Abstract
Several commercial methods exist for the molecular detection of Chlamydia trachomatis and Neisseria gonorrhoeae in clinical samples. Here we evaluated the performance characteristics of the newly FDA-cleared Abbott RealTime CT/NG assay (where “CT” stands for Chlamydia trachomatis and “NG” stands for Neisseria gonorrhoeae) that uses the automated m2000 molecular platform. Results were compared to those of the Roche Cobas Amplicor CT/NG assay. A total of 926 cervical swab, 45 female urine, 6 male urethral swab, and 407 male urine specimens from 1,384 patients were examined. After resolving all Roche N. gonorrhoeae-positive results with two additional real-time PCR assays, we found that the agreement between the assays was excellent. For urine samples, there was 99.6% positive agreement and 97.7% negative agreement for C. trachomatis, and for male urine samples, there was 100% positive agreement and 99.7% negative agreement for N. gonorrhoeae. For cervical swab samples, there was 98.8% positive agreement and 98.5% negative agreement for C. trachomatis, and there was 96.6% positive agreement and 99.8% negative agreement for N. gonorrhoeae. In limiting dilution analyses, we found that the Abbot assay was more sensitive than the Roche assay for both C. trachomatis and N. gonorrhoeae. In addition, there appeared to be an enhanced ability of the Abbott assay to detect dual infections, especially in the presence of large amounts of N. gonorrhoeae and small amounts of C. trachomatis organisms. In summary, we conclude that the Abbott RealTime CT/NG assay is an accurate and automated new addition to the available testing options for C. trachomatis and N. gonorrhoeae.
INTRODUCTION
The incidences of Chlamydia trachomatis and Neisseria gonorrhoeae infection continue to increase globally. A more intensive screening effort has been advocated by the U.S. Preventive Services Task Force (18), the Centers for Disease Control and Prevention (24), other public health agencies, and medical societies to bring this emerging epidemic under control. This includes the yearly screening of sexually active women under the age of 25 years for C. trachomatis. The goal of screening is to prevent transmission and the severe sequelae of unrecognized infection, such as pelvic inflammatory disease and associated infertility.
Rapid, automated, and sensitive nucleic acid amplification testing methods are needed to respond optimally to this public health mandate. In this regard, there have been limited published evaluations of the recently FDA-cleared Abbott RealTime CT/NG assay (where “CT” stands for Chlamydia trachomatis and “NG” stands for Neisseria gonorrhoeae) (9). There was a recent report of clinical trial data comparing the Abbott assay to Gen-Probe Aptima Combo 2 (AC2), BD ProbeTec ET CT/GC, and GC culture (where “GC” represents N. gonorrhoeae) (6). There was also one prior study from Canada using a CE-marked Abbott kit with different cutoff and interpretation algorithms (8) and a U.S. study performed by Abbott Laboratories (Des Plaines, IL) using a prototype version of the now FDA-cleared assay (9). The last-named study included only a small number of N. gonorrhoeae-positive samples. Taken together, these studies provide only minimal data regarding the performance of the Abbott assay in comparison to that of the well-established, FDA-cleared, Roche Cobas Amplicor CT/NG method.
In order to obtain further insight into the performance characteristics of this new commercial method, we performed a comparative evaluation with several interdependent goals. The first was to assess the Abbott RealTime CT/NG method's clinical and analytical performance in comparison with that of the well-established Roche Cobas Amplicor CT/NG assay in current use in our laboratory. The second was to examine the ability of the Abbott method to avoid false-positive N. gonorrhoeae results observed with some commercially available nucleic acid amplification test (NAAT) methods (22). These false-positive results have previously been linked to the spurious detection of nonpathogenic Neisseria species. The third was to examine the effect of the specimen transport medium on the analytical performance of the Abbott method. The Abbott test recommends collection of specimens in assay-specific transport medium containing the denaturant guanidinium thiocyanate. In contrast, the Roche method recommends the collection of swab specimens in M4-RT medium (MicroTest, Inc., Lilburn, GA), a general-purpose transport medium also used for culture of viruses and Chlamydia. During the study period, swab specimens were transported in M4-RT medium. Therefore, we sought to determine whether the use of M4-RT medium versus Abbott transport medium affected the analytical performance of the Abbott RealTime CT/NG assay. Results would confirm the reasonableness of comparing clinical test performances on samples collected in M4-RT medium.
MATERIALS AND METHODS
Clinical samples and testing.
