Abstract
The performance of hepatitis E virus (HEV) RNA nucleic acid amplification (NAT)-based assays has been investigated using a panel of HEV-containing plasma samples. The panel comprised 22 HEV-positive plasma samples representing 10-fold serial dilutions of HEV genotypes 3a, 3b, 3f, and 4c obtained from blood donors. Two negative-control plasma samples were included. All samples were blinded. The plasma samples were prepared as liquid/frozen materials and distributed to participants on dry ice. Laboratories were requested to test the panel using their routine HEV assays and to score samples as either positive or negative and could optionally return data in copies/ml for HEV RNA. Twenty laboratories from 10 different countries participated in the study. Data were returned by all participating laboratories; 10 laboratories returned quantitative data. All assays except one were developed in-house using conventional or real-time reverse transcriptase PCR (RT-PCR) methodologies. There was a 100- to 1,000-fold difference in sensitivity between the majority of assays, independent of the virus strain. Although the quantitative data were limited, for the samples in the range of ∼6 to 4 log10 copies/ml, the standard deviations of the geometric means of the samples ranged between 0.38 and 1.09. Except for one equivocal result, HEV RNA was not detected in the negative samples. The variability of assay sensitivity highlights the need for the standardization of HEV RNA NAT assays.
INTRODUCTION
Hepatitis E virus (HEV) is a major cause of acute hepatitis, being transmitted mainly via the fecal-oral route (17). The consequences of HEV infection may be severe for pregnant women and individuals with underlying liver disease, with increasing evidence of persistent infection in immunocompromised patients (9, 11, 17). In industrialized countries HEV infection is usually associated with travel to regions where HEV is endemic; however, autochthonous infections are being reported more frequently in Europe, Japan, and elsewhere. HEV is a nonenveloped, single-stranded, positive-sense RNA virus that is classified in the family Hepeviridae, with four main genotypes of HEV causing infections in humans. Genotype 1 has been found in people throughout Asia and in other parts of the world. Genotype 2 has been reported in Mexico and parts of West Africa. Viruses belonging to genotypes 3 and 4 have been detected not only in humans but also in swine and other animal species, including wild boar and deer, with sequence analysis demonstrating that viruses originating from the same geographic regions are genetically very similar to one another regardless of whether they have been found in humans or animals. The zoonotic spread of HEV occurs, with reports of food-borne transmission of genotypes 3 and 4 (reviewed in references 16 and 18). An additional route of transmission is via transfusion, which has been reported for several countries worldwide, including Japan, France, and the United Kingdom (3, 4, 14, 15).
Diagnostic testing for HEV is important for patients for which other causes of acute hepatitis have been excluded (19). Since HEV infection is clinically indistinguishable from other types of acute viral hepatitis, diagnosis is based upon the detection of specific antibodies, with several commercial assays available. However, the low sensitivity of certain serological tests has probably resulted in an underestimation of HEV seroprevalence (2), with HEV infection likely to be more common than previously believed in certain industrialized countries (7). For the optimal diagnosis of acute HEV infection, a combination of serological testing and nucleic acid amplification technique (NAT) assays is preferred (6). NAT-based assays have been developed for the detection of HEV RNA to confirm active HEV infection. These assays include conventional reverse transcriptase PCR (RT-PCR) as well as real-time RT-PCR and reverse transcription–loop-mediated isothermal amplification (10) for the detection of HEV RNA in serum and plasma or in fecal samples. While consensus assays have been developed for the detection of HEV genotypes 1 to 4 (5, 8), no studies have been performed so far to compare the abilities of laboratories to detect HEV RNA.
The World Health Organization (WHO) has established a number of International Standards (ISs) for NAT-based assays for several blood-borne viruses (reviewed in reference 13), and these were developed initially for their use in the field of blood and plasma safety, allowing the comparison of analytical sensitivities of assays by use of a common reporting unit, i.e., the international unit (IU). The ISs have also found use in clinical laboratories, with results for viral load testing being expressed in IU. At the WHO Expert Committee on Biological Standardization (ECBS) meeting in Geneva, Switzerland, in October 2009, proposals by the Paul-Ehrlich-Institut (PEI) to develop ISs for HEV RNA and hepatitis D virus RNA were endorsed. In this initial study we have evaluated several HEV strains for the purposes of determining a suitable virus preparation to develop into a WHO IS and to investigate for the first time performance of HEV RNA NAT assays used in different laboratories worldwide.
