Abstract
The rapidSTRIPE H1N1 test, based on a nucleic acid lateral-flow assay, has been developed for diagnosis of a swine-origin influenza A (H1N1) virus. This test is simple and cost-effective and allows specific detection of the S-OIV A (H1N1) virus from swab sampling to final detection on a lateral-flow stripe within 2 to 3 h.
In April 2009, a novel swine-origin influenza A (H1N1) virus (S-OIV A) was detected in specimens from several patients in the United States and Mexico. This virus spreads from person to person, probably in the same way that seasonal influenza viruses spread (16). On 11 June 2009, the World Health Organization declared an influenza pandemic caused by novel S-OIV A (H1N1) and raised the pandemic alert level to phase 6. Through rapid and frequent international travel, this virus spread worldwide, with more than 214 countries and overseas territories or communities reporting laboratory-confirmed cases of pandemic influenza A H1N1 virus, including at least 18,449 deaths by 6 August 2010 (17).
Rapid diagnosis of influenza is important for introduction of antiviral therapy and quarantine measures, since antiviral therapy should preferably be initiated within 24 h after appearance of the patient's first clinical symptoms (12). This article describes a nucleic acid lateral-flow (NALF) assay, called the rapidSTRIPE assay, used as a molecular-genetic rapid test for the diagnosis of the pandemic S-OIV A (H1N1) virus. This assay is based on rapid amplification/hybridization (RAH) technology (Analytik Jena AG, Jena, Germany). The aim of this study was to evaluate the rapidSTRIPE assay based on a rapid amplification/hybridization reaction coupled with instrument-independent detection of the amplification products by a user-friendly lateral-flow strip (LFS). Furthermore, the diagnostic sensitivity and specificity for the rapidSTRIPE assay were determined and compared to those of the real-time PCR method (11), which is widely considered a gold standard (4).
Two different standard preparations of H1N1 influenza viruses (A/California/04/2009 and A/Hamburg/04/2009) were provided by the European Network for Diagnostics of Imported Viral Diseases Collaborative Laboratory Response Network (ENVID-CLRN). Different representative influenza A and B subtype virus strains for specificity testing were provided by the National Reference Center for Influenza, Robert Koch Institute (RKI), Berlin, Germany. Viral RNA samples extracted from nasal swabs from patients during the 2009 H1N1 pandemic were kindly provided by the Medizinisches Labor Ostsachsen MVZ GbR, Dresden, Germany (MLO MVZ GbR). A total of 174 clinical specimens were tested by the rapidSTRIPE H1N1 assay and by reference quantitative real-time PCR.
The specimens were collected in different patient centers in Saxony, Germany, as pharyngeal or nasal swab samples, placed in 200 μl of virus transport medium, and stored at 4°C. The rapidSTRIPE H1N1 assay KF system consists of three modules: module 1 for nucleic acid extraction, module 2 for cDNA synthesis and RAH reaction, and module 3 for detection on LFS. Total RNA from swab collection and reference virus material was extracted by module 1 of the system using the Innuprep RNA virus KFFLX kit (Analytik Jena AG, Jena, Germany) and the Kingfisher FLX system (Thermo Scientific, Finland) according to the manufacturer's instructions. In addition, RNA samples from clinical specimens included in this study were tested with in-house glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reverse transcription-PCR (RT-PCR) to check the quality of the extracted RNA samples using the AffinityScript One-Step RT-PCR kit (Agilent Technologies, Santa Clara, CA) according to the manufacture's instructions. Each RNA was subjected to PCR with the primers GAPDHF (5′-CCATGGAGAAGGCTGGGGCT-3′) and GAPDHR (5′-GGTGGTGCAGGAGGCATTGCT-3′). Subsequently, the amplification products were analyzed by using a 2% ethidium bromide-stained agarose gel and were visualized under UV light.
cDNA synthesis was performed by module 2 of the system with 10 μl of viral RNA in a 15-μl final reaction volume according to the manufacturer's instructions. These samples of cDNA were used for real-time PCR and the rapidSTRIPE H1N1 assay. Additionally, cDNA synthesis was performed using 1 μM random hexamer primer for further specificity tests and stored at −80°C until further use.