During the study, 926 cervical swabs, 45 female urine samples, 6 male urethral swabs, and 407 male urine samples were collected from 1,384 patients for clinical diagnosis. At our institution we receive relatively few female urine specimens because of the lack of FDA approval for N. gonorrhoeae testing by the Roche assay for this specimen type. The prevalences of N. gonorrhoeae and C. trachomatis at our institution are also relatively low, at 0.2% and 2.2%, respectively. Therefore, in order to establish more confidently comparative performance on positive specimens, we selectively tested a larger percentage of positive specimens than we otherwise would have based on the prevalences of C. trachomatis and N. gonorrhoeae in our patient population. With this caveat, swab and first-catch urine specimens were otherwise selected randomly for comparative testing by the Abbott method without knowledge of the sex of the patient.
Cervical and urethral swabs were transported in 3 ml of M4-RT medium. Urine was collected into sterile tubes with no additives. Testing was performed using two methods: the Roche Cobas Amplicor CT/NG assay used for clinical testing in our hospital system and the Abbott RealTime CT/NG assay. Specimens were tested with the Roche system and either tested concurrently using the Abbott system or frozen at −20°C and later tested in batch runs with the Abbott method. For the Roche assay, 100 μl of swab sample and 500 μl of urine sample were extracted for testing. For the Abbott assay, 3 ml of each urine specimen was first mixed with 1.2 ml of Abbott transfer buffer. Four hundred microliters of this urine mixture or 400 μl of swab specimens (collected in M4-RT medium) was then extracted with the Abbott m2000 instrument according to the manufacturer's protocol. The Abbott method uses an automated nucleic acid extraction platform based on magnetic bead technology and robotic PCR setup, followed by the manual transfer of PCR mixtures to an m2000rt thermocycler for real-time PCR amplification and reading. Testing was performed according to the manufacturer's instructions, including recommended procedures for resolution of indeterminate results. This study was approved by our institutional review board.
Confirmation of N. gonorrhoeae-positive test results.
Because of known N. gonorrhoeae false positivity resulting from cross-reactivity of primer and probe sets of some commercial methods with nonpathogenic Neisseria species (22), all results that were N. gonorrhoeae positive by the Roche method were confirmed in our laboratory by two previously described, hybridization probe-based, real-time PCR tests that amplify alternative targets. Both tests were previously shown to have high specificity. The target of the first method, the porA pseudogene, is found only in N. gonorrhoeae and Neisseria meningitidis (5, 17, 20). Although both species yield positive amplification signals, they can be distinguished by their different melting curves resulting from nucleotide polymorphisms that differentially affect the melting temperature (Tm) of the hybridization probes. The second test targets the multicopy 16S rRNA gene, again distinguishing among the several Neisseria species amplified through melting-temperature analysis (3). Both methods were previously described as confirmatory methods for the Roche N. gonorrhoeae assay used in this study (3, 5).
For N. gonorrhoeae-positives samples, we extracted a portion of the original patient sample with High Pure columns (Roche Applied Sciences, Indianapolis, IN) as described previously (21). This differed from the Boom extraction cited previously for the 16S rRNA gene amplification method (3). Amplification was performed with a Roche LightCycler 2.0 (LC) instrument as described in the original publications, with the exception that the cycling protocol for the porA pseudogene was used for both assays (i.e., incorporating a longer denaturation time of 10 min and 55 amplification cycles) (3, 21). Primers and probes were obtained from Tib-Molbiol (Adelphia, NJ), and LightCycler FastStart DNA Master HybProbe master mix was obtained from Roche Applied Sciences (Indianapolis, IN). Limiting dilution studies showed that the confirmatory methods (here referred to as “LC”) had slightly lower to equivalent sensitivity compared to that of the primary Roche method (A. Cheng and J. E. Kirby, unpublished data). This likely resulted from a smaller volume of extracted sample being tested in the confirmatory real-time PCR assays. Therefore, to ensure adequate sensitivity, Roche N. gonorrhoeae-positive LC-negative samples were then concentrated using ultrafiltration with a Micron filter (Millipore, MA) with a 100,000-kDa nominal molecular mass cutoff. This led to an estimated 10- to 50-fold concentration of sample based on volume. Real-time PCR analysis of a limiting dilution series demonstrated that this concentration step increased sensitivity beyond that of unconcentrated samples tested by the Roche method. Therefore, the LC methodology combined with ultrafiltration could serve as a confirmatory test for samples with low levels of target.
As described in Results, our initial clinical verification of the LC confirmatory methods in our laboratory also included testing of samples that were N. gonorrhoeae positive by the Roche method using a second commercial method, specifically the ProbeTec ET Chlamydia trachomatis and Neisseria gonorrhoeae Amplified DNA assay. ProbeTec testing was performed at the Cleveland Clinic Clinical Microbiology Laboratory, which had previously verified the use of M4-RT medium-based specimens with this assay.
