Abstract
We evaluated the usefulness of PCR analysis of the 16S-23S rRNA gene internal transcribed spacer (ITS) and the CTX-M extended-spectrum β-lactamase (ESBL) followed by microchip gel electrophoresis (MGE) for direct identification and CTX-M detection of Gram-negative bacteria (GNB) from positive blood culture bottles. Of 251 GNB isolated from blood cultures containing a single bacterium, 225 (90%) were correctly identified at the species level directly from positive blood culture bottles by comparing the ITS-PCR patterns of the sample strain with those of the control strains. There were no cases of incorrect identification. Limitations encountered included the inability to detect mixed cultures (four bottles) as well as some species (Enterobacter species and Klebsiella oxytoca) demonstrating identical ITS-PCR patterns. A total of 109 ESBL-producing isolates from various clinical materials obtained between January 2005 and December 2008 were examined for blaCTX-M, blaSHV, and blaTEM genes by PCR and sequences of PCR products. CTX-M ESBL was detected in 105 isolates, and SHV ESBL was detected in two isolates. The remaining two isolates (K. oxytoca) were shown to harbor blaOXY. Twenty (19%) of 104 Escherichia coli isolates from blood cultures were suspected to produce ESBL by the combination disk method, and these isolates were shown to harbor CTX-M ESBL by PCR-MGE. The results were obtained within 1.5 h at a calculated cost of $6.50 per specimen. In conclusion, simultaneous detection of ITS length polymorphisms and blaCTX-M by single PCR followed by MGE is useful for rapid, cost-effective, and reliable species-level identification of CTX-M ESBL-producing GNB responsible for bloodstream infections.
INTRODUCTION
Bloodstream infections are a major cause of morbidity and mortality. Traditional identification of blood culture isolates requires subculture and overnight incubation to obtain isolated colonies. Rapid methods to differentiate bacteria directly from positive blood cultures have been developed (8, 9, 15), thus reducing the turnaround time compared to traditional testing. Rapid identification can significantly improve outcomes in infected patients by enabling rapid and appropriate antimicrobial therapy, leading to a decrease in mortality, shortened hospital stay, and lower hospitalization costs (3).
Several nucleic acid-based approaches have proven useful for identifying many microorganisms and are becoming more commonplace in the clinical microbiology laboratory (15, 21). Spacer regions separating the two prokaryotic rRNA genes are characterized by a high degree of sequence and length variation at both the genus and species levels (5, 9, 11, 14). The aim of the present study was to establish a rapid and accurate method for the identification of Gram-negative bacteria (GNB) from isolated colonies, as well as positive blood culture bottles with positive signals, by PCR amplification of the 16S-23S rRNA gene spacer region. As for rapid sizing of PCR products, Turenne et al. (25) used a capillary electrophoresis system which needs less than 30 min for analysis. Recently, electrophoretic analysis with a microchip was developed (6). To determine the length of PCR products more rapidly, we used a microchip gel electrophoresis (MGE) instrument. This instrument detects PCR products within 5 min, whereas agar gel electrophoresis used in our study needed 2 h to determine the length of PCR amplicons. The MGE instrument has enabled us to generate a profile of the expected amplicon sizes for a broad range of bacteria. The advantages of using such an instrument include speed and the elimination of slab gels and staining steps as well as of species-specific probes or DNA sequencing. Furthermore, minimal manual labor is required, thus making the fragment detection system accurate and cost-effective.
Earlier studies have shown that antimicrobial susceptibility testing does not significantly influence a physician's choice of appropriate antimicrobial therapy (19). However, with advances in technology, PCR-based detection of mecA and van directly from positive blood culture bottles is now available (8). On the other hand, extended-spectrum β-lactamase (ESBL)-producing strains, especially CTX-M ESBL-producing strains, are increasing worldwide (4, 10, 12, 16, 23). Although ESBLs are inactivated with β-lactamase inhibitors, ESBL-producing strains may be resistant to third- to fourth-generation cephalosporins such as cefotaxime, ceftazidime, and cefepime. Therefore, antibiotic options in the treatment of ESBL-producing organisms are limited (12, 24). In addition to rapid species identification of microorganisms, detection of the CTX-M gene in blood culture bottles was simultaneously examined by single PCR in the present study.
MATERIALS AND METHODS
Bacterial strains.
A total of 329 GNB (identified by a K prefix in Table 1) isolated from clinical samples between January 2005 and December 2007 at Kanazawa University Hospital and 39 reference strains were used to determine the internal transcribed spacer PCR patterns (ITS-PCR). The reference strains were obtained from the American Type Culture Collection (ATCC; Manassas, VA), NITE Biological Resource Center (NBRC) (Tokyo, Japan), Laboratory Culture Collection (IID) (University of Tokyo, Tokyo, Japan), and the Japan Collection of Microorganisms (JCM) (Wako, Japan). In addition, isolates of clinically relevant species not included in the reference strains were selected from the isolates from our laboratory and used as controls. When two or more ITS-PCR patterns were found among the same species, a strain isolated at Kanazawa University Hospital was selected randomly from each group of ITS-PCR patterns and was also used as a control.