Hemagglutinin (HA) gene sequences of S-OIV A (H1N1) virus were aligned by using the ClustalW2 software program (http://www.ebi.ac.uk/tools/clustalw2/index.html) to design the primers and probe for the LFS assay. The RAH reaction was carried out on a cycler or the Alpha SC cycler by using module 2 of the system according to the manufacturer's instructions. In brief, 3 μl of the cDNA was subjected to PCR in a 25-μl-final-volume reaction mixture containing 150 nm of primer HN1 (5′-TGGGAAATCCAGAGTGTGAATCACTCTC-3′), 300 nm of primer HN2 (5′-Biotin-CGTTCCATTGTCTGAACTAGRTGTTTCC-3′), and 300 nm of probe HN (5′-fluorescein isothiocyanate (FITC)-AGCAAGCTCATGGTCCTACATT-3′).
Final detection was carried out using module 3 of the system according to the kit instructions. Briefly, 15 μl of amplification/hybridization product was added to a sample pad on the lateral-flow strip and placed in the tube containing 150 μl of running buffer at room temperature. The result was read visually after 10 min of incubation. A test was considered positive when the detection line and the control line were visible. A test was considered negative when only the control line was visible.
As a method of comparison, a real-time PCR targeting the HA gene, developed at the Robert Koch Institute, was chosen (11). It was performed with 2 μl cDNA in 25-μl reaction volume. Thermal cycling was done on a Stratagene Mx3000 cycler instrument (Agilent Technologies, Inc., Santa Clara, CA) under the following conditions: 15 min at 95°C, and 45 cycles of 15 s at 95°C and 30 s at 58°C. To quantify the real-time PCR, a 10-fold serial dilution of the standard plasmid (10 to 106 copies/μl) was tested in duplicate within the same sample run and compared.
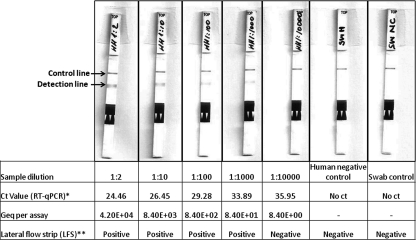
In order to determine the sensitivity of the rapidSTRIPE H1N1 assay KF system, serially diluted concentrations of the viral strain A/Hamburg/04/2009 were applied in the LFS assay and in reference quantitative real-time PCR after cDNA synthesis. As few as 8 genome equivalent (geq) copies/assay of S-OIV A (H1N1) were detected per assay by the reference method, real-time PCR, where as little as 84 geq copies/assay of S-OIV A (H1N1) was detected clearly per LFS assay (Fig. 1). In this case, the LFS assay showed 10-fold-lower sensitivity than the reference method, real-time PCR. Real-time PCR was performed in duplicate, while the LFS assay was performed with a single sample (Fig. 1). The same experiments were repeated 3 times independently for the rapidSTRIPE H1N1 assay and compared for reproducibility. All three times, as little as 84 geq copies/assay of S-OIV A (H1N1) was detected as positive on LFS by the rapidSTRIPE H1N1 assay (data not presented here).
Fig. 1.
Sensitivity of LFS assay. Serial dilutions of the viral strain A/Hamburg/04/2009 were used to evaluate the sensitivity of the LFA assay and compare it with the reference method, RT-quantitative PCR (qPCR). Test strip control lines were visible for all samples. Detection lines were visible below the test line for the first four dilutions of viral samples.
The specificity of the LFS assay was assessed by testing 10 different strains of subtypes of influenza virus A and influenza virus B and human negative-control swab material (Table 1). All tested influenza viruses but S-OIV A (H1N1) yielded negative results, demonstrating the high specificity of the LFS assay. The reference real-time PCR was also negative for all reference influenza A and influenza B virus strains except two strains of influenza A virus (Table 1). Both of the influenza A viruses were detected at the threshold of detection, with threshold cycle (CT) values of 38.01 and 38.8, respectively. To control the quality of the cDNA of reference influenza viruses, in-house real-time PCR assays for influenza A and B viruses (flu A+B) (Analytik Jena AG, Jena, Germany) were performed with all samples. All of the influenza viruses were positive in the real-time PCR flu A+B assay. Swabs from the human negative control and swab control were negative in all three assays.
Table 1.
