Abstract
Molecular analysis of recurrent tuberculosis has revealed that a second episode may be caused by a strain of Mycobacterium tuberculosis other than that involved in the first infection, thus indicating that exogenous reinfection plays a role in recurrence. We focused on two aspects of reinfection that have received little attention. First, we evaluated whether a lack of methodological refinement could lead to inaccurate assignment of mixed infections as exogenous reinfection, in which a differential selection of each of the coinfecting strains occurred over time. We used the mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) method to genotype 122 isolates from 40 patients with recurrent tuberculosis. We identified 11/40 (27.5%) cases with genotypic differences between the isolates involved in the sequential episodes. Major genotypic differences were found in 8/11 cases, suggesting exogenous reinfection; in the remaining 3 cases, subtle genotypic differences were observed, probably indicating microevolution from a parental strain. In all cases, only a single strain was detected for the isolate(s) from each episode, thus ruling out the possibility that reinfection could correspond to undetected mixed infection. Second, we analyzed the infectivity of a selection of 12 strains from six cases with genotypically different strains between episodes. No main differences were observed in an ex vivo model of infection between the strains involved in the first episodes and those involved in the recurrent episodes. In our setting, our results suggest the following: (i) the possibility of misassignment of mixed infection as exogenous reinfection is improbable, and (ii) bacterial infectivity does not seem to play a role in exogenous reinfection.
INTRODUCTION
The traditional assumptions in the analysis of infection by Mycobacterium tuberculosis are that each episode is caused by a single M. tuberculosis strain and that recurrence is considered to result from an endogenous reactivation of the strain involved in the first episode. In recent years, molecular analysis of M. tuberculosis isolates has revealed that a recurrence may also be caused by a strain other than that isolated in the first episode, suggesting that exogenous reinfection plays a role in recurrence of tuberculosis (5, 8, 26). More complex situations have also been observed, including simultaneous infection by two M. tuberculosis strains (5, 28) and infection by the same strain at different anatomical sites (13) or at different sites within the lung (14).
Exogenous reinfection has mainly been observed in immunocompromised patients (8, 21) and in settings with a high prevalence of tuberculosis (8, 26) or a high risk for transmission (5), thus suggesting that mainly socioepidemiological factors determine the occurrence of these phenomena. However, detection of exogenous reinfection in settings with moderate and low prevalence of tuberculosis and in both HIV-positive and HIV-negative patients (2, 4, 12) could indicate that specific bacterial factors may also be involved. Several in vivo and in vitro models of infection have shown higher infectivity for strains in which transmission and infection had been observed to be particularly efficient (3, 15, 25). We might therefore postulate that infectivity is also involved in reinfection.
Our study focused on two as-yet-unexplored aspects of M. tuberculosis infection: (i) the possibility that exogenous reinfection could be the result of a different chronological selection of strains in patients with undetected polyclonal infections and (ii) the relative contribution of bacterial factors (e.g., infectivity) to reinfection.
MATERIALS AND METHODS
Analysis of patients with recurrent tuberculosis. (i) Patients.
The study population comprised all patients with two or more culture-positive episodes of tuberculosis between 1998 and 2006 at Hospital Gregorio Marañón, Madrid, Spain. M. tuberculosis isolates were stored frozen at −70°C from isolation until analysis. When available, two different isolates from independent specimens within a single episode were selected for genotyping. Some of the patients had been studied elsewhere for different reasons (16), but were included in the present study because previous genotyping was rather limited compared with current standards for mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing. We defined an episode as the microbiological isolation of M. tuberculosis and recurrence as the presence of two episodes separated by more than 6 months in a single patient.
(ii) Microbiological procedures. (a) Culture and DNA extraction.
Selected isolates were subcultured in MGIT liquid media (Becton Dickinson, Sparks, MD) for 2 to 3 weeks at 37°C. One milliliter of culture was centrifuged at 13,000 rpm for 5 min, and the pellet was boiled in the presence of GeneProbe lysis reagent (1:16 diluted; bioMérieux, Geneva, Switzerland) for 7 min.
(b) MIRU-VNTR analysis.
The template for MIRU-VNTR analysis consisted of 5 μl of crude extract obtained after boiling of the subcultured isolates. We applied the 15-locus set (22) and ran the amplified products at 45 V for 17.5 h in an MS8 2% agarose gel (Pronadisa, Madrid, Spain). Fragment size was calculated with the ChemiDoc system (Bio-Rad, Hercules, CA) and the Diversity database (Bio-Rad), using a 100-bp ladder (Invitrogen, Carlsbad, CA) as a molecular weight marker. The number of repeats in each locus was calculated by applying the corresponding conversion table.