Testing algorithm and resolution of discrepancies.
As mentioned above, all samples that were N. gonorrhoeae positive by the Roche method were confirmed by LC. LC-confirmed specimens were considered Roche N. gonorrhoeae true positives. Any non-LC-confirmed specimen results were considered Roche false positives and are listed as being N. gonorrhoeae negative under the Roche results shown in Tables 1 and 2. Otherwise, the test results are listed in Tables 1 and 2 as determined on initial testing by each method. Any discrepant samples underwent repeat testing by both the Roche and Abbott methods. Furthermore, any Abbott N. gonorrhoeae-positive, Roche N. gonorrhoeae-negative sample was also tested by LC. Finally, when sufficient sample was available, deidentified coded samples were also further tested by using the Gen-Probe Aptima Combo 2 (AC2) DTS system. In this case, 1.5 to 2 ml of urine samples or 100 μl of swab samples in M4-RT medium was placed into Aptima urine collection tubes or an Aptima unisex swab collection tube containing 2 and 2.9 ml of transport medium, respectively, and further processed according to the manufacturer's instructions. Testing was performed at the Indiana University School of Medicine Infectious Diseases Department, which uses AC2 for clinical testing. Follow-up testing results for discrepant samples are described in Results.
Table 1.
Comparison of test results for urine specimensa
| Abbott assay result | No. of specimens with indicated Roche assay result |
|||
|---|---|---|---|---|
| C. trachomatis positive | C. trachomatis negative | N. gonorrhoeae positiveb | N. gonorrhoeae negative | |
| C. trachomatis positive | 238 | 5c | ||
| C. trachomatis negative | 1 | 208 | ||
| N. gonorrhoeae positive | 53 | 1 | ||
| N. gonorrhoeae negative | 0 | 353 | ||
For C. trachomatis there was 99.6% (95% score confidence interval, 97.7% to 99.9%) positive agreement and 97.7% (94.6% to 99.0%) negative agreement. For N. gonorrhoeae there was 100% (93.2% to 100%) positive agreement and 99.7% (98.4% to 100%) negative agreement.
The Roche N. gonorrhoeae test results were scored as positive in this table only if both porA and 16S rRNA real-time PCR confirmatory (LC) assays were also positive. Accordingly, 11 Roche N. gonorrhoeae-positive test results were considered false-positive results based on confirmatory LC testing and are tabulated among the Roche N. gonorrhoeae-negative test results. Forty-five female urine samples were tested only for C. trachomatis using the Roche method and are therefore not included in the N. gonorrhoeae tabulation. All were N. gonorrhoeae negative by the Abbott assay.
Three specimens were C. trachomatis negative by the Roche assay, and two were equivocal for C. trachomatis by the Roche assay.
Table 2.
Comparison of test results for cervical swabsa
| Abbott assay result | No. of swabs with indicated Roche assay result |
|||
|---|---|---|---|---|
| C. trachomatis positive | C. trachomatis negative | N. gonorrhoeae positiveb | N. gonorrhoeae negative | |
| C. trachomatis positive | 511 | 6 | ||
| C. trachomatis negative | 6 | 401 | ||
| N. gonorrhoeae positive | 56 | 2 | ||
| N. gonorrhoeae negative | 2 | 868 | ||
For C. trachomatis there was 98.8% ((95% score confidence interval, 97.5% to 99.5%) positive agreement and 98.5% (96.8% to 99.3%) negative agreement. For N. gonorrhoeae there was 96.6% (88.3% to 99.1%) positive agreement and 99.8% (99.2% to 99.9%) negative agreement.
Roche N. gonorrhoeae test results were scored as positive only if confirmed by LC assays. Accordingly, 185 initially Roche N. gonorrhoeae-positive samples were considered false-positive results based on LC and are tabulated as Roche N. gonorrhoeae-negative test results.
The analysis of discrepant results also made use of quantitative data available for the Abbott assay. As described previously (16), the Abbott software calculates a measure called the FCN, the cycle number at which the change in signal from one amplification cycle to the next is greatest. The FCN is used as an alternative to the cycle threshold. The software then calculates a delta cycle (DC), which is the difference in cycle number between the sample FCN and the cutoff control, with an additional number of buffer cycles added to establish a cutoff for positivity (1). The DC value correlates in a positive, log-linear fashion with the target concentration in the original sample.
Limiting dilution studies with M4-RT medium.