Specificity test with two reference real-time PCR assays and the rapidSTRIPE test with representative influenza virus subtypes
| Influenza virus |
CT valuea |
LFS assay resultb | |
|---|---|---|---|
| RKI real-time PCR assay | In-house flu A+B assay | ||
| A/Hamburg/04/2009 H1N1 | 24.46 | 23.69 | Positive |
| A/Brisbane/59/07 H1N1 | 38.02 | 29.3 | Negative |
| A/Caledonia/20/99 H1N1 | No CT | 23.5 | Negative |
| A/Brisbane/10/07 H3N2 | No CT | 28 | Negative |
| A/Wellington/1/04 H3N2 | No CT | 25.5 | Negative |
| A/dk/Germany R603/06 H5N1 | 38.8 | 24.4 | Negative |
| A/dk/Vietnam TG24-01/05/H5N1 | No CT | 21.3 | Negative |
| A/Italy/472/99/H7N1 | No CT | 20.3 | Negative |
| A/Germany/R11/01/H7N1 | No CT | 31.5 | Negative |
| B/Malasiya/2506/04 (Victoria lineage) | No CT | 21.2 | Negative |
| B/Iangsu/10/03 (Yamagata lineage) | No CT | 24.5 | Negative |
| Human virus-negative swab | No CT | No CT | Negative |
| Swab negative control | No CT | No CT | Negative |
| Influenza A+B negative control | No CT | Negative | |
Samples were analyzed in duplicate in the real-time PCR assay and flu A+B assay. CT, threshold cycle.
Samples were analyzed once in the LFS assay.
One hundred seventy-four viral RNA samples obtained from patient nasal swabs included in this study were positive by in-house GAPDH RT-PCR. The same RNA samples were also tested with the rapidSTRIPE H1N1 assay and real-time PCR. The CT values obtained by real-time PCR for positive samples ranged from 22.58 to 38.9 (data not shown here). One hundred five samples out of 174 samples (60.3%) were positive and 69 (39.7%) were negative by real-time PCR. Of the 105 samples that were detected as positive by real-time PCR, 92 tested positive by the rapidSTRIPE H1N1 assay, providing a sensitivity of 88% (95% confidence interval [CI], 80% to 92.6%) and a positive predictive value of 96%. Of the 69 samples that tested negative in real-time PCR, 65 tested negative by the rapidSTRIPE H1N1 assay, providing a specificity of 94% (95% CI, 86% to 97.7%) and a negative predicative value of 84%. The overall agreement between the two assays was 90.2% (157/174).
Our finding demonstrates the usefulness of the rapidSTRIPE H1N1 assay for the rapid detection of novel S-OIV H1N1 as an alternative to real-time PCR in a resource-poor laboratory setting. This assay showed no cross-reactivity either with other influenza A and B viruses or with human negative-control material, providing a good specificity profile, required for diagnostic accuracy. As little as 84 geq copies/assay could be detected by the rapidSTRIPE H1N1 assay, corresponding to 8,400 viral RNA copies in 100 μl RNA or 8,400 virus particles in the initial sample (experimental swab sample). The rapidSTRIPE test showed an overall sensitivity of 88% and specificity of 94% in comparison to real-time PCR, the widely preferred method for diagnosis of S-OIV A (H1N1) (4). The total cost of the rapidSTRIPE H1N1 test, including manual nucleic acid extraction, is about 10 euros per sample.
Several PCR-based assays for the detection of the S-OIV A (H1N1) were developed and published soon after the emergence of the pandemic 2009 H1N1 virus (1, 9, 10, 14, 15). Also, new rapid assays, such as real-time nucleic acid sequence-based amplification and multifluorescent real-time RT-PCR, have been developed to detect novel S-OIV A (H1N1) (2, 6). All of these molecular detection methods in the form of real-time RT-PCR have been broadly used in medical diagnostic laboratories because of their high sensitivities and specificities. Although real-time RT-PCR is widely regarded as a gold standard for diagnosis of influenza viruses (4), it is relatively expensive and requires trained laboratory expertise and extensive evaluation, which limits the broad use of in-house assays (13). Rapid antigen-antibody-based influenza tests as point-of-care tests have been used since they require only 10 to 15 min and minimal expertise for testing. They also provide a source of data for clinical management of the patients. However, a high virus concentration is required to yield a positive rapid test (3). Depending on the virus load of the respiratory sample, an overall sensitivity of 40% to 69% has been reported for rapid antigen-antibody-based influenza tests among different commercial tests (5, 7, 8).