MATERIALS AND METHODS
Virus strains.
Panels were prepared at the PEI, a WHO collaborating center for the quality assurance of blood products and in vitro diagnostic devices (IVDs), from HEV-positive human plasma samples obtained from blood donors. Three viremic HEV donations were provided by Keiji Matsubayashi of the Japanese Red Cross Hokkaido Blood Center. A fourth virus strain was obtained from a German plasmapheresis donor and was subsequently characterized at the Robert-Koch Institut in Berlin, Germany (1). The details of each virus strain are shown in Table 1. The strains tested negative for HIV-1/2 RNA, hepatitis B virus (HBV) DNA, and hepatitis C virus (HCV) RNA using the Cobas TaqScreen MPX test (Roche Molecular Systems Inc.).
Table 1.
Details of HEV strains evaluated in the study
| Genotype | Virus strain | HEV RNA level (copies/ml)a | GenBank accession no. | Anti-HEV IgM/IgG resultb | ALT level (IU/liter)c |
|---|---|---|---|---|---|
| 3a | HRC-HE104 | 1.6 × 107 | AB602891 | −/− | 36 |
| 3b | JRC-HE3 | 2.5 × 107 | AB434146 | +/− | 398 |
| 3f | RKI | 1.3 × 106 | FJ956757 | −/− | Negative |
| 4c | HRC-HE15 | 1.0 × 106 | AB602890 | −/− | 505 |
Concentrations determined by laboratories where samples were identified.
+/+, positive result; +/−, equivocal result; −/−, negative result.
ALT, alanine transaminase.
Phylogenetic analysis of virus strains.
In order to determine the subgenotypes of the HEV strains used in the study, sequence analysis was performed for the ORF2 region of the genome, as previously described (12). HEV RNA was amplified by using primers HEVORF2-5972 (5′-GTY ATG YTY TGC ATA CAT GGC T) and HEVORF2-6319 (5′-AGC CGA CGA AAT YAA TTC TGT C), corresponding to nucleotides 5972 to 5993 and 6298 to 6319, respectively, of the HEV Burma strain (GenBank accession number M73218). RT-PCR was performed by using the OneStep RT-PCR kit (Qiagen GmbH, Hilden, Germany), as follows: RNA was reverse transcribed at 50°C for 30 min, followed by denaturation at 95°C for 15 min. The PCR consisted of 45 cycles: a denaturation step at 94°C for 30 s, an annealing step at 48°C for 30 s, and an elongation step at 72°C for 1 min.
Two independent amplification products were purified (QIAquick PCR purification spin kit; Qiagen GmbH, Hilden, Germany), sequenced directly using the Big-Dye Terminator Cycle Sequencing Ready Reaction kit, and analyzed by using the Applied Biosystems 3730xl sequencing system (Applied Biosystems Deutschland GmbH, Darmstadt, Germany). Phylogenetic analysis was performed as previously described (12; Jacques Izopet, Institut National de la Santé et de la Recherche Médicale, Toulouse, France, personal communication).
Preparation of panels.
Each panel contained 24 coded samples representing HEV-containing human plasma donations diluted in pooled HEV-negative plasma and negative-control plasma. Twenty-two samples contained HEV RNA with approximate target levels ranging from ∼106 to ∼101 copies/ml. Serial 10-fold dilutions of the four viruses were prepared by using pooled human plasma nonreactive for anti-HEV IgG and IgM (Ulrich Mohn, Mikrogen GmbH, Neuried, Germany, personal communication) and tested negative for HEV RNA (data not shown). In addition, the plasma was negative for HIV-1/2 RNA, HBV DNA, and HCV RNA, and testing was performed as described above. The plasma diluent was centrifuged at 3,000 × g for 30 min prior to use. The two HEV-negative samples in the panel were prepared in a separate area where HEV-positive materials were not handled. Testing was run on sample panels by the coordinating laboratory. The panels were stored at −70°C.
Dispatching of panels.
All panels were shipped on dry ice, and participants were requested to store the samples at −70°C or below until analysis.
Study protocol.