Table 1.
Results of ITS-PCR
| Organism | Control strain | ITS-PCR type (no. of strains tested) | Size of amplified DNA fragment(s) (bp [mean ± SD])a |
|---|---|---|---|
| Achromobacter xylosoxidans | JCM 5490T | axy1 (2) | 436 ± 1, 683 ± 4 |
| JCM 9659T | axy2 (5) | 690 ± 4, 745 ± 6 | |
| Acinetobacter calcoaceticus-baumannii complexb | JCM 6841T | ||
| JCM 6842T | aba (12) | 685 ± 6 | |
| Acinetobacter junii | K6122 | aju (1) | 750 ± 4, 790 ± 6 |
| Acinetobacter lwoffii | JCM 6840T | alw (2) | 740 ± 7, 760 ± 7, 815 ± 10 |
| 850 ± 10, 910 ± 15 | |||
| Aeromonas hydrophila | JCM 1027T | ahy (9) | 562 ± 7, 618 ± 7, 626 ± 9, 642 ± 7, 663 ± 8 |
| Alcaligenes faecalis | JCM 1474T | afa (2) | 438 ± 3, 679 ± 5 |
| Bacteroides fragilis group | ATCC 25285T | bfr1 (18) | 241 ± 2, 583 ± 8, 645 ± 7 |
| K1845 | bfr2 (14) | 242 ± 3, 458 ± 3, 580 ± 4, 645 ± 8 | |
| K7144 | bfr3 (10) | 458 ± 3, 540 ± 3, 610 ± 3 | |
| Burkholderia cepacia | K9261 | bce (4) | 479 ± 8, 619 ± 5, 661 ± 5 |
| Buttiauxella agrestis | JCM 1090T | bag (1) | 486 ± 1, 653 ± 4 |
| Campylobacter jejuni | K9554 | cje (3) | 949 ± 8 |
| Cardiobacterium hominis | K5271 | cho (1) | 612 ± 4 |
| Cedecea davisae | JCM 1685T | cda (1) | 451 ± 1, 479 ± 2, 629 ± 2 |
| Chryseobacterium indologenes | NBRC 14944T | cin (2) | 743 ± 5, 1,765 ± 35 |
| Citrobacter freundii | K948 | cfr (4) | 324 ± 2, 483 ± 8, 629 ± 2 |
| Citrobacter koseri | K12195 | cko (10) | 443 ± 5, 475 ± 4, 498, 605 ± 3 |
| Cronobacter sakazakii | JCM 1233T | csa (1) | 450 ± 2, 517 ± 2 |
| Delftia acidovorans | JCM 5833T | dac (3) | 756 ± 5 |
| Edwardsiella tarda | JCM 1656T | eta (2) | 434 ± 5, 529 ± 6 |
| Elizabethkingia meningoseptica | NBRC 12535T | eme (1) | 688 ± 5 |
| Enterobacter aerogenes | K1235 | eae (8)c | 291 ± 6, 447 ± 5, 595 ± 4 |
| Enterobacter cloacae | ATCC 23355 | ecl1 (10)c | 294 ± 5, 440 ± 6, 591 ± 6 |
| K12309 | ecl2 (20) | 295 ± 4, 441 ± 6, 529 ± 6 | |
| K12880 | ecl3 (11) | 292 ± 4, 438 ± 7, 526 ± 7, 593 ± 8 | |
| Escherichia coli | ATCC 35218 | eco (22) | 422 ± 1, 444 ± 4, 455 ± 5, 478 ± 5, 529 ± 4, 540 ± 4, 553 ± 2 |
| Haemophilus influenzae | IID983 | hin (17) | 578 ± 5, 681 ± 2, 766 ± 3, 793 ± 5, 825 ± 9, 854 ± 8 |
| Klebsiella pneumoniae | ATCC 700603 | kpn1 (14) | 290 ± 4, 311 ± 3, 439 ± 5, 467 ± 5, 520 ± 5, 591 ± 7 |
| K14488 | kpn2 (2) | 315 ± 2, 477 ± 5, 623 ± 3 | |
| Klebsiella oxytoca | K7 | kox (15)c | 293 ± 3, 439 ± 4, 590 ± 5 |
| Morganella morganii | JCM 1672T | mmo1 (5) | 451 ± 2, 569 ± 6, 591 ± 7, 775 ± 10 |
| K4081 | mmo2 (4) | 562 ± 2, 586 ± 3, 744 ± 4, 778 ± 6 | |
| K5488 | mmo3 (3) | 589 ± 9, 760 ± 6, 779 ± 6 | |
| Neisseria meningitidis | K15850 | nme (3) | 732 ± 7 |
| Pantoea agglomerans | JCM 1236T | pag (2) | 470 ± 1, 487 ± 3, 674 ± 6 |
| Pasteurella multocida | K5105 | pmu (3) | 498 ± 3, 716 ± 4 |
| Proteus mirabilis | JCM 1669T | pmi1 (15) | 501 ± 6, 622 ± 5, 647 ± 7, 674 ± 8 |
| 830 ± 7 | |||
| K7446 | pmi2 (4) | 629 ± 7, 659 ± 5, 689 ± 4, 820 ± 9 | |
| Proteus vulgaris | JCM 1668T | pvu (6) | 608 ± 3, 633 ± 7, 657 ± 2, 784 ± 4 |
| 812 ± 5, 865 ± 3 | |||
| Providencia alcalifaciens | JCM 1673T | pal (1) | 514 ± 5, 672 ± 8 |