In summary, the rapidSTRIPE H1N1 assay offers a powerful tool for specific detection of S-OIV A (H1N1) in about 2 to 3 h, from swab sampling, nucleic acid isolation, cDNA synthesis, and rapid amplification/hybridization to final detection of the PCR products on an LFS. Furthermore, the rapidSTRIPE H1N1 assay KF system provides all reagents needed for molecular diagnostics, from nucleic acid isolation to final detection on LFS, in one single system. This rapid assay allows qualitative detection of S-OIV A (H1N1) with several advantages, such as quickness, cost-effectiveness, and long-term stability. Readout of the test is performed optically, which makes it independent from an instrument-specific analysis system. This system can easily be used as a high-throughput screening system for laboratories not equipped with real-time PCR instruments and in resource-poor diagnostic settings during an epidemic.
Acknowledgments
We thank Barbara Biere and Martin Schulz (Fachgebiet 17 Influenza/Respiratorische Viren, RKI, Berlin, Germany) for providing us with different influenza virus samples for the cross-reactivity test and plasmid standard. We also thank Sonja Linke, Oliver Dononso Mantke, and Cristina Domingo-Carrasco for their critical review of the manuscript.
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1. Carr M. J., et al. 2009. Development of a real-time RT-PCR for the detection of swine-lineage influenza A (H1N1) virus infections. J. Clin. Virol. 45:196–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong H., et al. 2010. Detection of human novel influenza A (H1N1) viruses using multi-fluorescent real-time RT-PCR. Virus Res. 147:85–90 [DOI] [PubMed] [Google Scholar]
- 3. Drexler J. F., et al. 2009. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg. Infect. Dis. 15:1662–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellis J., et al. 2009. Evaluation of four real-time PCR assays for detection of influenza A(H1N1) viruses. Euro Surveill. 14:pii.19230 [DOI] [PubMed] [Google Scholar]
- 5. Ganzenmueller T., et al. 2010. Comparison of the performance of direct fluorescent antibody staining, a point-of-care rapid antigen test and virus isolation with that of RT-PCR for the detection of novel 2009 influenza A (H1N1) virus in respiratory specimens. J. Med. Microbiol. 59:713–717 [DOI] [PubMed] [Google Scholar]
- 6. Ge Y., et al. 2010. Detection of novel swine origin influenza A virus (H1N1) by real-time nucleic acid sequence-based amplification. J. Virol. Methods 163:495–497 [DOI] [PubMed] [Google Scholar]
- 7. Herzum I., Lutz T., Koch F., Geisel R., Gehrt A. 2010. Diagnostic performance of rapid influenza antigen assays in patients infected with the new influenza A (H1N1) virus. Clin. Chem. Lab. Med. 48:53–56 [DOI] [PubMed] [Google Scholar]
- 8. Karre T., et al. 2010. Comparison of Becton Dickinson Directigen EZ Flu A+B test against the CDC real-time PCR assay for detection of 2009 pandemic influenza A/H1N1 virus. J. Clin. Microbiol. 48:343–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panning M., et al. 2009. Detection of influenza A(H1N1)v virus by real-time RT-PCR. Euro Surveill. 14:pii.9329 [PubMed] [Google Scholar]
- 10. Poon L. L., et al. 2009. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin. Chem. 55:1555–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schulze M., Nitsche A., Schweiger B., Biere B. 2010. Diagnostic approach for the differentiation of the pandemic influenza A(H1N1)v virus from recent human influenza viruses by real-time PCR. PLoS One 5:e9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van der Vries E., Schutten M. 2010. Satisfying the need for rapid diagnosis of new variant influenza A H1N1. Expert Rev. Mol. Diagn. 10:251–253 [DOI] [PubMed] [Google Scholar]
- 13. Vasoo S., Stevens J., Singh K. 2009. Rapid antigen tests for diagnosis of pandemic (swine) influenza A/H1N1. Clin. Infect. Dis. 49:1090–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang D. Y., et al. 2009. Establishment of a method for rapid detection of the nucleic acid of the novel A (H1N1) influenza virus. Bing Du Xue Bao 25(Suppl.):1–3 (In Chinese.) [PubMed] [Google Scholar]
- 15. Whiley D. M., et al. 2009. Detection of novel influenza A(H1N1) virus by real-time RT-PCR. J. Clin. Virol. 45:203–204 [DOI] [PubMed] [Google Scholar]
- 16. WHO 2009. Influenza A(H1N1). WHO, Geneva, Switzerland: http://www.who.int/csr/disease/swineflu/en/index.html [Google Scholar]
- 17. WHO 2010. Pandemic H1N1 2009 update 112. World Health Organization, Geneva, Switzerland: Accessed 6 August 2010 http://www.who.int/csr/don/2010_08_06/en/index.html [Google Scholar]