Serial 10-fold dilutions of AmpliTrol CT/GC reagent (Bio-Rad Laboratories, Hercules, CA) were made in M4-RT and Abbott-specific CT/NG transport media in siliconized Eppendorf tubes. The AmpliTrol reagent contains a mixture of C. trachomatis LGV type II strain 434 elementary bodies and a whole-cell lysate of N. gonorrhoeae ATCC 19424 (2). Initially, four replicates of each 10-fold dilution were tested to roughly define detection limits, followed by 20 replicates of a finer series of dilutions to define the 95% limit of detection (LOD). M4-RT medium dilutions were also tested in the Roche assay according to the manufacturer's instructions for swab-based specimens. Correlation analysis between samples diluted in M4-RT medium and Abbott transfer buffer, and tested by the Abbott method, was performed by using data points from the initial 10-fold dilution series.
For LOD analysis, Roche N. gonorrhoeae test results with values in the expanded indeterminate zone (A660 of 0.2 to 3.5) were scored as positive results. In clinical practice, these samples would normally be retested in duplicate and resolved as positive if A660 values were above 2.0 in two of the three test runs (19). Conversely, the sample would be scored as negative if two of three results were less than this cutoff. The expanded indeterminate zone is used during clinical testing to reduce the frequency of false-positive results due to low-level cross-reactivity with other Neisseria species (19). However, for our analysis, we made the conservative assumption that all signals above 0.2 were specific for N. gonorrhoeae, as cross-reacting species are presumably not present in the AmpliTrol control material. The Roche assay does not provide a procedure for resolving equivocal C. trachomatis results (A660 values of ≥0.2 and ≤2). As equivocal results would likely lead to further clinical follow-up, they were conservatively scored as being positive in the LOD analysis.
Statistical analysis.
Statistical comparisons were performed by using JMP, version 8.0.2 (SAS Institute, Cary, NC). DC and LOD data were compared by using the Wilcoxon ranked-sum test and Fisher's exact test, respectively. A P value of ≤0.05 was considered statistically significant. R2 correlation coefficients were determined with Excel 2008 (Microsoft, Redmond, WA). Percent agreement and 95% score confidence intervals (listed as a range in parentheses following the percent agreement) were calculated as recommended by CLSI document EP12-A2 (4).
RESULTS
N. gonorrhoeae confirmatory assay.
As the Roche Cobas N. gonorrhoeae test is known to yield false-positive test results arising from cross-reactivity with nonpathogenic Neisseria species (19, 22), positive Roche N. gonorrhoeae tests are confirmed as part of our standard laboratory practice through the use, with minor modifications (see Materials and Methods), of two previously described real-time PCR assays for the alternative porA pseudogene and 16S rRNA gene targets (3, 21). Roche N. gonorrhoeae-positive results were considered true-positive results (and scored as positive in Tables 1 and 2) only if confirmed by both of these LightCycler (LC)-based assays.
To verify the ability of the LC method to distinguish between Roche N. gonorrhoeae true- and false-positive results prior to comparisons of the Abbott and Roche methods, we retested all Roche N. gonorrhoeae-positive samples during a 1-year time period using both the LC and a second commercially available assay, the ProbeTec ET system (BD, Franklin Lakes, NJ). The latter assay detects the N. gonorrhoeae pilin gene-inverting protein homologue, in contrast to the N. gonorrhoeae cytosine methyltransferase gene detected by the Roche assay, and is known to cross-react with a presumably nonoverlapping set of nonpathogenic Neisseria species (22). Of 50 Roche-positive swab samples evaluated, 25 were confirmed to be negative by both the ProbeTec and LC methods. Similarly, of 28 Roche-positive urine samples, 3 were confirmed to be negative by both the ProbeTec and LC methods. All samples confirmed positive by the ProbeTec system were also confirmed positive by both LC assays, except for one ProbeTec-positive sample, which was confirmed by neither LC assay. Notably, in this sample, the 16S rRNA LC assay showed amplification but with a low Tm, suggesting the presence of nongonococcal Neisseria species. Unfortunately, no sample was left for further investigation. However, this observation raises the concern that this one strain may have given false-positive results in two commercial assays with different targets. Based upon this verification study and data from prior literature, we concluded that the two LC assays are both sensitive and specific for confirmation of Roche N. gonorrhoeae positivity (3, 14, 20, 21).
Urine specimens.