Participants were asked to test the panel using their routine assay for HEV RNA, without further dilution of the samples, in a single assay run. It was requested that results be reported in a qualitative way as either being positive, i.e., HEV RNA was detected, or negative, i.e., HEV RNA was not detected. As an optional exercise, laboratories using quantitative assays for HEV RNA were encouraged to return results in copies/ml.
Evaluation of results.
Qualitative data were scored as positive, negative, or equivocal. Quantitative data were scored as either positive or negative for comparisons of assay sensitivity with the qualitative assays. Statistical analysis of the quantitative data was performed with SAS/STAT software, version 9.2 (SAS System for Windows).
RESULTS
Phylogenetic analysis of HEV strains used in the panel.
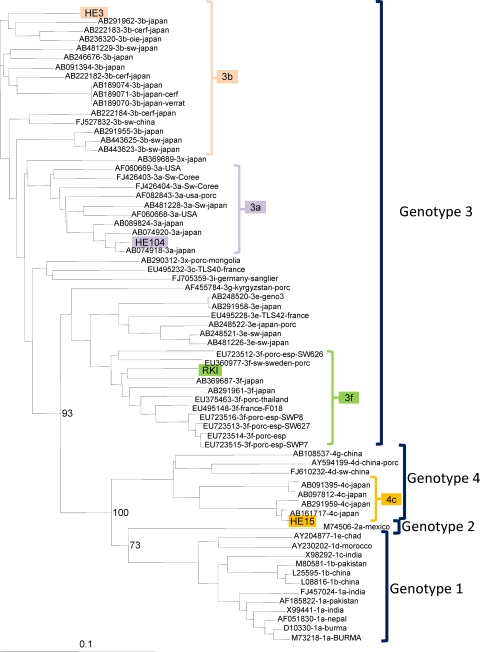
The subgenotyping analysis of the HEV strains used for the preparation of the panel is shown in Fig. 1. The designation of the subgenotypes was determined by sequence analysis of ORF2 and confirmed by an analysis of ORF1 sequences. The subgenotype of the German HEV strain was reported previously (1).
Fig. 1.
Phylogenetic analysis of HEV strains evaluated in the study. GenBank accession numbers are shown for reference strains; the numbers of the panel strains are shown in Table 1.
Data returned.
Twenty-three panels were distributed to 20 laboratories in 10 different countries. All participants returned results. Ten laboratories returned data from quantitative assays. Three laboratories returned data from two different types of assay. The participants represent clinical laboratories, particularly hepatitis reference laboratories, laboratories performing HEV research, organizations developing HEV vaccines, IVD manufacturers, blood banks, plasma fractionation organizations, and associated control laboratories.
The extraction methods and the amplification/detection techniques are listed in Table S1 in the supplemental material. Each laboratory was assigned a random code. With a single exception (laboratory 18a), all assays were developed in-house, some of which were based upon previously reported assays. The assays represented a mixture of both conventional and real-time RT-PCR.
Laboratory 10 performed virus genotyping by sequence analysis and correctly genotyped the samples that they found positive in the panel. However, in the case of the most diluted sample for HEV genotype 4c (sample 22), the laboratory found the sample to be positive in three separate runs (from extraction through to amplification/detection). Genotyping analysis by this laboratory suggested that the HEV strain was of genotype 1. It is most likely that upon initial sampling, the plasma became contaminated with genotype 1 HEV sequences present in the participating laboratory, since genotype 1 viruses were not included in the panel.
Two negative plasma samples were included in the panel (samples 7 and 24); these were all scored as negative, except for one laboratory, which reported an equivocal result for a single replicate in a real-time RT-PCR assay (data not shown).
Qualitative results for panel samples.
A summary of the qualitative results is given in Table 2. The qualitative data are presented in more detail in Tables S2 to S5 in the supplemental material for each of the respective virus strains represented in the panel. The most sensitive methods were those based upon real-time RT-PCR regardless of the virus strain analyzed. The least sensitive methods were nested PCRs, which were based upon sequences in ORF1 of the HEV genome. Indeed, one laboratory reported negative results for all HEV-positive samples in the panel; this was one of two assays used in the study that were designed to detect HEV ORF1, while the remaining assays targeted ORF2 and ORF3 regions. In the case of laboratory 15a, there is a 1,000-fold difference in the sensitivity of detection of the genotype 3a and the genotype 3b HEV strains, suggesting that the primers used with this method do not target well-conserved sequences in the virus genome. This assay failed to detect the genotype 3f virus. When these methods directed against the ORF1 sequences are excluded, there is a 100- to 1,000-fold difference in sensitivity between the majority of assays independent of the virus strain. Laboratory 12 was able to detect all the positive samples with the exception of the most diluted genotype 4c sample.
