities (46) and cannot decisively differentiate between active and cured cases (19). On the molecular side, several PCR tests have been developed (6, 30, 41), but issues of sensitivity and reproducibility (44) and the requirement for high-precision instrumentation have limited their use.
Early and accurate diagnosis of Gambian HAT is essential, since the drugs used for treatment, particularly those for the late stage, can cause unacceptably severe side effects. The first stage of disease is treated with pentamidine, while the second stage is treated with melarsoprol, which is associated with encephalopathy in about 10% of treated patients (39, 49). Eflornithine is an alternative drug for the second stage of Gambian HAT but is expensive and difficult to administer (48). The latest advance in treatment has been a combination of eflornithine and nifurtimox (40); this brings with it reduction of the treatment duration, as well as the number of eflornithine infusions, to the relief of nursing staff attending to the patients. This complexity in the treatment regime calls for a diagnostic test(s) that is accurate and that minimizes false positives to reduce overtreatment and exposure of patients to expensive and potentially toxic drugs whose efficacy may not be guaranteed.
In recent years, a DNA amplification platform called loop-mediated isothermal amplification (LAMP) has been developed (36). The technique has been used to develop LAMP tests specific for the subgenus Trypanozoon (18, 34, 45) and Trypanosoma brucei rhodesiense (35), but so far, there is no LAMP test for T. b. gambiense. The major advantages of LAMP include (i) rapidity and the use of six to eight primers providing high specificity; (ii) ability of the technique to amplify target DNA from partially processed template; (iii) requirement for only a simple heating device, such as a water bath; and (iv) results that can be inspected visually through the use of varied detection formats, such as turbidity (28), fluorescent dye (5), probes (1, 27), lateral-flow dipstick (LFD) format (31), and microfluidic chips (13). LAMP has attracted much interest as an easily applicable yet highly sensitive molecular tool with great potential for diagnosis in resource-poor rural settings, where HAT typically occurs. This is demonstrated by the large number of publications on the technique (26).
The molecular characterization of human-infective trypanosomes indicates that the majority of Gambian HAT cases are caused by a genetically homogeneous group, group 1 T. b. gambiense (12, 17, 37), with group 2 T. b. gambiense accounting for an insignificant percentage (25). In addition, studies have identified a T. b. gambiense-specific glycoprotein (TgsGP) gene (2) that is specific to group 1 T. b. gambiense and absent in isolates from group 2 (14). The 3′ end of the TgsGP gene has been used to develop the only reliable PCR tests for T. b. gambiense (41). In this study, we designed a rapid and sensitive LAMP test based on the 3′ end of the TgsGP gene and evaluated it using Trypanozoon isolates and clinical samples from HAT patients with a view to obtaining data for a more comprehensive field study.
MATERIALS AND METHODS
Ethical clearance.
Institutional ethical clearance for the collection of human samples in Uganda was obtained from the Uganda National Council of Science and Technology (UNCST), Kampala, Uganda, as reported previously (34), and the use of samples from a HAT patient diagnosed in Australia was approved by Royal Perth Hospital, Western Australia, Australia, through Christopher Heath.
Reference DNA.
Well-characterized T. b. gambiense DNA samples were used in this study, as shown in Table 1. The DNA was prepared either using a Qiagen DNA extraction kit (Qiagen, Victoria, Australia) or by the published method (43). The samples were chosen to ensure wide geographical representation. DNA from other trypanosome species, tsetse fly, bovine, human, and Plasmodium falciparum were included to check the test specificity.
Table 1.
Trypanosome isolates used in the study
| Species/subspecies | Identification codea | Isolate origin | Yr of isolation | Original host | Resultb |
||
|---|---|---|---|---|---|---|---|
| RIME LAMPc | TgsGP PCR | TgsGP LAMPd | |||||
| T. b. gambiense | MOS | (Mbam), Cameroon | 1974 | Human | + | + | + |
| T. b. gambiense | B014 | (Fontem), Cameroon | 1988 | Human | + | + | + |
| T. b. gambiense | Font I | (Fontem), Cameroon | 1993 | Human | + | + | + |
| T. b. gambiense | PT16 | Ivory Coast | 1992 | Human | + | + | + |
| T. b. gambiense | PT41 | Ivory Coast | 1992 | Human | + | + | + |
| T. b. gambiense | Boula | Bouenza, Congo | 1989 | Human | + | + | + |
| T. b. gambiense | NW2 | Uganda | 1992 | Human | + | + | + |
| T. b. gambiense | NW5 | Uganda | 1992 | Human | + | + | + |
| T. b. gambiense | Da 972 | Daloa, Ivory Coast | 1978 | Human | + | + | + |
| T. b. gambiense | Mba | Daloa, Ivory Coast | 1978 | Human | + | + | + |
| T. b. gambiense | JE16 | Adjuman, Uganda | 1992 | Human | + | + | + |