MIRU-VNTR enables rapid identification of polyclonal infection by simultaneous detection of more than one amplification product for the specific locus (or loci) in which the M. tuberculosis strains or clonal variants show differences.
Characterization of infectivity of M. tuberculosis strains. (i) Cell cultures.
The human promonocytic cell line THP-1 was obtained from the American Type Culture Collection (catalog no. TIB-202; Manassas, VA). Cell cultures were maintained in modified RPMI 1640 medium plus l-glutamine (Gibco, BRL) supplemented with 10% fetal bovine serum (FBS) (Biochrom AG), 10 mM HEPES, and 50 μg/ml of gentamicin (Gibco, BRL). Cultures were maintained at 7 × 105 to 10 × 105 cells/ml and incubated at 37°C in 5% CO2 in a humidified incubator. THP-1 cells were differentiated as adherent macrophages by addition of 200 nM phorbol myristate acetate (PMA) (Sigma) for 3 days at 37°C in 5% CO2.
(ii) Cell infection and measurement of bacterial intracellular growth.
Cells were infected as described elsewhere (24), with slight modifications. Briefly, differentiated THP-1 cells seeded in 24-well flat-bottom tissue culture plates were washed, and the medium was replaced to remove PMA and gentamicin 2 h before the addition of bacteria. Cells were infected with a multiplicity of infection of 2 to 10 bacteria per cell and incubated for 3 h at 37°C in 5% CO2. After incubation, monolayers were thoroughly washed with phosphate-buffered saline (PBS) to remove extracellular bacteria, and fresh medium was added. Bacterial growth was evaluated by aspirating the supernatants and lysing the monolayers with 0.5% Nonidet P-40 (Roche) at 3 h and 1, 4, and 7 days after infection. Serial 10-fold dilutions of cellular lysates were plated on Middlebrook 7H11 plates and incubated for 3 weeks at 37°C in 5% CO2 before colonies were counted.
(iii) Cytokine analysis.
Culture supernatants from infected THP-1 cells were harvested at 3 h and 1, 4, and 7 days after infection; frozen at −70°C; and assayed with an enzyme-linked immunosorbent assay kit according to the manufacturer's instructions (BD Biosciences, Lincoln Park, NJ) to measure levels of tumor necrosis factor alpha (TNF-α) and interleukin-10 (IL-10).
(iv) Statistical analysis.
For each patient, intracellular growth and cytokine production were measured in three independent analyses for both the first and second clinical episodes involving the tuberculosis strain. Due to the low number of specimens, the Wilcoxon signed-rank test for paired samples was used to assess differences in growth ratios and cytokine levels between episodes.
RESULTS
Analysis of patients with genotypically different strains between recurrences.
Our primary objective was to evaluate whether patients initially considered as having exogenous reinfection could actually be coinfected by more than one strain whose representativeness, and therefore detectability, could vary at different points during infection.
Therefore, we first sought candidates for exogenous reinfection among patients with recurrent tuberculosis. We identified 60 patients with recurrent tuberculosis. The difference between episodes ranged from 182 to 1,963 days (median, 388 days). Isolates were available for genotyping in 40 (66.66%) patients (2 to 4 episodes) (Table 1). More than one isolate per episode was available for analysis in 16 cases.
Table 1.