Table 2.
Qualitative analysis of the four HEV strains
| Virus strain (genotype) | Nominal concn (log10 copies/ml) | % positivea |
|---|---|---|
| HRC-HE104 (3a) | 6.2 | 96 |
| 5.2 | 96 | |
| 4.2 | 92/88 | |
| 3.2 | 75/67 | |
| 2.2 | 38/25 | |
| 1.2 | 13/8 | |
| JRC-HE3 (3b) | 6.4 | 96 |
| 5.4 | 92 | |
| 4.4 | 92/88 | |
| 3.4 | 75/67 | |
| 2.4 | 58/33 | |
| 1.4 | 21/9 | |
| RKI (3f) | 5.1 | 92 |
| 4.1 | 75/71 | |
| 3.1 | 71/63 | |
| 2.1 | 33/25 | |
| 1.1 | 4 | |
| HRC-HE15 (4c) | 5.0 | 92/88 |
| 4.0 | 83 | |
| 3.0 | 50/38 | |
| 2.0 | 33/25 | |
| 1.0b | 4/0 |
For the number of positive test results, a best-case/worst-case percentage is reported for results reported as being simply positive or equivocal.
Data were returned for a total number of 24 tests for each sample. However, in the case of sample 22 (genotype 4c), a false-positive result (as evidenced by the detection of an HEV genotype not represented in the panel) was reported by one laboratory and was not included during the calculation of percent positive results. A breakdown of the original data is shown in Tables S2 to S5 in the supplemental material.
Analysis of CT values for virus dilutions.
Thirteen sets of data were returned where it was possible to analyze the CT (threshold cycle) values. While the CT values alone provide no quantitative measure of the panel samples, the returned data indicated that in almost all cases, with the increasing dilution of each of the HEV strains, there was a corresponding increase in the CT value, and consecutive samples were ranked appropriately in these assays (data not shown).
Quantitative results for panel samples.
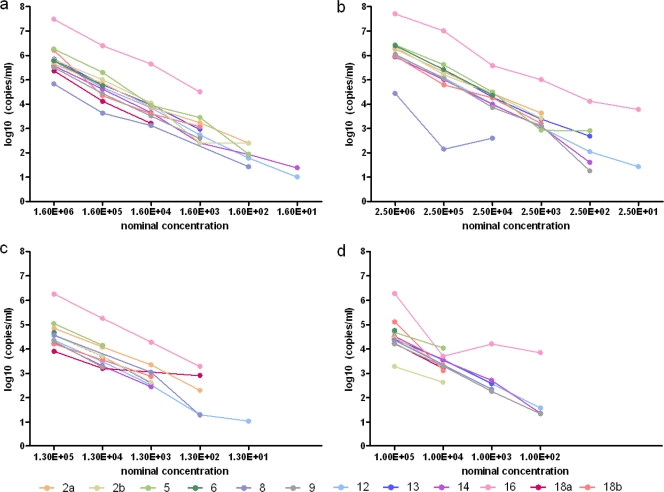
A total of 12 data sets were returned from 10 participating laboratories. The geometric means, median values, and ranges reported for the viral loads (log10 copies/ml of HEV RNA) for the samples are summarized in Table 3. The laboratories that returned quantitative results were able to consecutively rank the samples according to dilution (Fig. 2), similar to analyses of the CT values. Although there was only a limited number of quantitative data sets, for the samples in the range of approximately 6 to 4 log10 copies/ml, the standard deviations of the geometric means of the samples ranged between 0.38 and 1.09. For the highest concentrations of each virus strain (in the range of ∼5 to 6 log10 copies/ml), at least two-thirds of the data sets fell within ±0.5 log10 copies/ml of the median value for the different HEV strains.
Table 3.