| T. b. gambiense | JE17 | Adjuman, Uganda | 1992 | Human | + | + | + |
| T. b. gambiense | KETRI 2565 | Sudan | 1982 | Human | + | + | + |
| T. b. brucei | LUMP 266 | Kiboko, Kenya | 1969 | G. pallidipes | + | − | − |
| T. b. brucei | B8/18 | (Nsukka), Nigeria | 1962 | Pig | + | − | − |
| T. b. brucei | J10 | Luangwa Valley, Zambia | 1973 | Hyena | + | − | − |
| T. b. brucei | TSW187/78E | Ivory Coast | 1978 | Pig | + | − | − |
| T. b. brucei | Katerema | Uganda | 1990 | Cow | + | − | − |
| T. b. rhodesiense | WB 58 | Uganda | Human | + | − | − | |
| T. b. rhodesiense | 058 | Luangwa Valley, Zambia | 1974 | Human | + | − | − |
| T. b. rhodesiense | UTRO 2509 | Uganda | Human | + | − | − | |
| T. b. rhodesiense | KETRI 2492 | Lambwe Valley, Kenya | 1980 | Tsetse fly | + | − | − |
| T. b. rhodesiense | KETRI 3639 | Busia, Kenya | 1999 | Human | + | − | − |
| T. b. rhodesiense | TMRS 58 | Mpanda, Tanzania | 2006 | Human | + | − | − |
| T. b. rhodesiense | Gambella II | Ethiopia | 1968 | Human | + | − | − |
| T. evansi | SA17 | Isiolo, Kenya | 2003 | Camel | + | − | − |
| T. evansi | KETRI 3093 | Colombia, South America | 1979 | Horse | + | − | − |
| T. congolense forest | Cam 22 | Mbetta, Cameroon | 1984 | Goat | − | − | − |
| T. c. Kilifi | WG5 | Kenya | 1980 | Sheep | − | − | − |
| T. simiae | Ken 4 | Keneba, The Gambia | 1988 | Fly | − | − | − |
| T. simiae tsavo | KETRI 1864 | Kenya | Fly | − | − | − | |
| Bovine, human, tsetse fly, P. falciparum | NA | Kenya | 1998–2003 | − | − | − | |
Clinical samples.
Ten DNA samples prepared from blood and cerebrospinal fluid (CSF) from confirmed T. b. gambiense patients in Uganda, as previously reported (35), were used. The OM series samples were purified using a Gentra (Minneapolis, MN) DNA purification kit. Additionally, a variety of samples (the RPH series) from a T. b. gambiense patient diagnosed in Australia were also included (Table 2). The samples were prepared as follows. First, the buffy coat (BC) was prepared from a pool of 10 heparinized blood capillaries and made up to 210 μl with ultrapure-grade water; then, equal amounts (210 μl) of the BC, the bone marrow aspirate (BMA), and CSF were divided into three equal portions for (i) direct use, (ii) extraction of DNA using the commercial kit, and (iii) supernatant processing as described previously (34). To reduce the chances of false positives (33), the collected supernatant was double centrifuged at 14,000 rpm for 5 min. The prepared samples were then stored at −80°C until they were needed. The template for a 50-μl reaction mixture was 4 to 5 μl for supernatant and 1 to 2 μl for DNA, direct BC, BMA, or CSF samples (Table 2).
Table 2.
Results of the analysis of various clinical samples from HAT patients
| Source | Sample identifier | Template | Origin | Yr of isolation | PCR test resultsi |
LAMP test resultsi |
Species/subspecies | ||
|---|---|---|---|---|---|---|---|---|---|
| TBRa | TgsGPb | RIMEc | TgsGPf | ||||||
| Blood | OM55c | DNA | Northwest Uganda | 2004 | − | − | + | + | T. b. gambiense |
| Blood | OM56c | DNA | Northwest Uganda | 2004 | − | − | + | − | T. b. gambiense |
| Blood | OM66c | DNA | Northwest Uganda | 2004 | − | − | − | − | T. b. gambiense |
| Blood | OM62c | DNA | Northwest Uganda | 2004 | − | − | − | − | T. b. gambiense |
| Blood | OM51c | DNA | Northwest Uganda | 2004 | + | − | + | + | T. b. gambiense |
| Blood | OM52c | DNA | Northwest Uganda | 2004 | − | − | + | − | T. b. gambiense |
| Blood | RPH1d | DNA | Australia | 2008 | − | − | − | − | T. b. gambiense |
| Blood | RPH2d | Supernatantg | Australia | 2008 | − | − | − | − | T. b. gambiense |
| BC | RPH3d | DNA | Australia | 2008 | − | − | − | − | T. b. gambiense |
| BC | RPH4d | Supernatant | Australia | 2008 | − | − | − | − | T. b. gambiense |
| BC | RPHbc | BC | Australia | 2008 | NDh | ND | − | − | T. b. gambiense |
| BMA | RPH5d | DNA | Australia | 2008 | + | − | + | − | T. b. gambiense |
| BMA | RHP6d | Supernatant | Australia | 2008 | − | − | + | + | T. b. gambiense |
| BMA | RHPbma | BMA | Australia | 2008 | ND | ND | − | − | T. b. gambiense |
| CSF | OM54c | DNA | Northwest Uganda | 2004 | + | − | + | + | T. b. gambiense |
| CSF | OM64c,e | Supernatant | Northwest Uganda | 2004 | ND | ND | + | + | T. b. gambiense |
| CSF | OM64c,e | DNA | Northwest Uganda | 2004 | + | − | + | + | T. b. gambiense |
| CSF | RPH7d | DNA | Australia | 2008 | + | − | + | + | T. b. gambiense |
| CSF | RPH8d | Supernatant | Australia | 2008 | ND | ND | + | + | T. b. gambiense |
| CSF | RPH9d | Native | Australia | 2008 | − | − | + | + | T. b. gambiense |
| Blood | JE2c | DNA | Tororo, Uganda | 1991 | − | − | + | − | T. b. rhodesiense |
| Blood | TMRS10Bc | Supernatant | Tanzania | 2007 | ND | − | + | − | T. b. rhodesiense |
| CSF | JE8c | DNA | Tororo, Uganda | 2001 | − | − | + | − | T. b. rhodesiense |
| CSF | JE9c | Supernatant | Tororo, Uganda | 2001 | + | − | + | − | T. b. rhodesiense |
| Serum | TMRS11Sc | DNA | Tanzania | 2007 | ND | − | + | − | T. b. rhodesiense |
Design of LAMP primers.