After identifying Roche N. gonorrhoeae true-positive results by confirmatory LC assays, there was 99.6% (95% score confidence interval, 97.9% to 99.9%) positive agreement and 97.7% (94.6% to 99.0%) negative agreement for C. trachomatis and 100% (93.2% to 100%) positive agreement and 99.7% (98.4% to 100%) negative agreement for N. gonorrhoeae between the Abbott and Roche methods, respectively. As shown in Table 1, there were only six discrepant C. trachomatis results, five of which were Abbott C. trachomatis positive and Roche C. trachomatis negative (n = 3) or equivocal (n = 2). Upon repeat Abbott testing, one of the five samples changed from C. trachomatis positive to negative. Interestingly, all of these specimens were positive for N. gonorrhoeae by both methods. Four of the five discrepant samples were also tested by the Gen-Probe Aptima Combo 2 (AC2) assay. Two samples were confirmed as being C. trachomatis positive and N. gonorrhoeae positive, while only N. gonorrhoeae was detected in the remaining two samples. In addition, the median DC (a measure of the cycle number difference between the specimen and the negative control that varies in a positive log-linear fashion with the target concentration) for C. trachomatis was significantly lower (P = 0.0007) for discrepant samples (DC = 2.7) than for nondiscrepant samples (DC = 9.4). Furthermore, DC values of N. gonorrhoeae were greater than those of C. trachomatis for each discrepant sample (average ratio of N. gonorrhoeae to C. trachomatis of 4; range, 1.4 to 8.0). Taken together, these observations suggested that at least for some samples the Roche assay might not always detect dual infections, especially when there is a small amount of C. trachomatis and excess N. gonorrhoeae present. Finally, there was a single Roche C. trachomatis-positive/Abbott C. trachomatis-negative sample. The sample was C. trachomatis negative by both methods on repeat testing.
Among discrepant results for N. gonorrhoeae, there was one Abbott N. gonorrhoeae-positive/Roche N. gonorrhoeae-negative sample, which was C. trachomatis positive by both methods. It initially tested Roche N. gonorrhoeae positive but was scored as N. gonorrhoeae negative based on negative LC confirmatory testing. Notably, the Abbott N. gonorrhoeae-positive DC value was barely above the cutoff for positivity (DC = 0.05), and repeat Abbott N. gonorrhoeae testing was negative. The C. trachomatis signal in this sample was very strong (DC = 11). AC2 detected only C. trachomatis. It is possible that this sample also represented a dual infection, with the N. gonorrhoeae level near the threshold for detection.
Swab specimens.
After identifying Roche N. gonorrhoeae true-positive samples by confirmatory LC assays, there was 98.8% (95% score confidence interval, 97.5% to 99.5%) positive agreement and 98.5% (96.8% to 99.3%) negative agreement for C. trachomatis and 96.6% (88.3% to 99.1%) positive agreement and 99.8% (99.2% to 99.9%) negative agreement for N. gonorrhoeae between the Abbott and Roche methods, respectively, among cervical swab specimens. As shown in Table 2, there were 12 discrepant C. trachomatis results. Among the 6 Roche C. trachomatis-positive, Abbott C. trachomatis-negative samples, all were C. trachomatis negative upon repeat Roche testing, and one was weakly C. trachomatis positive upon repeat Abbott testing (DC = 1.43). Furthermore, all of these samples were C. trachomatis negative by the AC2 assay.
In addition, there were 6 Roche C. trachomatis-negative, Abbott C. trachomatis-positive samples. All of these were low-level Abbott-positive samples (average DC value of 1.8; range, 0.9 to 3.6). One of four samples available for confirmatory testing by AC2 testing was C. trachomatis positive. Two additional samples were Roche C. trachomatis negative, Abbott C. trachomatis equivocal, and N. gonorrhoeae negative by both methods. However, there was an insufficient amount of sample for the resolution of the equivocal Abbott C. trachomatis results by repeat testing as suggested in the manufacturer's package insert. Therefore, these specimen results were not included in the C. trachomatis tabulation.
In comparison to C. trachomatis results, N. gonorrhoeae agreement was almost complete when Roche N. gonorrhoeae-positive results were resolved by LC. Among the Abbott N. gonorrhoeae-positive, Roche N. gonorrhoeae-negative discrepant results, one specimen was repeatedly weakly N. gonorrhoeae positive by the Abbott assay (average DC = 1.6). Notably, the specimen was strongly C. trachomatis positive by the Abbott assay (DC = 14.84), the Roche assay, and AC2 (961 relative light units). The Roche internal amplification control was initially negative; however, it was positive upon diluting the sample 1:10 and retesting. Nevertheless, the sample remained N. gonorrhoeae negative and was also negative by testing either a 1:10 dilution or a neat sample with LC or by testing a neat sample by AC2. A second discrepant sample was repeatedly Roche N. gonorrhoeae negative and Abbott N. gonorrhoeae low positive (DC = 1.9). The porA pseudogene LC test was negative. However, the 16S rRNA test showed amplification with a melting temperature of 56.6°C (versus the expected 61°C and/or 67°C), suggesting the potential presence of a nongonococcal Neisseria species (3). Unfortunately, no sample remained for the further investigation of this possibility.