Viral load data for the four HEV strains
| Virus strain | Sample | Nominal concn (log10 copies/ml) | No. of samples | Observed viral load (log10 copies/ml) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Geometric mean | SD | 95% CIa | Median | Minimum | Maximum | ||||
| HRC-HE104 | 13 | 6.2 | 12 | 5.84 | 0.64 | 5.43, 6.24 | 5.77 | 4.82 | 7.48 |
| 11 | 5.2 | 12 | 4.74 | 0.68 | 4.31, 5.17 | 4.72 | 3.63 | 6.40 | |
| 3 | 4.2 | 11 | 3.85 | 0.67 | 3.40, 4.30 | 3.84 | 3.11 | 5.64 | |
| 18 | 3.2 | 9 | 3.04 | 0.66 | 2.53, 3.54 | 2.96 | 2.40 | 4.49 | |
| 17 | 2.2 | 6 | 1.98 | 0.37 | 1.59, 2.37 | 1.94 | 1.43 | 2.40 | |
| 19 | 1.2 | 4 | 0.10 | 1.27 | −1.93, 2.12 | 0.00 | −1.00 | 1.38 | |
| JRC-HE3 | 20 | 6.4 | 12 | 6.16 | 0.72 | 5.70, 6.61 | 6.15 | 4.43 | 7.70 |
| 23 | 5.4 | 12 | 5.07 | 1.09 | 4.38, 5.76 | 5.14 | 2.15 | 7.00 | |
| 4 | 4.4 | 12 | 4.21 | 0.67 | 3.79, 4.63 | 4.27 | 2.60 | 5.58 | |
| 12 | 3.4 | 10 | 3.40 | 0.60 | 2.97, 3.83 | 3.20 | 2.92 | 5.00 | |
| 10 | 2.4 | 6 | 2.43 | 1.03 | 1.35, 3.52 | 2.36 | 1.26 | 4.11 | |
| 6 | 1.4 | 3 | 1.40 | 2.39 | −4.53, 7.34 | 1.43 | −1.00 | 3.78 | |
| RKI | 1 | 5.1 | 12 | 4.63 | 0.59 | 4.26, 5.01 | 4.57 | 3.91 | 6.26 |
| 8 | 4.1 | 10 | 3.77 | 0.61 | 3.33, 4.21 | 3.63 | 3.20 | 5.26 | |
| 2 | 3.1 | 9 | 2.83 | 0.69 | 2.30, 3.37 | 2.63 | 1.77 | 4.28 | |
| 16 | 2.1 | 6 | 1.68 | 1.54 | 0.06, 3.30 | 1.80 | −1.00 | 3.28 | |
| 21 | 1.1 | 3 | −0.32 | 1.18 | −3.25, 2.61 | −1.00 | −1.00 | 1.04 | |
| HRC-HE15 | 5 | 5.0 | 12 | 4.56 | 0.69 | 4.12, 5.00 | 4.44 | 3.28 | 6.28 |
| 14 | 4.0 | 10 | 3.40 | 0.38 | 3.13, 3.67 | 3.44 | 2.63 | 4.04 | |
| 9 | 3.0 | 8 | 1.83 | 1.85 | 0.28, 3.38 | 2.46 | −1.00 | 4.20 | |
| 15 | 2.0 | 6 | 1.02 | 1.83 | −0.90, 2.94 | 1.35 | −1.00 | 3.85 | |
| 22 | 1.0 | 3 | −1.00 | 0.00 | −1.00 | −1.00 | −1.00 | ||
95% CI, 95% confidence interval for the geometric mean.
Fig. 2.
Analysis of viral loads (log10 copies/ml) by laboratory and sample. (a) HRC-HE104 genotype 3a. (b) JRC-HE3 genotype 3b. (c) RKI genotype 3f. (d) HRC-HE15 genotype 4c.
DISCUSSION
This study has for the first time compared the performances of assays for the detection of HEV RNA and investigated the suitability of different HEV strains for development into a candidate WHO IS. Qualitative analysis of the data has demonstrated significant variability in assay sensitivity irrespective of the HEV strains examined. For the majority of assays, there was a 100- to 1,000-fold difference in sensitivity independent of the virus strain. The overall specificity was very good, with the negative samples being correctly scored, except for a single equivocal result on a replicate sample. In the case of one laboratory, cross-contamination was observed, as evidenced by the detection of an HEV genotype that was not represented in the panel. Although there were only a limited number of quantitative data sets returned, the data were in reasonable agreement for the higher-titer samples.