A total of five primer sets recognizing six distinct sections of TgsGP (accession number AJ277951) were designed using Primer Explorer version 3 software (http://primerexplorer.jp/lamp3.0.0/index.html). They included forward and backward outer primers (F3 and B3) and forward and backward inner primers (FIP and BIP) (Table 3). Additionally, two loop primers, loop forward (LF) and loop backward (LB), were manually designed for each set. The 3′ end of the TgsGP gene was chosen for amplification because of its reported specificity to T. b. gambiense (14, 41). Primer specificity was checked with the basic local alignment search tool (BLAST) against human DNA and other human-infectious pathogens. The primer sets were analyzed with “must-detect samples,” i.e., T. b. gambiense isolates, and “must-not-detect samples,” i.e., T. b. rhodesiense, Trypanosoma brucei brucei, and Trypanosoma evansi (Table 1). The sets of primers that passed these criteria were then analyzed using a 10-fold serial dilution of T. b. gambiense DNA from isolate PT41 under the standard LAMP conditions (36). The most sensitive primer set (Table 3) was then chosen for further analysis.
Table 3.
Nucleotide sequences of the optimized LAMP primers targeting the TgsGP gene
| Primer name | Sequence (5′–3′) | Length (baes) | Amplicon sizea | Target sequence |
|---|---|---|---|---|
| TgsGP-F3 | GTTCGGAGAGCTCAGACAG | 19 | 150 | TgsGP gene |
| TgsGP-B3 | CCAACCGTTCCCAGTGTTG | 19 | ||
| TgsGP-FIP | TTGCTCCTTATCGCCGCCAGGCAAGAGCACAAAACCACAG | 40 | ||
| TgsGP-BIP | TGACGGGGACAACGGCTATCTATTTAACGCAGACACCGCC | 40 | ||
| TgsGP-LF | CCGCCCTGATCCCGCCTG | 18 | ||
| TgsGP-LB | GCAACTGCACAGGAACGGCG | 20 |
Length between F2 and B2c.
LAMP reactions.
To improve the sensitivity of TgsGP LAMP primers, four reaction mixture components (magnesium sulfate, FIP/BIP primers, deoxynucleoside triphosphates [dNTPs], and betaine) were subjected to rigorous optimization using the modified Taguchi method, followed by regression analysis to determine the concentration optima for each reaction mixture component (9). Briefly the forward inner primer (FIP) and backward inner primer (BIP) concentrations were varied from 20 to 80 pmol, dNTPs (Promega, NSW, Australia) from 1 to 4 mM, betaine (Sigma-Aldrich, St. Louis, MO) from 0.5 to 2.0 M, and magnesium sulfate (New England BioLabs, MA) from 0 to 6 mM. The 1× ThermoPol reaction buffer contained 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, and 0.1% Triton X-100. The Bst DNA polymerase (large fragment; New England BioLabs, MA) was at 1 μl (8 units), while SYTO-9 fluorescent dye at 3.0 μM (Molecular Probes, OR) was added for each real-time reaction. The template was ∼1 ng of purified trypanosome DNA from the T. b. gambiense isolate PT41. The LAMP reaction was performed for 30 to 60 min at 62°C using the Rotor Gene 6000 (Qiagen, Victoria, Australia), and data were acquired on the HRM channel (460 to 510 nm), followed by inactivation of the reaction at 80°C for 4 min. After the optimized conditions were determined, they were compared with the standard LAMP conditions, followed by trials using a normal water bath that maintained the temperature at ∼62 to 63°C. For comparison purposes, the mobile genetic element (RIME) LAMP (34) specific for the subgenus Trypanozoon was carried out.
Detection and confirmation of LAMP products.
The formation of LAMP product was first monitored in real time through fluorescence of Syto-9 dye, after which the product was divided into two equal portions of ∼10 μl each. One portion was analyzed using electrophoresis in 2.0% agarose gels stained with SYBR safe DNA gel stain and the other by visual inspection after the addition of a 1/10 dilution of SYBR green I. Two approaches were used to confirm that the TgsGP LAMP test amplified the correct target, namely, (i) the acquisition of melting curves using 1°C steps, with a hold of 30 s, from 62°C to 96°C and (ii) through cloning of the uppermost single band into a Topo-TA vector, transformation in Escherichia coli, and sequencing. The resulting sequence was manually compared with the expected target sequence.
Sensitivity of TgsGP LAMP and PCR.