Characteristics of the patients analyzed and genotypes obtained for each isolate after MIRU-VNTR typinga
| Patient | Sex | Risk factor(s) | Adherence to antituberculosis therapy | Episode no. | No. of days between episodes | Type of isolate | MIRU typeb |
|---|---|---|---|---|---|---|---|
| 1 | Female | No | No | 1 | Sputum | 252353232232425 | |
| 2 | 568 | Sputum | 252353232232425 | ||||
| 3 | 469 | Sputum | 252353232232425 | ||||
| 4 | 356 | Sputum | 252353232232425 | ||||
| 2 | Male | IVDUc | No | 1 | Sputum | 245423342123217 | |
| 2 | 762 | Sputum | 245423342123217 | ||||
| 3 | Male | No | Not available | 1 | Sputum | 233543242212427 | |
| 2 | 870 | Sputum | 233543242212427 | ||||
| 4 | Male | HIV infection, IVDU, prison | No | 1 | Sputum | 253533233423533 | |
| 2 | 225 | Urine | 253533233423533 | ||||
| 5 | Male | No | No | 1 | Sputum | 253533333433426 | |
| 2 | 182 | Sputum | 253533333433426 | ||||
| 3 | 253 | Sputum | 253533333433426 | ||||
| 4 | 310 | Sputum | 253533333433426 | ||||
| 6 | Male | Alcoholism | Not available | 1 | Sputum | 25442342221243.11 | |
| 2 | 993 | Sputum | 25442342221243.11 | ||||
| 7 | Male | Alcoholism | No | 1 | Sputum | 25453342221243.11 | |
| 2 | 1,120 | Sputum | 25453342221243.11 | ||||
| 8 | Male | HIV infection, IVDU | No | 1 | Sputum | 2′52453242232225 | |
| 2 | 753 | Sputum | 2′52453242232225 | ||||
| 3 | 567 | Sputum | 2′52453242232225 | ||||
| 9 | Female | HIV infection, IVDU | Not available | 1 | Sputum | 2515_3333433726 | |
| 2 | 714 | Sputum | Not available | ||||
| 10 | Male | HIV infection, IVDU | Not available | 1 | Urine | 252433422212437 | |
| 2 | 1,367 | Sputum | 2534_3342133137 | ||||
| 11 | Male | HIV infection, alcoholism | Not available | 1 | Sputum | 252533233432425 | |
| 2 | 602 | Sputum | 252333233432425 | ||||
| 12 | Male | HIV infection, alcoholism, prison | Not available | 1 | Sputum | 253333243232325 | |
| 2 | 1,492 | Sputum | 252333243232325 | ||||
| Sputum | 252333243232325 | ||||||
| 13 | Male | No | Not available | 1 | Sputum | 252343232232125 | |
| 2 | 863 | Sputum | 252343232232125 | ||||
| 14 | Male | HIV infection, alcoholism | Yes | 1 | Sputum | 255323143232525 | |
| 2 | 1,888 | Sputum | 255433340211434 | ||||
| 3 | 327 | Urine | 255433340211434 | ||||
| 15 | Male | HIV infection | Not available | 1 | Sputum | 253323422212236 | |
| 2 | 288 | Sputum | 253323422212236 | ||||
| 16 | Male | Silicosis, lupus | Yes | 1 | Sputum | 252433243443347 | |
| Sputum | 252433243443347 | ||||||
| 2 | 1,963 | Sputum | 252433243443347 | ||||
| 17 | Male | HIV infection, IVDU, hepatitis B, | Yes | 1 | Sputum | 253333242232425 | |
| hepatitis C | Sputum | 253333242232425 | |||||
| 2 | 409 | Blood | 253333242232425 | ||||
| 18 | Male | HIV infection, IVDU, hepatitis B, | No | 1 | Sputum | 254223422213236 | |
| hepatitis C | 2 | 238 | CSFd | 254223422213236 | |||
| 19 | Male | No | Not available | 1 | Sputum | 333333444232416 | |
| Sputum | 333333444232416 | ||||||
| 2 | 489 | Sputum | 243433422212434 | ||||
| 20 | Male | Alcoholism, smoking | Yes | 1 | Sputum | 251433542122438 | |
| 2 | 202 | Sputum | 251433542122438 | ||||
| 21 | Male | No | Not available | 1 | Sputum | 263432233431244 | |
| Sputum | 263432233431244 | ||||||
| 2 | 263 | Sputum | 252343242232325 | ||||
| 22 | Female | Alcoholism, smoking | No | 1 | Sputum | 253423422212334 | |
| 2 | 387 | Sputum | 253423422212334 | ||||
| 23 | Male | HIV infection, alcoholism | Not available | 1 | Sputum | 253423422212437 | |
| Sputum | 253423422212437 | ||||||
| 2 | 610 | Sputum | 253423422212437 | ||||
| 24 | Male | HIV infection, IVDU, hepatitis B, | Yes | 1 | Sputum | 243433442212424 | |
| hepatitis C | Sputum | 243433442212424 | |||||
| 2 | 321 | Sputum | 243433442212424 | ||||
| Sputum | 243433442212424 | ||||||
| 25 | Male | HIV infection, alcoholism, | No | 1 | Sputum | 253633233433635 | |
| homelessness | 2 | 981 | Sputum | 253532233333235 | |||
| 26 | Male | No | Not available | 1 | Sputum | 255433442212334 | |
| Sputum | 255433442212334 | ||||||
| 2 | 252 | Sputum | 255433442212334 | ||||
| Sputum | 255433442212334 | ||||||
| 3 | 1,203 | Sputum | 255433442212334 | ||||
| 27 | Female | No | Not available | 1 | Sputum | 251343142232225 | |