Among the two Roche N. gonorrhoeae-positive, Abbott N. gonorrhoeae-negative samples (Table 2), one sample was weakly N. gonorrhoeae positive by the Abbott test upon repeat testing (DC = 0.43). Initially, only the 16S rRNA LC confirmatory test was positive; however, upon repeat testing, both the 16S rRNA and porA pseudogene LC assays were positive. The second sample was LC positive for the 16S rRNA target alone but only after the concentration of the extracted sample by ultrafiltration. The specimen was found to be a quantity not sufficient (QNS) for repeat Abbott and Roche testing. These results are consistent with samples having an amount of the N. gonorrhoeae target near the detection threshold for all methods.
There were also six swab specimens from males in the data set, all of which had concordant positive test results, 5 for N. gonorrhoeae and 1 for C. trachomatis.
Quantitative data analysis.
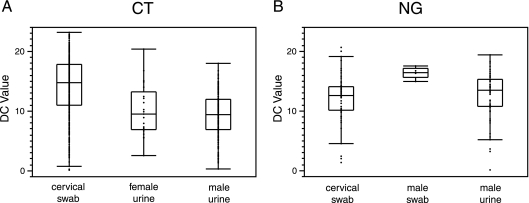
The inherent quantitative nature of real-time PCR enabled quantitative data to be culled from the Abbott CT/NG qualitative assay. Interestingly, the median DC value for C. trachomatis-positive cervical swab specimens (DC = 14.7) was significantly greater than the median DC values for C. trachomatis-positive female (DC = 9.4) and male (DC = 9.3) urine specimens (P ≤ 0.0002 in pairwise comparisons between cervical swabs and male and female urine samples, respectively) (Fig. 1A). This is a difference of 5.3 cycles, or an approximately 39-fold-greater median amount of C. trachomatis target in cervical swab specimens (assuming 100% PCR efficiency). In contrast, median DC values for N. gonorrhoeae-positive cervical swab and male urine samples were not statistically different (P = 0.18). However, the median DC value for N. gonorrhoeae-positive male swab specimens (DC = 16.4) was significantly greater than the median DC values for N. gonorrhoeae-positive male urine (DC = 13.4) and cervical swab (DC = 12.5) samples (P = 0.013 and P = 0.004 in pairwise comparisons, respectively) (Fig. 1B). There were no Abbott N. gonorrhoeae-positive female urine specimens for comparison.
Fig. 1.
Plot of DC values for Abbott N. gonorrhoeae (NG) and C. trachomatis (CT) amplification reactions from different specimen types. (A) DC values for C. trachomatis amplification reactions. Box and whisker plots are shown. The outer edges of the boxes represent the 25th and 75th percentiles, respectively, and the dividing line between them is the median. The whiskers extend from the lower and upper quartiles to the lowest and highest data points, respectively, still within a region bounded by the interquartile range multiplied by 1.5. (B) DC values for N. gonorrhoeae amplification reactions.
Limiting dilution analysis.
The relative analytical sensitivity of the Roche and Abbott methods was also assessed by comparing the abilities to detect C. trachomatis and N. gonorrhoeae in serial dilutions of the commercially available AmpliTrol CT/GC control material in M4-RT medium. Note that an assigned C. trachomatis/N. gonorrhoeae target concentration is not available for the control material from the manufacturer, and therefore, we were able to establish relative but not absolute sensitivities for each method. In this analysis (Table 3), we found that the Abbott method had a lower LOD (detecting a greater dilution of control material) for C. trachomatis (1:2,000 < LOD ≤ 1:500 for the Abbott method and 1:500 < LOD ≤ 1:100 for the Roche method; P < 0.05 for the difference in detection at 1:500 and 1:2,000 dilutions). The Abbott method also had a lower LOD for N. gonorrhoeae (LOD ≤ 1:5,000 for the Abbott method and 1:2,000 < LOD ≤ 1:500 for the Roche method; P < 0.05 for the difference in detection at 1:2,000 and 1:5,000 dilutions). To assess whether the transport buffer might affect the analytical performance of the Abbott assay, control material was also diluted in parallel in Abbott transport medium and tested by the Abbott method (Table 3). No statistically significant difference in detection was observed when performing Abbott testing on samples diluted in M4-RT medium versus Abbott transport medium. Furthermore, DC values over the range of dilutions tested for these two transport media were highly correlated (R2 = 0.944 for C. trachomatis and R2 = 0.985 for N. gonorrhoeae) (data not shown). Therefore, the two transport media appeared to perform equivalently in analytical testing.