A 10-fold serial dilution of ∼100 ng of T. b. gambiense isolate PT41 DNA was used. Second, DNA and supernatant were prepared from archived mouse blood that had been mixed with T. b. gambiense. Briefly, cultured T. b. gambiense parasites were mixed with mouse blood, adjusted to achieve approximately 1.0 × 106 trypanosomes/ml, and divided into two portions. One portion was used for DNA extraction using the Qiagen kit, and the other was boiled for supernatant as described previously (34). Tenfold serial dilutions were then prepared from the two DNA stocks, and the supernatant was used to determine the analytical sensitivity of TgsGP LAMP, TgsGP PCR (41), and nested TgsGP PCR (30). The LAMP test was carried out under optimized conditions, and the PCR tests followed the respective published procedures. The resulting LAMP and PCR products were electrophoresed in a 2.0% Tris-acetate-EDTA (TAE) agarose gel stained with SYBR safe DNA gel stain (Invitrogen, Victoria, Australia). The gel images were documented using the Gel-Doc-XR system (Bio-Rad Laboratories).
RESULTS
Optimum conditions for TgsGP LAMP.
The TgsGP primers chosen (Table 3) target a 220-bp section of TgsGP sequence that is between the specific sections amplified by the TgsGP PCR test (41). Surprisingly, 3 of the 5 LAMP primer sets amplified T. b. brucei isolates B8/18 from Nigeria and TSW187/78E from Ivory Coast, raising the question of the specificity of the T. b. gambiense 3′ end. A fourth primer set had a detection limit of 103 trypanosomes/ml, while the TgsGP primer set for this study showed a detection limit of 102 trypanosomes/ml using the 10-fold serial dilution of isolate PT41. The Taguchi method determined the final/optimal FIP and BIP primer concentrations at 40 pmol of each, 3 mM for each deoxynucleoside triphosphate, 1.2 M betaine, and 4 mM extra magnesium sulfate. The concentrations of other reagents were as previously reported (36). The optimum temperature for TgsGP LAMP was determined to be 62°C, and 35 min was the assay cutoff point.
TgsGP-LAMP product.
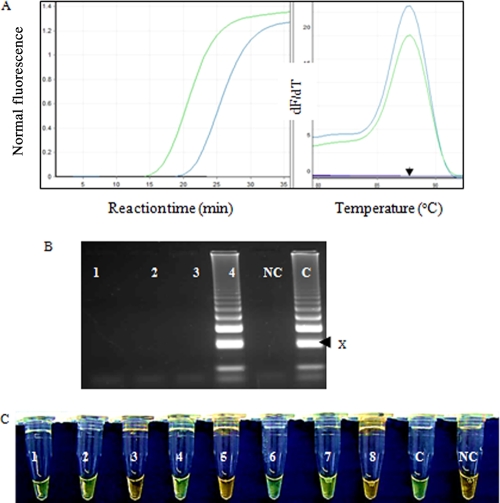
Positive LAMP reactions showed exponential amplification curves, as visualized in the real-time PCR machine. The advantage of optimization was demonstrated by the reduction in the cycle threshold (CT) value by an average of 5 cycles for all 10-fold serial dilutions (Fig. 1A) and a 10-fold increase in sensitivity. T. b. gambiense isolates from different geographical regions showed identical melting curves, with a melting temperature (Tm) of 87°C (Fig. 1A), suggesting amplicons with similar sequences. The positive LAMP products showed the predicted ladderlike pattern on the agarose gel, indicating the formation of stem-loops with inverted repeats (Fig. 1B). Further addition of a 1/10 dilution of SYBR green I showed a green color with positive reactions and orange with negative reactions (Fig. 1C). Sequencing of the uppermost LAMP band (Fig. 1B) revealed the predicted sequence from the F2-to-B2 primer region flanked by sequences from regions F1 and B1c on the 3′ and 5′ ends (Fig. 2).
Fig. 1.
(A) Amplification and postamplification melting curves obtained with 1 ng of T. b. gambiense DNA isolate PT41 under optimized (green line) and standard (blue line) LAMP conditions. The LAMP reactions were more efficient when the optimized conditions were used, with a reduction of an average of 5 cycles with every 10-fold serial dilution. The standard and optimized conditions showed identical melting curves, with a Tm of ∼87°C (arrow), indicating similar products. dF/dT, derivative of the fluorescence with respect to temperature. (B) Electrophoresis of TgsGP LAMP products. Lane 1, T. b. brucei; lane 2, T. b. rhodesiense; lane 3, T. evansi; lane 4, OM64 (DNA prepared from CSF sample RPH9 [Table 2]); lane C, positive control (T. b. gambiense PT41); lane NC, negative control. The arrow (x) indicates the sequenced band. (C) Visual appearance of TgsGP LAMP amplification products after addition of 1/10 dilution of SYBR green I dye. The dye fluoresces strongly when bound to the double-stranded DNA, and the resulting DNA-dye complex gives a green color, while fluorescence is minimal when the dye is free in the solution and gives an orange/brown color. The reactions were carried out in water baths at 62 to 63°C for 40 min. 1, OM55; 2, OM51; 3, OM52; 4, RHP6; 5, OM62; 6, OM54; 7, RHP9; 8, OM56; C, PT41; and NC, water.
Fig. 2.