| 2 | 1,036 | Sputum | 354323343222518 | ||||
| 28 | Male | No | Not available | 1 | Sputum | 251323142122234 | |
| Sputum | 251323142122234 | ||||||
| 2 | 221 | Sputum | 251323142122234 | ||||
| 29 | Male | No | Not available | 1 | Sputum | 254233242232425 | |
| 2 | 258 | Sputum | 254233242232425 | ||||
| 30 | Male | HIV infection, IVDU, hepatitis C | Yes | 1 | Sputum | 253533242232425 | |
| Sputum | 253533242232425 | ||||||
| 2 | 285 | Sputum | 253533242232425 | ||||
| 31 | Male | HIV infection, IVDU, hepatitis B, | Unknown | 1 | Sputum | 251423442122337 | |
| hepatitis C | 2 | 312 | Sputum | 251423442122337 | |||
| 32 | Female | HIV infection, IVDU, hepatitis C | No | 1 | Sputum | 252343242232325 | |
| 2 | 186 | Sputum | 252343242232325 | ||||
| 33 | Male | HIV infection, IVDU, hepatitis B, | No | 1 | Sputum | 253533233433426 | |
| hepatitis C, prison | Sputum | 253533233433426 | |||||
| 2 | 277 | Sputum | 253533233433426 | ||||
| 34 | Male | HIV infection, IVDU, alcoholism | No | 1 | Sputum | 251423442122337 | |
| Sputum | 251423442122337 | ||||||
| 2 | 217 | Blood | 244333043_3_525 | ||||
| 35 | Male | HIV infection, IVDU, hepatitis C | Yes | 1 | Sputum | 234333242232325 | |
| Sputum | 234333242232325 | ||||||
| 2 | 222 | Sputum | 234333242232325 | ||||
| 36 | Male | Alcoholism, smoking, hepatitis C | No | 1 | Sputum | 252433342123237 | |
| 2 | 880 | Sputum | 252433342123237 | ||||
| 37 | Male | HIV infection, multidrug user | No | 1 | Sputum | 261423342132235 | |
| 2 | 425 | Sputum | 261423142132235 | ||||
| Sputum | 261423142132235 | ||||||
| 38 | Male | No | Yes | 1 | Sputum | 351433233423436 | |
| 2 | 1,400 | Sputum | 351433233423436 | ||||
| 39 | Male | HIV infection, hepatitis C, | Not available | 1 | Sputum | 251323442122438 | |
| IVDU, smoking | Sputum | 251323442122438 | |||||
| 2 | 1,472 | Sputum | 251623442122337 | ||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| Sputum | 251623442122337 | ||||||
| 40 | Male | HIV infection, hepatitis C, | No | 1 | Sputum | 263335444432647 | |
| IVDU, smoking | Sputum | 263335444432647 | |||||
| 2 | 304 | Sputum | 263335444432647 | ||||
| Sputum | 263335444432647 | ||||||
| Sputum | 263335444432647 | ||||||
| Sputum | 263335444432647 | ||||||
| 3 | 382 | Sputum | 263335444432647 | ||||
| Sputum | 263335444432647 | ||||||
| Urine | 263335444432647 | ||||||
| Urine | 263335444432647 | ||||||
| Urine | 263335444432647 | ||||||
| Pleural fluid | 263335444432647 | ||||||
| Pleural fluid | 263335444432647 |
Those cases in which clonal complexity was detected are highlighted in gray.
Loci at which the isolates show differences in their MIRU types are in boldface. The lack of an allelic value at an MIRU locus indicates that no amplified product was obtained. In patients 6 and 7, the allele in the last locus in the MIRU type corresponded to a value of 11.
IVDU, intravenous drug user.
CSF, cerebrospinal fluid.
We identified 8/40 (20%) cases (cases 10, 14, 19, 21, 25, 27, 34, and 39) in which major genotypic differences (involving at least three loci) were found between the M. tuberculosis strains isolated from the sequential episodes, thus suggesting exogenous reinfection (Table 1). Four patients were HIV positive. All isolates were susceptible, except for the last one from case 14, which was resistant to rifampin and ethambutol. First, we used MIRU-VNTR to rapidly detect polyclonal infections. We ruled out the presence of coinfecting strains in the first or recurrent episodes, based on the detection of one allele in all 15 loci studied. Second, in order to ensure that reinfection did not correspond to undetected mixed infection, we refined the analysis by searching for coinfecting strains in independent isolates from the same episodes. Unfortunately, additional isolates were available for MIRU-VNTR typing in only four of the eight cases with reinfection, but in each of these, only a single strain was identified in the different isolates from the same episode (Table 1). These results indicate that no data were found to support the hypothesis that reinfection could be due to mixed infection by more than one M. tuberculosis strain whose representativeness, and therefore detectability, varied at different times during the infection.