| Providencia rettgeri | JCM 1675T | pre1 (4) | 702 ± 8, 732 ± 10, 756 ± 4, 787 ± 3 |
| 819 ± 8, 890 ± 12 | |||
| K8723 | pre2 (3) | 670 ± 5, 761 ± 9 | |
| Pseudomonas aeruginosa | K10836 | pae (15) | 318 ± 3, 560 ± 4 |
| Pseudomonas alcaligenes | JCM 5967T | pal (2) | 576 ± 4 |
| Pseudomonas pseudoalcaligenes | JCM 5968T | pps (2) | 606 ± 3 |
| Pseudomonas fluorescens | JCM 5963T | pfl (6) | 421 ± 1, 608 ± 3 |
| Pseudomonas putida | JCM 6156 | ppu1 (7) | 374 ± 6, 383 ± 1, 558 ± 2, 595 ± 6 |
| JCM 6158 | ppu2 (1) | 401 ± 3, 589 ± 3 | |
| Pseudomonas stutzeri | ATCC 17588 | pst (1) | 591 ± 3 |
| Ralstonia pickettii | JCM 5969T | rpi (3) | 594 ± 7 |
| Salmonella entericad | ATCC 14028 | sen (9) | 449 ± 4, 512 ± 5, 624 ± 4 |
| Serratia marcescens | JCM 1239T | smar1 (12) | 501 ± 4, 568 ± 5 |
| ATCC 8100 | smar2 (2) | 504 ± 4, 547 ± 1, 568 ± 3 | |
| K7701 | smar3 (4) | 548 ± 3, 566 ± 3 | |
| K12077 | smar4 (3) | 546 ± 4, 564 ± 6, 615 ± 3 | |
| Sphingobacterium spiritivorum | JCM 1277T | ssp (1) | 474 ± 4 |
| Stenotrophomonas maltophilia | JCM 1976 | smal1 (16) | 606 ± 8, 627 ± 5, 647 ± 6 |
| JCM 1975T | smal2 (3) | 624 ± 5 | |
| Vibrio vulnificus | JCM 3725T | vvu (1) | 540 ± 3, 601 ± 3, 661 ± 4, 787 ± 9 |
Mean amplicon sizes were obtained from the results of three experiments for each control strain. The underlining indicates the sharp and intense fragments that were consistently observed.
Acinetobacter calcoaceticus-baumannii complex included two reference strains, Acinetobacter calcoaceticus JCM6841T and Acinetobacter baumannii JCM6842T.
ITS-PCR patterns of Enterobacter aerogenes strains and some Enterobacter cloacae strains were similar to those of Klebsiella oxytoca.
Nine strains of the genus Salmonella included six serotypes: S. enterica serotype Paratyphi A, S. enterica serotype Paratyphi B, S. enterica serotype Choleraesuis, S. enterica serotype Typhimurium, S. enterica serotype Typhi, and S. enterica serotype Enteritidis.
A total of 255 blood culture bottles (259 isolates) that gave positive results on testing for GNB from January 2007 to December 2009 were used for molecular identification of bacteria by ITS-PCR and for detection of CTX-M ESBL by PCR. Our laboratory uses the BacT/Alert 3D blood culture system (bioMérieux Inc., Durham, NC) with SA aerobic and SN anaerobic bottles. For each positive blood culture, 5% sheep blood agar, Drigalski agar, and brucella HK (hemin, vitamin K1) agar (only for isolation of anaerobes) were inoculated with a drop of broth for isolation. All strains were identified by colony morphology, Gram stain characteristics, and an oxidase test and by using commercially available kits, such as API 20E (bioMérieux) for Enterobacteriaceae, ID test NF-18 (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) for glucose nonfermenting GNB, and ID test HN-20 (Nissui) for Haemophilus and Neisseria species. The Bacteroides fragilis group was identified presumptively by growth on both Bacteroides agar plate (Nissui) and Bacteroides bile esculin agar plate (Kyokuto Pharmaceutical Ind., Co., Ltd., Tokyo), and Fusobacterium species was identified by growth on a modified FM agar plate (Nissui).