Representative sequence obtained after cloning and sequencing of the uppermost bands from the TgsGP LAMP product. The sequence was identical to the expected target sequence of 195 bp (15 bp from F2 to B2, plus F1c, 20 bp, and B1, 22 bp). An identical sequence was obtained for the LAMP product acquired using the native CSF sample RPH9 (Table 3). Shading is used to distinguish the primer sections. Note that the sequence differs depending on the band sequenced and the inner primer initiating the reaction. F, forward primers; B, backward primers; L, loop primers; C, complementary sequence.
PCR and LAMP sensitivities.
The PCR results are shown in Tables 1 and 2. The positive samples showed the predicted ∼308-bp amplicon using TgsGP PCR and an ∼270-bp amplicon using nested TgsGP PCR. The analytical sensitivity was equivalent to 103 trypanosomes/ml and 10 trypanosomes/ml for the classical and nested TgsGP PCRs, respectively (Table 4). The TgsGP LAMP showed a detection limit of approximately 10 trypanosomes/ml of 10-fold serial dilutions of T. b. gambiense PT41 and approximately 1 trypanosome/ml when supernatant prepared from mouse blood was used (Table 4). The TgsGP LAMP sensitivity results were identical when either a Rotorgene 6000 thermocycler or a water bath was used as a source of heat. The LAMP assay was specific, and no cross-reactivity was recorded with nontarget DNA (Table 1).
Table 4.
Analytical sensitivity of TgsGP LAMP assay compared with TgsGP PCR tests using templates from 10-fold serial dilution of T. b. gambiense isolate PT41 and various templates prepared from mouse blood mixed with T. b. gambiense
| Test | Template | Expected specificity | Result at 10-fold dilution ofc: |
Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neat | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | 10−7 | 10−8 | ||||
| TgsGP LAMP | Supernatanta | T. b. gambiense | + | + | + | + | + | + | + | − | − | This study |
| TgsGP LAMP | DNAb | T. b. gambiense | + | + | + | + | + | + | − | − | − | This study |
| TgsGP (nested) | DNA | T. b. gambiense | + | + | + | + | + | + | − | − | − | (30) |
| TgsGP | DNA | T. b. gambiense | + | + | + | + | − | − | − | − | − | (41) |
Supernatant was prepared from mouse blood mixed with trypanosomes.
DNA from T. b. gambiense PT41 and from mouse blood prepared from 1.0 × 106 trypanosomes/ml.
10−1, ∼1.0 × 105 trypanosomes/ml; 10−2, ∼1.0 × 104 trypanosomes/ml; 10−8, ∼0.01 trypanosomes/ml; neat, approximately 100 ng; +, positive; −, negative.
Results for clinical samples.
Results for various clinical samples from patients diagnosed with T. b. gambiense are shown in Table 2. After eliminating the use of direct buffy coat (RPHbc) and bone marrow aspirate (RPHbma) samples where inhibition of the LAMP reaction was expected, the highly sensitive RIME LAMP detected 12/18 samples and the TgsGP LAMP assay detected 9/18 and 9/12 of the RIME LAMP-positive samples. Analysis of various sample formats from single and different patients using LAMP tests and Trypanozoon-specific PCR (29) showed better results with BMA and CSF than with blood (Table 2).
DISCUSSION
Case detection, followed by successful treatment, is a prerequisite for prevention and control of Gambian HAT. This strategy has faced significant problems due to a lack of sensitive diagnostic tests. Moreover, the diagnosis and staging of T. b. gambiense disease remains challenging, because the clinical features of the disease are not specific (4, 20). Therefore, research into the advancement of HAT diagnostic capabilities is still a priority. In this study, we have demonstrated specific amplification of T. b. gambiense DNA using a LAMP assay based on the TgsGP gene. The test is rapid, and amplification is achieved within 30 min using a real-time PCR machine at 62°C (Fig. 1A) and 40 min using a normal water bath and detecting the product through addition of fluorescent dye (Fig. 1C). Although the use of a normal water bath simplifies the need for instrumentation, nevertheless, the requirement for power to heat the water is still a drawback. As such, other sources of heat, like packaged exothermic reactions, need to be explored. The TgsGP LAMP was specific and exhibited analytical sensitivity of ∼10 trypanosomes/ml, which was equal to that of nested TgsGP PCR (Table 4); therefore, to date, TgsGP LAMP is the most sensitive single-step T. b. gambiense DNA-based test. However, as was observed with the serum resistance-associated (SRA) gene-based LAMP tests (35), this specific laboratory-based sensitivity may not be reproducible under field conditions, and thus, rigorous TgsGP LAMP field evaluations will be the next most important step.
The potential usefulness of TgsGP LAMP as a point-of-care test is demonstrated by the ability of the new assay to amplify target DNA from various templates, such as native CSF and double-centrifuged supernatant (Table 2). More promising is the ability of TgsGP LAMP to achieve 10-fold-higher sensitivity from supernatant than from DNA prepared from the same sample, meaning that DNA extraction may not be necessary, which would also shorten the assay time. Similar results were recorded with RIME LAMP (34). It is suggested that a significant amount of parasite DNA is lost during the extraction process, resulting in a lower detection limit when purified DNA is used as a template. However, before supernatant can be relied upon as a template for LAMP reactions, protocols for template purification and buffers that stabilize DNA in the supernatant need to be developed. This is because false-positive results (albeit rare) have previously been recorded with single centrifuged supernatants (33), while an initially positive supernatant turns negative after 3 weeks of storage at −20°C, suggesting degradation of the target DNA (Z. K. Njiru, unpublished data).