Analysis of recurrent cases also enabled us to identify three additional cases (cases 11, 12, and 37) in which subtle differences (involving a single locus) were found between the strains involved in the sequential isolates, thus suggesting microevolution from a parental strain (Table 1). The three patients were HIV positive. All isolates were susceptible, except for the second one from case 11, which was resistant to isoniazid. The same methodological procedure applied for the analysis of the representatives of exogenous reinfection was applied to the microevolution candidates. We proceeded to evaluate whether the new clonal variants generated could coexist with their parental strains. Simultaneous presence of clonal variants was not observed, and only one of the two clonal variants was detected in each of the episodes analyzed, even for those cases in which more than one isolate per episode was available (Table 1).
Characterization of infectivity in cases with genotypically different strains in sequential episodes.
We selected four pairs of M. tuberculosis strains isolated from four patients with exogenous reinfection (patients 10, 19, 21, and 34) to analyze infectivity. We also included two additional pairs of clonal variants isolated from two patients with microevolution between the first episode and the recurrent episode (patients 11 and 37) (Table 1).
Intracellular growth of the selected M. tuberculosis strains in THP-1 cells.
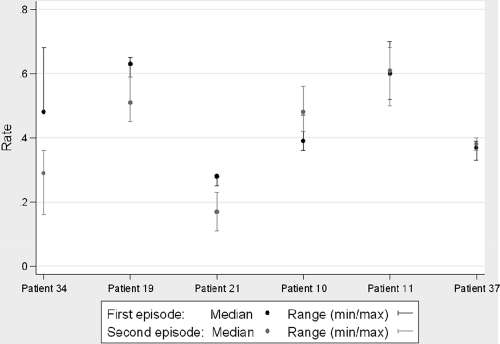
We screened for potential differences in infectivity between the strains isolated from the first and second episodes of patients with reinfection/microevolution and found no marked differences (Fig. 1). No differences were found after comparing the medians of the growth rates of all strains isolated from the first episode with those of the strains isolated from the second episode (z value, 0.34). When each patient was analyzed separately, differences in growth rates were recorded for two cases from the reinfection group (patients 21 and 34), with higher values for the strains isolated from the first episode. No differences were detected for the representatives of microevolution.
Fig. 1.
In vitro intracellular growth rates (median and range) for the two isolates from the sequential episodes of each patient.
IL-10 production by THP-1 cells infected by M. tuberculosis.
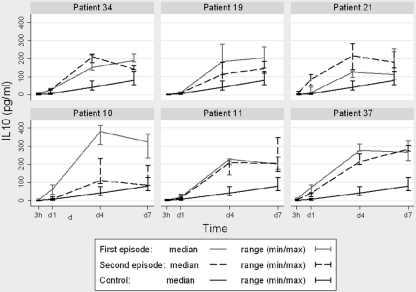
IL-10 levels were measured at 3 h and 1, 4, and 7 days after infection. At each time point, the median of the IL-10 levels produced by THP-1 cells infected by the set of strains isolated from the first episodes was compared with the median of the IL-10 levels produced by THP-1 cells infected by the set of strains isolated from the second episodes. No differences were found. When the results obtained for each patient were compared separately, higher levels of IL-10 were observed after infection by strains from the first episodes in patients 10 (reinfection) and 37 (microevolution). These differences were recorded at days 4 and 7 in patient 10 and at day 4 in patient 37 (Fig. 2).
Fig. 2.
IL-10 levels (median and range) produced by THP-1 cells infected by the M. tuberculosis strains isolated from each patient. IL-10 levels obtained in noninfected controls are shown as a reference.
TNF-α production by THP-1 infected by MTB.
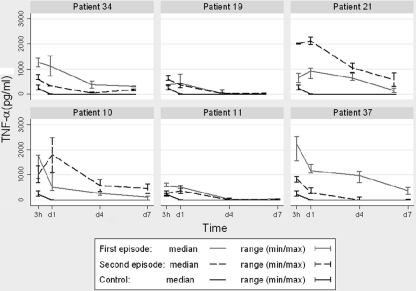
TNF-α levels were also measured at 3 h and 1, 4, and 7 days after infection, and the same comparisons as for IL-10 were performed. In the first case (comparison between the medians of the TNF-α levels detected in the first and second episodes), no differences were observed. When the results obtained for each pair of strains isolated from each patient were compared separately, in patients 34 (reinfection case) and 37 (microevolution case), higher levels of TNF-α were detected for the THP-1 cells infected by the M. tuberculosis strains isolated from the first episode at all time points analyzed. However, in patient 21 (reinfection), the highest levels of TNF-α were observed at 3 h and day 1 in the THP-1 cells infected by the strain isolated from the second episode (Fig. 3).