With the exception of a single assay, all methods were developed in-house. The real-time RT-PCR methods were generally proven to be more sensitive than the nested RT-PCR assays. Clearly, the availability of a standard for HEV RNA would allow a comparison of assay sensitivity, which has been shown to vary widely even in laboratories with broad experience in HEV molecular diagnostics. The lack of sensitivity of assays may result in the misdiagnosis of acute HEV infection in patients presenting with hepatitis of unknown etiology or in the failure to identify viremic blood/plasma donations where testing is implemented. Standardization would also be helpful in monitoring HEV loads in chronically infected patients undergoing antiviral therapy.
The HEV strains included in the panel represented viruses that were obtained from blood donors. Three virus strains (genotypes 3a, 3b, and 4c) were obtained from the Japanese Red Cross Society, where the samples were identified in an epidemiological survey of HEV infection in blood donors. The fourth HEV strain (genotype 3f) was identified in a German plasma donor (1), who after donation presented with acute hepatitis and was subsequently found to be infected with HEV. From the data generated in the study, any of the HEV strains could potentially be developed into an IS; however, genotype 3 viruses have the widest distribution worldwide and represent the virus genotype identified in chronic infections, and one strain will be taken forward for the preparation of a candidate WHO IS.
Supplementary Material
ACKNOWLEDGMENT
We are grateful for assistance from Gudrun Winskowsky during the organization of the study.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Adlhoch C., Kaiser M., Pauli G., Koch J., Meisel H. 2009. Indigenous hepatitis E virus infection of a plasma donor in Germany. Vox Sang. 97:303–308 [DOI] [PubMed] [Google Scholar]
- 2. Bendall R., Ellis V., Ijaz S., Ali R., Dalton H. 2010. A comparison of two commercially available anti-HEV IgG kits and a re-evaluation of anti-HEV IgG seroprevalence data in developed countries. J. Med. Virol. 82:799–805 [DOI] [PubMed] [Google Scholar]
- 3. Boxall E., et al. 2006. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus. Med. 16:79–83 [DOI] [PubMed] [Google Scholar]
- 4. Colson P., et al. 2007. Transfusion-associated hepatitis E, France. Emerg. Infect. Dis. 13:648–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gyarmati P., et al. 2007. Universal detection of hepatitis E virus by two real-time PCR assays: TaqMan and Primer-Probe Energy Transfer. J. Virol. Methods 146:226–235 [DOI] [PubMed] [Google Scholar]
- 6. Huang S., et al. 2010. Profile of acute infectious markers in sporadic hepatitis E. PLoS One 5:e13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ijaz S., et al. 2009. Indigenous hepatitis E virus infection in England: more common than it seems. J. Clin. Virol. 44:272–276 [DOI] [PubMed] [Google Scholar]
- 8. Jothikumar N., Cromeans T. L., Robertson B. H., Meng X. J., Hill V. R. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 131:65–71 [DOI] [PubMed] [Google Scholar]
- 9. Kamar N., et al. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358:811–817 [DOI] [PubMed] [Google Scholar]
- 10. Lan X., et al. 2009. Reverse transcription-loop-mediated isothermal amplification assay for rapid detection of hepatitis E virus. J. Clin. Microbiol. 47:2304–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Legrand-Abravanel F., et al. 2010. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J. Infect. Dis. 202:835–844 [DOI] [PubMed] [Google Scholar]
- 12. Legrand-Abravanel F., et al. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madej R. M., et al. 2010. International standards and reference materials for quantitative molecular infectious disease testing. J. Mol. Diagn. 12:133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsubayashi K., et al. 2004. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44:934–940 [DOI] [PubMed] [Google Scholar]
- 15. Matsubayashi K., et al. 2008. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 48:1368–1375 [DOI] [PubMed] [Google Scholar]
- 16. Meng X. J. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153–161 [DOI] [PubMed] [Google Scholar]
- 17. Purcell R. H., Emerson S. U. 2008. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48:494–503 [DOI] [PubMed] [Google Scholar]
- 18. Purcell R. H., Emerson S. U. 2010. Hidden danger: the raw facts about hepatitis E virus. J. Infect. Dis. 202:819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waar K., Herremans M. M., Vennema H., Koopmans M. P., Benne C. A. 2005. Hepatitis E is a cause of unexplained hepatitis in the Netherlands. J. Clin. Virol. 33:145–149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.