A field-based TgsGP LAMP will require a detection format(s) that is cheap and simple and that allows visual inspection of the results. However, most of these formats do not offer the option of confirming the LAMP product. Therefore, it is imperative to ensure that the developed test is specific and amplifies the predicted target. False positives in LAMP reactions are not necessarily absent and can result from amplicon contamination, unprocessed templates, the quality and composition of primers (poorly designed primers, frequent freezing and thawing, and AT-rich primers). In this study, the amplification of the target sequence from purified DNA, supernatant, and native CSF was unequivocally confirmed through postamplification acquisition of the melting curves, which showed a consistent Tm of ∼87°C (Fig. 1A), indicating similar sequences, and through sequencing of the uppermost LAMP band (Fig. 1C), which showed the predicted target sequence (Fig. 2). Therefore, the use of nonspecific dye in this study, supported by both positive and negative controls, increases our confidence.
Early and accurate diagnosis of T. b. gambiense is essential in reducing the risk of progression of infection to the late stage, which is difficult and dangerous to treat compared to the early stage (22). The challenge of T. b. gambiense diagnosis is well demonstrated in this study by low detection of parasite DNA in the most used patient sample, blood (samples OM and RPH1 to -4), using the available DNA detection tests. TgsGP LAMP detected 75% of the RIME LAMP-positive samples (Table 2). The sensitivity of the RIME LAMP is expected to be higher, since the test is a based on a multicopy gene (500 copies per haploid genome) (3) while TgsGP is based on a low-copy-number target. Unfortunately, RIME LAMP cannot differentiate between T. b. gambiense and T. b. rhodesiense, which is crucial, since the two parasites have different treatment regimens. Moreover, in East Africa, the introduction of T. b. rhodesiense into the T. b. gambiense region is certain to occur due to the closeness of the two disease foci and the continuous movement of livestock. We initially designed a test specific for T. b. rhodesiense (36), and the TgsGP LAMP designed in this study has the potential to contribute to T. b. gambiense diagnosis.
Our results show that the CSF is a better source of template for diagnosis of stage II diseases than blood. This is supported by the analysis of varied samples (native, supernatant, and DNA) from different patients (Table 2). Similar superior detection has been recorded using CSF PCR (16). Since the presence of T. b. gambiense had been confirmed in all the patients (34), it is suggested that the levels of parasitemia in the blood were too low and/or a great deal of DNA was lost during the extraction process. This concept is supported by similar low detection results observed using T. brucei (TBR) PCR and RIME LAMP tests specific for the multicopy and highly sensitive subgenus Trypanozoon (34). Further studies need to be carried out using only confirmed stage 1 patients to elucidate the sensitivity of the TgsGP LAMP test versus other molecular tests.
The application of nucleic acid-based tests in the diagnosis of Gambian sleeping sickness has been limited in the areas of endemicity, since most of the tests still require standardization and clinical validation (8). Furthermore, they are laborious and expensive and require elaborate visualization methods. In this study, the potential usefulness of the TgsGP LAMP test was demonstrated by its ease of applicability, rapidity, and higher sensitivity than the classical PCR targeting the same gene (Table 4). Moreover the ability of the test to detect parasite DNA in CSF is expected to contribute to diagnosis of the late-stage disease. Indeed, the staging of the late-stage HAT by the presence of trypanosomes in CSF and/or an elevated white blood cell (WBC) count above 5 cells/cm2 is not reliable (11,) nor are the existing CSF parasite detection methods sufficiently sensitive (23). However, a positive LAMP CSF result needs to be interpreted with caution, since it may not necessarily indicate living trypanosomes in the CSF but rather their DNA. It has been previously suggested that the presence of DNA in the CSF may result from leakage of circulating DNA in the blood through the blood brain barrier, or DNA can originate from nonsurviving parasites as a consequence of the suboptimal CSF survival environment (38). The issue of DNA rather than live parasites complicating interpretation should be studied using a primate model for HAT, which will elucidate the role of TgsGP LAMP and other HAT LAMP tests in determining cure. For now, LAMP results need to be interpreted in comparison to those of other tests and/or clinical symptoms.
Since the TgsGP LAMP test designed in this study is based on the TgsGP gene, it does not detect T. b. gambiense group 2, like TgsGP PCR. However, the vast majority of T. b. gambiense patients across all foci have group 1 infections, and the overall percentage of isolates from group 2 is so low as to be negligible, and only a few of them exist in laboratories. It is difficult to get a specific marker for group 2, since it is heterogeneous and genetically indistinguishable from T. brucei brucei. The lack of a universal diagnostic marker for T. b. gambiense and the reduced detection in blood affirms the need to continue evaluating other biomarkers. In practice, if the TgsGP LAMP test (CSF) is to be introduced, it may need to be combined with a test like CATT (blood) until the issues of LAMP sensitivity using blood are resolved. The TgsGP LAMP test developed here must be evaluated to investigate whether it can be used as a xenomonitoring tool and in detection of the T. b. gambiense parasite in suspected reservoir hosts (32).