DNA preparation.
A colony grown on modified Drigalski agar was suspended in 0.2 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) at a density equivalent to a 0.5 McFarland standard. Aliquots of 2 μl of proteinase K solution (20 mg/ml; Takara Shuzo Co., Ltd., Ohtsu, Japan) were added, followed by incubation at 60°C for 5 min. After 7 min of incubation in a boiling water bath, the lysate was centrifuged at 9,000 × g for 2 min, and the supernatant was used as the template DNA. Processing of positive blood culture bottles for PCR was performed as described previously (9) with slight modifications. Briefly, ∼10 ml of 0.1% sodium dodecyl sulfate was added to 0.1 to 0.2 ml of blood culture fluids, and the mixtures were centrifuged at 4,000 × g for 10 min. The pellets were suspended in 1.5 ml of distilled water and centrifuged again at 9,000 × g for 1 min. Then, 0.1 ml of TE buffer and 2 μl of proteinase K solution were added to the pellet, and the mixture was incubated at 60°C for 5 min. After 7 min of incubation in a boiling water bath, the lysate was centrifuged at 9,000 × g for 2 min, and the supernatant was used as the template DNA. DNA was extracted from isolated colonies in ∼20 min, and DNA from GNB-positive blood culture bottles was obtained in ∼30 min.
Identification of GNB by ITS-PCR.
ITS-PCR was performed using the bacterium-specific universal primers IX (5′-GGTGAAGTCGTAACAAG-3′) and II (5′-TGCCAAGGCATCCACC-3′) (9). PCR was performed with 1.5 μl of the template DNA in a total reaction volume of 30 μl consisting of 0.75 U of Z-Taq DNA polymerase (Takara Bio Inc., Shiga, Japan), 0.1 μM each primer, 0.2 mM each deoxynucleoside triphosphate, and 3 mM MgCl2. Reactions were run in a thermocycler (Biometra Co., Göttingen, Germany). The PCR program consisted of 95°C for 10 s (first cycle only) and 25 cycles of 99°C for 8 s, 52°C for 12 s, and 72°C for 20 s, followed by a final extension step at 72°C for 5 min. The ESBL genes were also amplified by PCR. The PCR products were separated with an MGE instrument (model SV1200; Hitachi Electronics Engineering Co., Ltd., Tokyo, Japan) as described previously (9). Briefly, a mixture of 9 μl of PCR product and 1 μl of loading buffer containing the internal standards was loaded into one of the sample wells of the microchip, and the mixture was run for 5 min. The mixture of ITS-PCR and CTX-M PCR products was applied to an MGE gel, and the electrophoresis patterns were analyzed. The database for ITS-PCR patterns of GNB was constructed by using the commercially available database software FileMaker Pro (FileMaker Inc. Santa Clara, CA). When PCR products of one to five different lengths, beginning with the shortest one, were input into the database, the names of the organisms which demonstrated ITS-PCR patterns similar to the pattern of the test strain were shown automatically. Then, the ITS-PCR pattern of the test strain was compared visually with the patterns of the control strains shown on the computer screen. Identifications were assigned to those isolates with ITS-PCR patterns that matched any of the control strains. The PCR procedure required ∼40 min, and MGE took ∼5 min. Therefore, the overall turnaround time of the PCR-MGE was ∼1.5 h with positive blood culture bottles.
Detection of ESBLs.
A total of 116 of 2,216 Enterobacteriaceae strains isolated from various clinical samples between January 2005 and December 2008 were shown to be nonsusceptible (intermediate or resistant) to cefotaxime, cefoperazone, or ceftazidime by the disk diffusion method. These antibiotic-nonsusceptible isolates were further subjected to analysis for ESBLs. Phenotypic testing for ESBL production was performed using combination disks according to the recommended method of the Clinical and Laboratory Standards Institute (CLSI) (7). The disks used were cefotaxime, cefpodoxime, and ceftazidime with and without clavulanate (Eiken Chemical Co., Ltd., Tokyo). Klebsiella pneumoniae ATCC 70063 was used as a control SHV ESBL-positive strain, and Escherichia coli ATCC 35218 was used as a blaTEM-1-positive strain. Fifty-three ESBL-nonproducing Enterobacteriaceae strains isolated in our laboratory were also used as negative controls. PCR was performed under the same conditions as described for ITS-PCR. The primers of CTX-F (5′-ATTCCRGGCGAYCCGCGTGATACC-3′) and CTX-R (5′-ACCGCGATATCGTTGGTGGTGCCAT-3′) used to detect CTX-M ESBLs were developed in our laboratory. The isolates were also examined for SHV and TEM ESBLs by PCR amplification and sequencing with primer pairs described previously (2). The CTX-M ESBL type was determined by sequence analysis using the primer pairs (1). The sequences were determined by direct sequencing of PCR products, and sequencing was performed on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems Japan Ltd., Tokyo).