Table 3.
LOD comparison between the Roche Cobas Amplicor and Abbott RealTime CT/NG testsa
| Dilution | % detection (no. of equivocal or indeterminate results) |
|||||
|---|---|---|---|---|---|---|
|
C. trachomatis |
N. gonorrhoeae |
|||||
| Roche assay with M4-RT medium | Abbott assay with M4-RT medium | Abbott assay with ATB | Roche assay with M4-RT medium | Abbott assay with M4-RT medium | Abbott assay with ATB | |
| 1:100 | 100 (1) | ND | ND | 100 (1) | ND | ND |
| 1:500 | 70 (1) | 100 | 100 | 95 (13) | 100 | 100 |
| 1:2,000 | 25 | 65 | 60 | 30 (6) | 100 | 100 |
| 1:5,000 | 15 | 10 | 25 | 30 (6) | 90 | 100 |
Values represent percentages of 20 replicates tested by the Abbott and Roche methods that were positive at each dilution of the AmpliTrol CT/GC control material. Samples tested by the Abbott method were diluted in either M4-RT medium or Abbott transport buffer (ATB). The numbers of test results in the Roche equivocal range (C. trachomatis) or expanded indeterminate range (N. gonorrhoeae) are indicated in parentheses. Note that these results were scored as positive results for the calculation of the percentages presented in this table (see Materials and Methods for the rationale). ND, testing not done.
DISCUSSION
The agreement between the Abbott RealTime CT/NG test and the Roche Cobas Amplicor CT/NG test was very high, after the resolution of Roche N. gonorrhoeae-positive results using alternative LC methods. Analysis of discrepant results, quantitative data from clinical samples, and limiting dilution studies led to several instructive observations.
First, our data suggested that the Roche method had difficulty in detecting dual infections, generally with male urine specimens, when there were relatively large amounts of N. gonorrhoeae and small amounts of C. trachomatis target based on DC analysis. Notably, all of the Abbott C. trachomatis-positive/Roche C. trachomatis-negative or C. trachomatis-equivocal urine discrepant results (Table 1) were N. gonorrhoeae positive by both methods. The corresponding C. trachomatis DC values were low, indicating that there were only small amounts of C. trachomatis in the samples, and the average ratio of N. gonorrhoeae to C. trachomatis was approximately 16-fold (based on a DC ratio of 4). Two of the four urine specimens available for further analysis were confirmed as being dually positive by AC2. Therefore, we infer that all of these discrepant results were due to true dual infections, below the C. trachomatis detection threshold for the Roche method and potentially in some cases for AC2 as well. However, it should be noted that AC2 testing was performed after freeze-thawing of urine samples, and it is possible that the sample target may therefore have suffered degradation prior to AC2 testing. Furthermore, we recognize that we cannot conclude with absolute certainty that all of the discrepant Abbott dual-positive samples in our study should be resolved in Abbott's favor, a previously noted limitation of discrepant analyses (11).
However, assuming the Abbott dual detection to be accurate, the Roche assay failed to reliably detect C. trachomatis in 5 of 13 (38%) dually infected male urine specimens identified during our study. In contrast, it appeared to miss N. gonorrhoeae in only 1 of 22 (5%) dually infected cervical swab samples. In this sample, the Roche assay was N. gonorrhoeae negative and C. trachomatis positive, and the Abbott C. trachomatis/N. gonorrhoeae ratio was very high at 9, or roughly a 500-fold difference between C. trachomatis and N. gonorrhoeae target concentrations. We hypothesize that the significantly more frequent observation of detection failures for dually infected male urine versus cervical swab specimens (P = 0.02, Fisher's exact test) relates to the relative amounts of N. gonorrhoeae and C. trachomatis generally found in these specimen types. Specifically, there was a significantly lower median DC value for C. trachomatis than for N. gonorrhoeae in male urine specimens (DC of 9.3 versus 13.4, respectively; P < 0.0001) versus a significantly greater median DC value for C. trachomatis than for N. gonorrhoeae in cervical swab specimens (DC of 14.7 versus 12.5, respectively; P < 0.0001). Moreover, the disparity between C. trachomatis and N. gonorrhoeae was generally greater and the absolute amount of C. trachomatis was generally lower in male urine than in cervical swabs specimens. Mechanistically, we therefore postulate that conditions where a much greater amount of N. gonorrhoeae masks a small amount of C. trachomatis were more likely to occur with male urine specimens and led to the failure of the C. trachomatis component of the assay.