Fig. 3.
TNF-α levels (median and range) produced by THP-1 cells infected by the M. tuberculosis strains isolated from each patient. TNF-α levels obtained in noninfected controls are shown as a reference.
DISCUSSION
Although clonally complex events are gradually being recognized in infection by M. tuberculosis (1, 5, 19), few studies analyze their relative importance, and most are based on difficult or cumbersome methodological approaches, such as interpretation of low-intensity bands in the restriction fragment length polymorphism (RFLP) patterns (6) or analysis of multiple independent colonies (13). The new PCR-based technique MIRU-VNTR has proven to be a faster, simpler, and more efficient approach for the identification of clonal complexity (9). Combination of MIRU-VNTR with analysis of several serial isolates per patient has been recommended to improve detection of clonal heterogeneity and to minimize underestimation of clonally complex infections (19).
Unlike the studies mentioned above, which analyzed reinfection and mixed infection independently, our objective was to determine whether exogenous reinfection could, in some cases, be the result of mixed infection with different selections of strains at different time points during sequential episodes of recurrent tuberculosis. Some authors have speculated about the possibility of undetected mixed infection being responsible for a relapse after apparent “curative” treatment (17), although no major studies have been undertaken to attempt to prove this hypothesis.
Clonal heterogeneity between M. tuberculosis isolates from the first and recurrent episodes was frequent (27.5%). In 20% of recurrent cases, major genotypic differences were found between the isolates, pointing to exogenous reinfection. Although high, this percentage is similar to that recorded by Bandera et al., who observed 16% of recurrences were due to exogenous reinfection in an area with a low incidence of tuberculosis. In our sample, we searched for the presence of more than one M. tuberculosis strain in any episode. We used MIRU-VNTR to directly identify cases infected by more than one strain and enhanced our screening by analyzing more than one isolate per episode, when available. This refinement tried to minimize the fact that not all of the infecting M. tuberculosis strains are necessarily equally represented in independent clinical specimens (18). In all cases, we detected only a single strain in each episode. Therefore, it was not possible to prove the hypothesis that exogenous reinfection could correspond to undetected polyclonal infections in which the representativeness of coinfecting strains varied at different time points. However, we cannot completely rule out the possibility of undetected polyclonal infection, since we did not genotype more than one isolate per episode in all cases. It is also possible that mixed infection remained undetected by MIRU; however, such a scenario seems improbable, since the percentage of one of the strains would have to be lower than 1% (9). Despite these limitations, to the best of our knowledge, this is the first study to experimentally address the hypothesis of possible misassignment of reinfections due to undetected polyclonal infection.
In addition to cases of reinfection, our analysis of recurrence allowed us to identify another interesting set of patients, namely, those in whom M. tuberculosis clonal variants with subtle genotypic differences were isolated from sequential episodes. Again, only one of the variants was identified in each episode, suggesting that the microevolved variant displaced the parental strain.
Exogenous reinfection has been observed mainly in settings with either a high incidence of tuberculosis (8, 20, 21, 26) or where the risk of overexposure is increased (5). Moreover, several studies have shown that immunocompromised patients are at a higher risk for exogenous reinfection (21), leading to the assumption that exogenous reinfection is associated with clinical and socioepidemiological factors, which would be consistent with Vynnycky and Fine, who found that exogenous reinfection in the analysis of recurrence increased in parallel with the risk of infection (27).
However, exogenous reinfection has also been observed in HIV-negative patients and in settings with a low incidence of tuberculosis (2, 4, 12), even in circumstances where this phenomenon is not expected, such as in patients who did not adhere to therapy during their first episode. Therefore, bacterial factors might also play a role in exogenous reinfection, although only one study has suggested this possibility (12).
For this reason, we also evaluated whether, in addition to socioepidemiological factors, bacterial factors could be contributing to exogenous reinfection. In addition, we thought it would be particularly interesting to analyze possible differences in the infectivities of the different clonal variants detected in recurrent cases, since generation of a clonal variant from a parental M. tuberculosis strain has been proposed as a mechanism of adaptation to the new host environment (6), and subtle genetic variations have been found to be responsible for modifications in the ability to infect different tissues (11). Moreover, we previously found a correlation between the in vivo and ex vivo infectivities of M. tuberculosis strains and their efficiencies at infecting extrarespiratory sites (10). Outside these findings, the specific role of differences in virulence in reinfection or in clonally complex infections has not been explored in depth. Therefore, we characterized the ex vivo infectivity of the M. tuberculosis strains involved in some of the clonally complex infections detected in our first approach. To do this, we used the model of infection based on the THP-1 cell line, since it has been shown to be suitable for the analysis of intracellular growth of M. tuberculosis (24). The model has also proven successful in identifying different intracellular growth rates and rates of cytokine production in cells infected by M. tuberculosis strains with different transmission dynamics (23, 25).