RESULTS
ITS-PCR patterns of GNB.
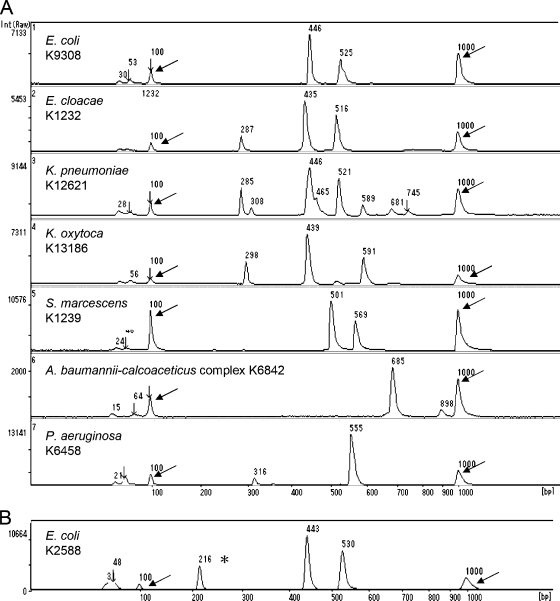
MGE analysis of the amplified products obtained from 368 isolates comprising 45 GNB species consistently showed one to four intense and sharp bands (major fragments) for each sample, ranging from 241 to 1,765 bp (Table 1). A single fragment of the same size (685 ± 6 bp) was obtained from Acinetobacter calcoaceticus JCM6842T and Acinetobacter baumannii JCM6841T. Two ITS-PCR patterns were observed among Achromobacter xylosoxidans, K. pneumoniae, Proteus mirabilis, Providencia rettgeri, Pseudomonas putida, and Stenotrophomonas maltophilia; three patterns were found among the B. fragilis group, Enterobacter cloacae, and Morganella morganii; and four patterns were found among Serratia marcescens. Nine Salmonella isolates belonging to six serotypes showed common strong bands at 449 and 624 bp and one weak band at 512 bp. There were no significant differences among Salmonella serotype isolates. Although most of the strains studied had unique ITS-PCR patterns at the species level, identical ITS-PCR patterns were found among Enterobacter aerogenes isolates, some E. cloacae strains, and Klebsiella oxytoca isolates. In addition, Pseudomonas stutzeri and Ralstonia pickettii yielded a single fragment of 591 ± 3 and 594 ± 7 bp, respectively. For A. xylosoxidans, two ITS-PCR patterns were observed: type axy1 from A. xylosoxidans subspecies denitrificans JCM 5490T and type axy2 from A. xylosoxidans subspecies xylosoxidans JCM 9659T. Figure 1A shows the seven representative ITS-PCR patterns of clinically relevant GNB.
Fig. 1.
Results of MGE of ITS-PCR products of clinically relevant Gram-negative organisms in positive blood cultures (A) and the simultaneous detection of PCR products of the CTX-M ESBL gene and ITS regions obtained from E. coli K2588 (B). The asterisk indicates the PCR product (216 bp) of the CTX-M ESBL gene, and arrows indicate DNA size markers of 100 bp (left) and 1,000 bp (right). Int, intensity of fluorescence.
ESBL-producing isolates.
Of 116 isolates that were intermediate or resistant to the third-generation cephalosporins, 109 were suspected as ESBL producers by the combination disk method. A total of 107 isolates (97 E. coli, 5 K. pneumoniae, 2 K. oxytoca, and 3 Proteus mirabilis) were positive for CTX-M genes; the remaining 2 isolates were positive for SHV-12 ESBL (1 isolate each of E. coli and K. oxytoca). Of 107 CTX-M-positive isolates determined by PCR, 89 (83%) isolates appeared to carry the CTX-M-9 gene, 7 carried the CTX-M-15 gene, 3 each carried the CTX-M-2 and CTX-M-14 genes, 2 carried the CTX-M-3 gene, and 1 carried the CTX-M-27 gene. Two CTX-M-positive K. oxytoca isolates were shown to harbor the blaOXY gene by sequencing analysis of CTX-M gene amplification products. No isolates harboring TEM ESBL genes were found among those tested, and there were no cases of false-positive results. Overall, 98 (92%) of 107 ESBL-producing isolates were E. coli.