The elimination of HAT as a public health problem in sub-Saharan Africa is a realistic objective. However, this will rely in part on the availability of affordable, sensitive, and field-applicable diagnostic technologies. In summary, this work shows (i) that the TgsGP LAMP is more sensitive than TgsGP PCR, (ii) that the use of supernatant increases the test sensitivity by 10-fold, (iii) that the use of CSF increases the chance of parasite DNA detection for stage II disease compared to blood (or its buffy coat from the same patient), and (iv) that the test is robust and amplification can be achieved using a normal water bath without compromising the test sensitivity. The LAMP test designed in this work and other recent technologies, such as the dipstick (10) and a sensitive semiquantitative card agglutination test, Latex/IgM (21), offer new prospects for improved detection of Gambian HAT. The next major step for the TgsGP LAMP test will be focused on field evaluation with the aim of generating data for development of a kit.
ACKNOWLEDGMENTS
This work was funded through a University of Queensland postgraduate grant to Z.K.N.
We contributed the samples analyzed, and we also acknowledge the provision of extra samples by Wendy Gibson, University of Bristol, Bristol, United Kingdom, and Christopher Health, Royal Perth Hospital, Western Australia.
The views expressed here do not necessarily reflect the views of our respective institutions.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Aonuma H., et al. 2010. A single fluorescence-based LAMP reaction for identifying multiple parasites in mosquitoes. Exp. Parasitol. 125:179–183 [DOI] [PubMed] [Google Scholar]
- 2. Berberof M., Pérez-Morga D., Pays E. 2001. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol. Biochem. Parasitol. 113:127–138 [DOI] [PubMed] [Google Scholar]
- 3. Bhattacharya S., Barke A., Bhattacharya A. 2002. Mobile genetic elements in protozoan parasites. J. Genet. 8:73–86 [DOI] [PubMed] [Google Scholar]
- 4. Bisoffi Z., et al. 2005. African trypanosomiasis gambiense, Italy. Emerg. Infect. Dis. 11:1745–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boehme C. C., et al. 2007. Operational feasibility of using loop-mediated isothermal amplification (LAMP) for the diagnosis of pulmonary TB in microscopy centers of developing countries. J. Clin. Microbiol. 45:1936–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bromidge T., Gibson W., Hudson K., Dukes P. 1993. Identification of Trypanosoma brucei gambiense by PCR amplification of variant surface glycoprotein genes. Acta Trop. 53:107–119 [DOI] [PubMed] [Google Scholar]
- 7. Cecchi G., et al. 2009. Towards the atlas of human African trypanosomiasis. Int. J. Health Geogr. 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chappuis F., Loutan L., Simarro P., Lejon V., Büscher P. 2005. Options for field diagnosis of human African trypanosomiasis. Clin. Microbiol. Rev. 18:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cobb B., Clarkson J. M. 1994. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 22:3801–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deborggraeve S., et al. 2006. Molecular dipstick test for diagnosis of sleeping sickness. J. Clin. Microbiol. 44:2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doua F., Miezan T. W., Sanon Singaro J. R., Boa Yapo F., Baltz T. 1996. The efficacy of pentamidine in the treatment of early late stage Trypanosoma brucei gambiense trypanosomiasis. Am. J. Trop. Med. Hyg. 55:586–588 [DOI] [PubMed] [Google Scholar]
- 12. Enyaru J. C., Allingham R., Bromidge T., Kanmogne G. D., Carasco J. F. 1993. The isolation and genetic heterogeneity of Trypanosoma brucei gambiense from north-west Uganda. Acta Trop. 54:31–39 [DOI] [PubMed] [Google Scholar]
- 13. Fang X., Liu Y., Kong J., Jiang X. 2010. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal. Chem. 82:3002–3006 [DOI] [PubMed] [Google Scholar]
- 14. Gibson W., Nemetschke L., Ndung'u J. 2010. Conserved sequence of the TgsGP gene in Group 1 Trypanosoma brucei gambiense. Infect. Genet. Evol. 10:453–458 [DOI] [PubMed] [Google Scholar]
- 15. Inojosa W. O., et al. 2006. Diagnosing human African trypanosomiasis in Angola using a card agglutination test: observational study of active and passive case finding strategies. BMJ 332:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jamonneau V., et al. 2003. Stage determination and therapeutic decision in human African trypanosomiasis: value of polymerase chain reaction and immunoglobulin M quantification on the cerebrospinal fluid of sleeping sickness patients in Côte d'Ivoire. Trop. Med. Int. Health 8:589–594 [DOI] [PubMed] [Google Scholar]
- 17. Kanmogne G. D., Stevens J. R., Asonganyi T., Gibson W. C. 1996. Characterization of Trypanosoma brucei gambiense isolates using restriction fragment length polymorphisms in 5 variant surface glycoprotein genes. Acta Trop. 61:239–254 [DOI] [PubMed] [Google Scholar]
- 18. Kuboki N., et al. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 41:5517–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lejon V., Ngoyi D. M., Boelaert M., Büscher P. 2010. A CATT negative result after treatment for human African trypanosomiasis is no indication for cure. PLoS Negl. Trop. Dis. 4:e590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lejon V., Büscher P. 2005. Cerebrospinal fluid in human African trypanosomiasis: a key to diagnosis, therapeutic decision and post-treatment follow-up. Trop. Med. Int. Health 10:395–403 [DOI] [PubMed] [Google Scholar]