We found no differences between the median growth rate for representative strains of exogenous reinfection involved in the first episodes and that of the strains involved in the recurrent episode. However, when we analyzed each pair of strains isolated from each patient independently, differences were observed in two cases from the exogenous reinfection group (patients 21 and 34), and in both cases, the strains involved in the first episode showed a higher growth rate. When we compared median cytokine levels in the supernatants from the cells infected by strains involved in first episodes with those from second episodes, once again, we found no differences. As for the TNF-α and IL-10 levels of the two pairs of strains showing differences in growth rates, TNF-α production varied in both cases. Lower levels of TNF-α were detected in the second episode in patient 21. The ability of infected cells to induce production of TNF-α has proven to be characteristic of “slow-growth” phenotypes, since initial control of M. tuberculosis infection depends on macrophage activation, which includes production of this cytokine by infected cells (25). Theus et al. have shown a strong correlation between suppression of TNF-α or early production of IL-10 (or both) and a higher growth rate for some M. tuberculosis strains in THP-1 cells (23, 25); however, we did not observe these behaviors in our study. The highest levels of TNF-α were detected in the first episode in patient 34, suggesting that, in this case, mechanisms other than the ability to reduce production of TNF-α could explain the higher growth rate observed in the strain from the first episode.
No differences were found in intracellular replication levels for the clonal variants involved in microevolution. Analysis of cytokine production revealed differences in TNF-α levels in THP-1 cells infected by M. tuberculosis strains isolated from patient 37, with higher levels detected in cells infected by the clonal variant from the first episode. The ability to contain production of TNF-α might be an advantage for the clonal variant detected in the second episode. This could explain why the initial clone is not detected once this second clonal variant emerges.
Despite the small number of strains assayed, we conclude that no bacterial factors (e.g., higher infectivity or the ability to control the host immune response) seem to be related to reinfection/microevolution events. We must remember that most patients (8/11) showing clonal heterogeneity in this study were HIV positive and that immunocompromised persons are at a higher risk for exogenous reinfection (21); therefore, the immunocompromised status of our hosts could bias the lack of association between bacterial factors and reinfection/microevolution.
Although we found no differences in ex vivo infectivities when comparing the two groups of M. tuberculosis strains, we did observe some differences in the growth rates of two pairs. In both cases, the strains involved in the first episode showed a higher intracellular growth rate, and the time between the first episode and the recurrent episode was relatively short. One of the patients had no risk factors for developing tuberculosis. Although rather speculative, a possible explanation could be that the host was severely immunocompromised after infection with the first M. tuberculosis strain, thus making reinfection by a new strain easier, even when the infectivity of the second strain was lower. Other authors have also suggested this possibility (7), which could explain the short time between the first and second episodes. Therefore, in this case, bacterial factors accounting for the ability to reduce the host response could indicate a certain susceptibility to reinfection on exposure to a new M. tuberculosis strain. Further studies will be required to investigate this hypothesis.
In the context of a low incidence of tuberculosis, it seems unlikely that polyclonal infections which went undetected by standard analytical methods could be responsible for misassignment of reinfections. In addition, no role was observed for bacterial factors such as infectivity in clonally complex recurrences.
ACKNOWLEDGMENTS
We are indebted to Thomas O'Boyle for proofreading and editing the manuscript. We are grateful to Beatriz Pérez for statistical analysis.
A.M. is the recipient of a grant from the Comunidad de Madrid, cofinanced by the European Social Fund (order no. 5297/2006). Y.N. holds a grant from CIBERES 4a Convocatoria de Becas de Iniciación a la Investigación. This study was partially financed by Fondo de Investigaciones Sanitarias (FIS S09/02205) and by Instituto de Salud Carlos III (CIBER Enfermedades Respiratorias CB06/06/0058 and Spanish Network for Research in Infectious Diseases REIPI RD06/0008).