Identification of GNB from positive blood culture bottles.
A total of 259 bacteria were isolated from 255 GNB-positive blood cultures; 251 bottles (98%) contained a single bacterial species, while 4 (2%) contained polymicrobial growth. ITS-PCR analyses accurately identified 225/259 (87%) organisms in positive blood culture bottles to the species level, while 22 isolates that showed the ITS-PCR pattern of K. oxytoca were identified as belonging to the Enterobacter/K. oxytoca group (Table 2). Four isolates (three Bacteroides fragilis group and one Acinetobacter lwoffii isolate) were not identified because they showed novel ITS-PCR patterns that were not included in our database. Furthermore, eight organisms in mixed culture bottles were not identified because of complex ITS-PCR patterns. Overall, 228 (91%) of 251 GNB isolated from bottles that contained a single bacterial species were correctly identified at the species level directly from blood culture bottles. There were no cases of incorrect identification. CTX-M ESBL genes were detected from 20 bottles by PCR, and all ESBL-positive strains were identified as E. coli by ITS-PCR patterns and the culture-based method. Figure 1B shows the amplification product of the CTX-M ESBL gene and ITS-PCR pattern of CTX-M ESBL-producing E. coli.
Table 2.
Results of ITS-PCR analysis and ESBL PCR with positive blood culture bottles for Gram-negative bacteria
| Organism (total no. of bottles)a | No. (%) of strains identified correctly at species level by ITS-PCR pattern | No. (%) of the CTX-M ESBL-producing isolates |
|---|---|---|
| E. coli (104) | 104 (100) | 20 (19) |
| P. aeruginosa (30) | 30 (100) | 0 |
| K. pneumoniae (29) | 29 (100) | 0 |
| S. maltophilia (18) | 18 (100) | 0 |
| E. cloacae (12) | 6 (50)b | 0 |
| K. oxytoca (10) | 0 (0)b | 0 |
| S. marcescens (8) | 8 (100) | 0 |
| A. calcoaceticus-baumannii complex (6) | 6 (100) | 0 |
| B. fragilis (6) | 3 (50) | 0 |
| E. aerogenes (6) | 0 (0)b | 0 |
| A. xylosoxidans (4) | 4 (100) | 0 |
| C. koseri (4) | 4 (100) | 0 |
| C. freundii (3) | 3 (100) | 0 |
| P. mirabilis (3) | 3 (100) | 0 |
| H. influenzae (2) | 2 (100) | 0 |
| A. lwoffii (2) | 1 (50) | 0 |
| B. cepacia (1) | 1 (100) | 0 |
| E. tarda (1) | 1 (100) | 0 |
| N. meningitidis (1) | 1 (100) | 0 |
| P. putida (1) | 1 (100) | 0 |
| Polymicrobial (4) | 0 (0) | 0 |
| Total (255) | 225 (88%) | 20 (19) |
When two or more samples obtained from the same patient were positive for the same organism, only one sample was included. C. koseri, Citrobacter koseri; C. freundii, Citrobacter freundii; H. influenzae, Haemophilus influenzae, B. cepacia, Burkholderia cepacia; E. tarda, Edwardsiella tarda; N. meningitidis, Neisseria meningitidis.
Six E. cloacae isolates and six E. aerogenes isolates showed ITS-PCR patterns similar to those of K. oxytoca. Therefore, the isolates having the ITS-PCR pattern of K. oxytoca were identified as belonging to the Enterobacter/K. oxytoca group.
Cost analysis was performed for reagents for conventional testing and molecular testing. The average cost for conventional identification of GNB ranged from $10 to $12. With molecular testing, testing cost is significantly lower when more than one test is processed at the same time. The cost of identifying GNB and detecting CTX-M ESBL from one sample alone would represent the maximum cost per sample, which was calculated to be $6.50. We have calculated the cost of running two samples for identification to be $4.80 per sample.
DISCUSSION
The PCR assay to detect polymorphisms in the 16S-23S rRNA gene spacer region is potentially useful for both species-level identification of coagulase-negative Staphylococcus species and GNB (9, 14) and strain discrimination of Staphylococcus aureus (9) and Clostridium difficile (5). One issue that is critical for the successful detection of spacer variation is the choice of PCR primers. Primers were selected to target highly conserved regions in the 16S and 23S rRNA genes. To date, four primers in the 16S rRNA gene and six primers in the 23S rRNA gene have been proposed, and some primers failed to detect spacer variation at the species or strain level (11). Jensen et al. (14) first demonstrated that PCR-amplified rRNA gene spacer polymorphisms using primers G1 (positions 1491 to 1504 based on the sequence of the E. coli 16S rRNA gene) and L1 (positions 21 to 35 based on the sequence of the E. coli 23S rRNA gene) were useful for identification of GNB. We used the similar primers IX and II targeting positions 1487 to 1503 of the E. coli 16S rRNA gene and positions 23 to 38 of the E. coli 23S rRNA gene, respectively. Based on the results obtained with the 368 strains tested here, length polymorphisms of the ITS region showed significant promise for identification of clinically relevant GNB.