- 21. Lejon V., et al. 2002. IgM quantification in the cerebrospinal fluid of sleeping sickness patients by a latex card agglutination test. Trop. Med. Int. Health 7:685–692 [DOI] [PubMed] [Google Scholar]
- 22. Lejon V., Boelaert M., Jannin J., Moore A., Büscher P. 2003. The challenge of Trypanosoma brucei gambiense sleeping sickness diagnosis outside Africa. Lancet Infect. Dis. 3:804–808 [DOI] [PubMed] [Google Scholar]
- 23. Lejon V., Buscher P. 2001. Stage determination and follow-up in sleeping sickness. Med. Trop. (Mars) 61:355–360 [PubMed] [Google Scholar]
- 24. Magnus E., Vervoort T., Van Meirvenne N. 1978. A card-agglutination test with stained trypanosomes (C.A.T.T.) for the serological diagnosis of T. b. gambiense trypanosomiasis. Ann. Soc. Belg. Med. Trop. 58:169–176 [PubMed] [Google Scholar]
- 25. Mehlitz D., Zillmann U., Scott C. M., Godfrey D. G. 1982. Epidemiological studies on the animal reservoir of Gambiense sleeping sickness. Part III. Characterization of Trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmed. Parasitol. 33:113–118 [PubMed] [Google Scholar]
- 26. Mori Y., Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mori Y., Hirano T., Notomi T. 2006. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mori Y., Nagamine K., Tomita N., Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154 [DOI] [PubMed] [Google Scholar]
- 29. Moser D. R., Kirchhoff L. V., Donelson J. E. 1989. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J. Clin. Microbiol. 27:1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morrison L. J., et al. 2008. Trypanosoma brucei gambiense type 1 populations from human patients are clonal and display geographical genetic differentiation. Infect. Genet. Evol. 8:847–854 [DOI] [PubMed] [Google Scholar]
- 31. Nimitphak T., Kiatpathomchai W., Flegel T. W. 2008. Shrimp hepatopancreatic parvovirus detection by combining loop-mediated isothermal amplification with a lateral flow dipstick. J. Virol. Methods 154:56–60 [DOI] [PubMed] [Google Scholar]
- 32. Njiokou F., et al. 2010. Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite 17:61–66 [DOI] [PubMed] [Google Scholar]
- 33. Njiru Z. K. 2011. Rapid and sensitive detection of Human African Trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn. Microbiol. Infect. Dis. 69:205–209 [DOI] [PubMed] [Google Scholar]
- 34. Njiru Z. K., et al. 2008. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int. J. Parasitol. 38:589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Njiru Z. K., et al. 2008. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS. Negl. Trop. Dis. 2:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paindavoine P., Zampetti-Bosseler F., Coquelet H., Pays E., Steinert M. 1989. Different allele frequencies in Trypanosoma brucei brucei and Trypanosoma brucei gambiense populations. Mol. Biochem. Parasitol. 32:61–71 [DOI] [PubMed] [Google Scholar]
- 38. Pentreath V. W., Owolabi A. O. 1992. Survival of Trypanosoma brucei brucei in cerebrospinal fluid. Ann. Trop. Med. Parasitol. 86:29–34 [DOI] [PubMed] [Google Scholar]
- 39. Pépin J., et al. 1994. Gambiense trypanosomiasis: frequency of, and risk factors for, failure of melarsoprol therapy. Trans. R. Soc. Trop. Med. Hyg. 88:447–452 [DOI] [PubMed] [Google Scholar]
- 40. Priotto G., et al. 2009. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomized, phase III, non-inferiority trial. Lancet 374:56–64 [DOI] [PubMed] [Google Scholar]
- 41. Radwanska M., et al. 2002. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am. J. Trop. Med. Hyg. 67:289–295 [DOI] [PubMed] [Google Scholar]
- 42. Robays. J., Bilengue M. M., Van der Stuyft P., Boelaert M. 2004. The effectiveness of active population screening and treatment for sleeping sickness control in the Democratic Republic of Congo. Trop. Med. Int. Health 9:542–550 [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Solano P., et al. 2002. Comparison of different DNA preparation protocols for PCR diagnosis of human African trypanosomosis in Cote d'Ivoire. Acta Trop. 82:349–356 [DOI] [PubMed] [Google Scholar]
- 45. Thekisoe O. M., et al. 2007. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 102:182–189 [DOI] [PubMed] [Google Scholar]
- 46. Truc P., et al. 2002. Evaluation of the micro-CATT, CATT/Trypanosoma brucei gambiense, and LATEX/T b gambiense methods for serodiagnosis and surveillance of human African trypanosomiasis in West and Central Africa. Bull. World Health Organ. 80:882–886 [PMC free article] [PubMed] [Google Scholar]
- 47. Van Nieuwenhove S. 2000. Gambiense sleeping sickness: re-emerging and soon untreatable? Bull. World Health Organ. 78:1283. [PMC free article] [PubMed] [Google Scholar]
- 48. Van Nieuwenhove S., et al. 1985. Treatment of gambiense sleeping sickness in the Sudan with oral DFMO (DL-alpha-difluoromethylornithine), an inhibitor of ornithine decarboxylase; first field trial. Trans. R. Soc. Trop. Med. Hyg. 79:692–698 [DOI] [PubMed] [Google Scholar]
- 49. WHO 1998. Control and surveillance of African trypanosomiasis. Technical report series 881. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]