The authors have no potential conflicts of interest to declare.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Baldeviano-Vidalon G. C., et al. 2005. Multiple infection with resistant and sensitive M. tuberculosis strains during treatment of pulmonary tuberculosis patients. Int. J. Tuberc. Lung Dis. 9:1155–1160 [PubMed] [Google Scholar]
- 2. Bandera A., et al. 2001. Molecular epidemiology study of exogenous reinfection in an area with a low incidence of tuberculosis. J. Clin. Microbiol. 39:2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barczak A. K., et al. 2005. In vivo phenotypic dominance in mouse mixed infections with Mycobacterium tuberculosis clinical isolates. J. Infect. Dis. 192:600–606 [DOI] [PubMed] [Google Scholar]
- 4. Caminero J. A., et al. 2001. Exogenous reinfection with tuberculosis on a European island with a moderate incidence of disease. Am. J. Respir. Crit. Care Med. 163:717–720 [DOI] [PubMed] [Google Scholar]
- 5. Chaves F., Dronda F., Alonso-Sanz M., Noriega A. R. 1999. Evidence of exogenous reinfection and mixed infection with more than one strain of Mycobacterium tuberculosis among Spanish HIV-infected inmates. AIDS 13:615–620 [DOI] [PubMed] [Google Scholar]
- 6. de Boer A. S., et al. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS6110 restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38:4478–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. du Plessis D. G., Warren R., Richardson M., Joubert J. J., van Helden P. D. 2001. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS 6110 and other repetitive element-based DNA fingerprinting. Tuberculosis (Edinb.) 81:211–220 [DOI] [PubMed] [Google Scholar]
- 8. Fitzpatrick L. K., et al. 2002. An investigation of suspected exogenous reinfection in tuberculosis patients in Kampala, Uganda. Int. J. Tuberc. Lung Dis. 6:550–552 [DOI] [PubMed] [Google Scholar]
- 9. Garcia de Viedma D., Alonso Rodriguez N., Andres S., Ruiz Serrano M. J., Bouza E. 2005. Characterization of clonal complexity in tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat typing. J. Clin. Microbiol. 43:5660–5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia de Viedma D., et al. 2005. Association between the infectivity of Mycobacterium tuberculosis strains and their efficiency for extrarespiratory infection. J. Infect. Dis. 192:2059–2065 [DOI] [PubMed] [Google Scholar]
- 11. Garcia de Viedma D., et al. 2006. Complex clonal features in an mycobacterium tuberculosis infection in a two-year-old child. Pediatr. Infect. Dis. J. 25:457–459 [DOI] [PubMed] [Google Scholar]
- 12. Garcia de Viedma D., et al. 2002. Tuberculosis recurrences: reinfection plays a role in a population whose clinical/epidemiological characteristics do not favor reinfection. Arch. Intern. Med. 162:1873–1879 [DOI] [PubMed] [Google Scholar]
- 13. Garcia de Viedma D., Marin M., Ruiz Serrano M. J., Alcala L., Bouza E. 2003. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J. Infect. Dis. 187:695–699 [DOI] [PubMed] [Google Scholar]
- 14. Kaplan G., et al. 2003. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect. Immun. 71:7099–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manca C., et al. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740–6746 [PubMed] [Google Scholar]
- 16. Martin A., Herranz M., Serrano M. J., Bouza E., Garcia de Viedma D. 2007. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palomino J. C., Leão S. C., Ritacco V. (ed.). 2007. Tuberculosis 2007. From basic science to patient care. www.TuberculosisTextbook.com
- 18. Post F. A., et al. 2004. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J. Infect. Dis. 190:99–106 [DOI] [PubMed] [Google Scholar]
- 19. Shamputa I. C., et al. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen G., et al. 2006. Recurrent tuberculosis and exogenous reinfection, Shanghai, China. Emerg. Infect. Dis. 12:1776–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sonnenberg P., et al. 2001. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 358:1687–1693 [DOI] [PubMed] [Google Scholar]
- 22. Supply P., et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theus S. A., et al. 2006. Differences in the growth of paired Ugandan isolates of Mycobacterium tuberculosis within human mononuclear phagocytes correlate with epidemiological evidence of strain virulence. Infect. Immun. 74:6865–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Theus S. A., Cave M. D., Eisenach K. D. 2004. Activated THP-1 cells: an attractive model for the assessment of intracellular growth rates of Mycobacterium tuberculosis isolates. Infect. Immun. 72:1169–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Theus S. A., Cave M. D., Eisenach K. D. 2005. Intracellular macrophage growth rates and cytokine profiles of Mycobacterium tuberculosis strains with different transmission dynamics. J. Infect. Dis. 191:453–460 [DOI] [PubMed] [Google Scholar]
- 26. van Rie A., et al. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174–1179 [DOI] [PubMed] [Google Scholar]
- 27. Vynnycky E., Fine P. E. 1997. The natural history of tuberculosis: the implications of age-dependent risks of disease and the role of reinfection. Epidemiol. Infect. 119:183–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warren R. M., et al. 2004. Patients with active tuberculosis often have different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169:610–614 [DOI] [PubMed] [Google Scholar]