It is not surprising that some strains belonging to different genera or species showed similar ITS-PCR patterns. In this study, Enterobacter species and K. oxytoca showed the same patterns. As these two species have been reported to show similar antibiotic susceptibility patterns (more than 10% of strains are resistant to piperacillin, third-generation cephalosporins, and tobramycin) (26), rapid species-level identification of these species may not be required for planning antibiotic treatment. When isolates tested yielded single bands of similar lengths, ITS-PCR patterns alone could not be used for general differentiation of glucose-nonfermenting GNB. For example, both P. stutzeri and R. pickettii (591 to 594 bp) and Pseudomonas alcaligenes and Pseudomonas pseudoalcaligenes (576 to 606 bp) showed bands of similar sizes. Although these four species strains are rarely isolated from blood cultures, as shown here and in a previous study (15), the physician would be able to implement therapy for either Pseudomonas (excluding P. aeruginosa) or Enterobacteriaceae infection much sooner with the ITS-PCR result than with the results of Gram staining, which would indicate only the presence of Gram-negative bacteria. To differentiate strains based on ITS-PCR products which revealed similar single bands, conventional phenotypic methods should be performed in parallel. On the other hand, four isolates were not identified as these strains showed novel ITS-PCR patterns. Therefore, it is necessary to continue adding new ITS-PCR patterns of isolates from clinical materials to the database, resulting in increased sensitivity.
Because our PCR-based method with universal primers amplified all kinds of bacteria, the major limitation of our methods for the direct detection of bacteria from clinical samples is mixed flora. Polymicrobial bacteremia due to GNB has been reported in 2 of 405 (1.0%) blood culture bottles (17). In our study, 4 of 255 bottles (1.6%) had two different members of the Enterobacteriaceae identified in each sample by conventional methods. When electrophoresis of a culture shows an unusual pattern with two or more fragments, the presence of mixed flora should be taken into consideration, and the identification of bacteria may be difficult. Overall, these observations indicated that ITS-PCR followed by MGE is useful for rapid, cost-effective, and reliable species-level identification of GNB responsible for bloodstream infections. The approach described here seems to be useful also for the recognition of potentially novel infection-associated pathogens.
In recent years, CTX-M ESBL has been recognized as a growing family possessing a high level of hydrolyzing activities, especially against cefotaxime. The present study showed that 105 (98%) of 107 ESBL producers isolated from various clinical samples were CTX-M-type ESBLs. The same results were reported by Suzuki et al. (23). They reported that 130 (92%) of 142 E. coli isolates suspected of producing ESBL were confirmed to be harboring blaCTX-M by PCR and sequencing analysis. There are more than 80 different variants of CTX-M enzymes, which can be further classified into five different subgroups based on their amino acid sequences: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 (4). Of these, the CTX-M-9, CTX-M1, and CTX-M-2 groups have been reported as the most common groups in Japan (23). In this study, the most common CTX-M was the CTX-M-9 group (88%), followed by the CTX-M-1 and CTX-M-2 groups. The predominance of CTX-M-9 in our hospital is not a consistent finding worldwide as different CTX-M-type enzymes have emerged as the predominant ESBLs in other locations (4). K. oxytoca may give a false-positive result in phenotypic tests for ESBL production due to hyperproduction of the chromosomally encoded K1 β-lactamase (20). Monstein et al. (18) have shown that the universal blaCTX-M PCR amplification primers also target the almost identical K1 DNA sequence. The same results were obtained in the present study. Therefore, reports regarding CTX-M ESBL production from K. oxytoca by PCR should be excluded until sequencing results have been determined.
As the CTX-M gene amplicon size was very small (216 bp) and PCR products obtained from ITS-PCR ranged from 241 bp to 1,765 bp, PCR-MGE enabled simultaneous detection of the CTX-M gene and ITS-PCR patterns, which was both cost-effective and saved time. Poor outcomes occur when patients with serious infections (bacteremia, hospital-acquired pneumonia, or peritonitis) due to ESBL producers are treated with cephalosporins to which the responsible organism is resistant (13, 22). Providing PCR results regarding bacterial identification and CTX-M ESBL at the initial physician contact could result in prudent therapy. The methods described here may also be useful for detecting novel CTX-M-producing species because CTX-M-producing P. aeruginosa and S. maltophilia have been reported previously (1). There are epidemiological implications for the detection of ESBL producers in that this resistance may not be as apparent if the organisms are simply reported as resistant to individual cephalosporins.
In conclusion, detection of ITS length polymorphisms by PCR-MGE is useful for rapid, reliable, and relatively cost-effective species-level identification of GNB responsible for bloodstream infections. In addition, PCR-MGE seems to be a useful tool for simultaneous detection of CTX-M ESBL and ITS-PCR patterns of GNB. The main limitations of the ITS-PCR identification method are its reliance on the database and interlaboratory variations. We are preparing a database with a high level of integrity that can be applied comprehensively to pathogenic bacteria, and our database may be shared to allow other laboratories to identify their strains via a simple Web-based tool.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. al Naiemi N., Duim B., Bart A. 2006. A CTX-M extended-spectrum β-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J. Med. Microbiol. 55:1607–1608 [DOI] [PubMed] [Google Scholar]
- 2. al Naiemi N., et al. 2005. Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J. Clin. Microbiol. 43:4862–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barenfanger J., Drake C., Kacich G. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Canton R., Coque T. M. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475 [DOI] [PubMed] [Google Scholar]
- 5. Cartwright C. P., Stock F., Beekmann S. E., Williams E. C., Gill V. J. 1995. PCR amplification of rRNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J. Clin. Microbiol. 33:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng J., et al. 1998. Degenerate oligonucleotide primed-polymerase chain reaction and capillary electrophoretic analysis of human DNA on microchip-based devices. Anal. Biochem. 257:101–106 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2005. Performance standards for antimicrobial susceptibility testing; 15th information supplement. CLSI M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Eigner U., Weizenegger M., Fahr A.-M., Witte W. 2005. Evaluation of a rapid direct assay for identification of bacteria and the mecA and van genes from positive blood cultures. J. Clin. Microbiol. 43:5256–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujita S., Senda Y., Iwagami T., Hashimoto T. 2005. Rapid identification of staphylococcal strains from positive-testing blood culture bottles by internal transcribed spacer PCR followed by microchip gel electrophoresis. J. Clin. Microbiol. 43:1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goossens H., Grabein B. 2005. Prevalence and antimicrobial susceptibility data for extended-spectrum β-lactamase- and AmpC-producing Enterobacteraceae from the MYSTIC program in Europe and the United States (1997–2004). Diagn. Microbiol. Infect. Dis. 53:257–264 [DOI] [PubMed] [Google Scholar]
- 11. Gürtler V., Stanisich V. A. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3–16 [DOI] [PubMed] [Google Scholar]
- 12. Hawser S. P., et al. 2009. Emergence of high levels of extended-spectrum-β-lactamase-producing Gram-negative bacilli in the Asia-Pacific region: data from the study for monitoring antimicrobial resistance trends (SMART) program, 2007. Antimicrob. Agents Chemother. 53:3280–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho P. L., Chan W. M., Tsang K. W., Wong S. S., Young K. 2002. Bacteremia caused by Escherichia coli producing extended-spectrum beta-lactamase: a case-control study of risk factors and outcomes. Scand. J. Infect. Dis. 34:567–573 [DOI] [PubMed] [Google Scholar]
- 14. Jensen M. A., Webster J. A., Straus N. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jordan, Jones-Laughner J. A. J., Durso M. B. 2009. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. J. Clin. Microbiol. 47:368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lewis I. I., Herrera J. S. M., Wickes B., Patterson J. E., Jorgensen J. H. 2007. First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marlowe E. M., et al. 2003. Application of an rRNA probe matrix for rapid identification of bacteria and fungi from routine blood cultures. J. Clin. Microbiol. 41:5127–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monstein H.-J., et al. 2007. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115:1400–1408 [DOI] [PubMed] [Google Scholar]
- 19. Munson E. L., Diekema D. J., Beekmann S. E., Chapin K. C., Doern G. V. 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paterson D. L., Bonomo R. A. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peters R. P. H., et al. 2006. Faster identification of pathogens in positive blood cultures by fluorescence in situ hybridization in routine practice. J. Clin. Microbiol. 44:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schiappa D. A., et al. 1996. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli bloodstream infection: a case-control and molecular epidemiologic investigation. J. Infect. Dis. 174:529–536 [DOI] [PubMed] [Google Scholar]
- 23. Suzuki S., et al. 2009. Changes in the prevalence of extended-spectrum β-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72–79 [DOI] [PubMed] [Google Scholar]
- 24. Tumbarello M., et al. 2007. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob. Agents Chemother. 51:1987–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turenne C. Y., Sanche S. E., Hoban D. J., Karlowsky J. A., Kabani A. M. 1999. Rapid identification of fungi by using the internal transcribed spacer 2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wisplinghoff H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]