Interestingly, in one previous study, the AC2 assay was also found to miss either N. gonorrhoeae or C. trachomatis in approximately 14% of dual infections in a female study population (7). The percentage may in fact have been an underestimate, as the comparators were the Roche Cobas Amplicor assay, the Abbott Ligase Chain Reaction (LCx) assay, and/or culture, all potentially less sensitive methods for the detection of dual infection. The majority of these presumed dual-infection misses were with urine specimens, similar to the observations reported in this study.
Notably, the 2006 CDC sexually transmitted diseases treatment guidelines recommend treatment for N. gonorrhoeae alone when the sample is positive for N. gonorrhoeae and found to be negative for C. trachomatis by a nucleic acid amplification method (NAAT) (23). However, our data raise the concern that the Roche method and potentially the AC2 method as well may not adequately detect low-level C. trachomatis infection, especially in male urine samples from patients coinfected with C. trachomatis and N. gonorrhoeae. Based on current treatment recommendations and extrapolating to patient populations in other health care settings, a significant number of dually infected patients may therefore receive inadequate treatment as a result of the suboptimal ability of some NAATs to detect coinfection. It is therefore imperative that manufacturers optimize and clinical laboratories thoroughly evaluate multiplex PCR tests for adequacy in detecting dual infections. Furthermore, clinicians should be aware of the potential for inadequate detection of dual infections with some of the currently used NAAT methods. Clinicians may therefore potentially wish to either treat patients empirically for dual infection or screen patients at a suitable interval after treatment (e.g., after the 4 weeks generally needed for clearance of the nucleic acid target [7, 13]) to both document successful treatment and rule out infection with initially NAAT-negative C. trachomatis or N. gonorrhoeae.
Second, although the Abbott assay did not initially receive FDA clearance for the detection of C. trachomatis in cervical swab samples, its comparative clinical performance for C. trachomatis detection from this sample type was excellent. Specifically, it was found that the performance characteristics of the Abbott method for cervical swabs appeared at least as good as those of the Roche method. Among our data set, the six Roche C. trachomatis-positive, Abbot C. trachomatis-negative swab specimen discrepant results were likely due to specimen degradation in the cervical specimens, as the 6 specimens that were initially Roche C. trachomatis positive and Abbott C. trachomatis negative were all negative upon repeat testing by the Roche assay and the AC2 assay. Furthermore, the 6 Roche C. trachomatis-negative, Abbott C. trachomatis-positive discrepant results may have resulted from potentially superior detection by the Abbott method. In support of this notion, all samples had low DC values, suggesting low bacterial loads, which likely stress tested the lower limits of detection of both the Abbott and Roche assays. One sample was also N. gonorrhoeae positive and might have been a Roche dual-infection detection failure, as noted above. Finally, one of four discrepant specimens tested by AC2 was positive for C. trachomatis. Although the remaining three samples were negative by AC2, it should be noted that cervical swab samples were diluted 30-fold in AC2 transport buffer prior to nucleic acid extraction, thereby reducing AC2 sensitivity.
Furthermore, DC analysis indicated that cervical swab samples had a greater median level of C. trachomatis target than urine samples (which initially were the only female sample type approved for use with the Abbott assay) and therefore should presumably allow enhanced detection of infection in patients with a very low level of the organism. However, one potential limitation of DC analysis should be noted. It is possible, but we think unlikely, that the much higher C. trachomatis load observed for cervical swab samples may have been specific to our patient population, e.g., if patients screened clinically for C. trachomatis alone using urine samples (based on the Roche test being cleared for the detection of only C. trachomatis and not N. gonorrhoeae for this specimen type) differed from those screened for both C. trachomatis and N. gonorrhoeae in cervical swab samples. Furthermore, it is possible that first-catch urine specimens were not always obtained, thereby leading to dilution of C. trachomatis in an undefined subset of samples.
Third, we similarly noted a higher median DC value for N. gonorrhoeae in male swab samples than in urine samples. However, in this case, it seemed plausible that only men with more symptomatic infection (e.g., obvious exudate) and, therefore, a higher burden of infection were likely to be tested via swab samples, helping to account for these differences. In support of this notion, all of the male swab samples analyzed during the course of this study were positive for N. gonorrhoeae or C. trachomatis. However, the higher DC values are also consistent with previously reported observations with paired collections from asymptomatic men in which swab specimens were clinically more sensitive than urine specimens (10).
Fourth, in limiting dilution studies with C. trachomatis/N. gonorrhoeae control material, the Abbott method appeared to be substantially more sensitive than the Roche method for the detection of N. gonorrhoeae. This difference may relate in part to the detection of the multicopy Opa gene (1) (up to 11 copies per cell) by the former verses the single-copy M.Ngo PII gene, encoding cytosine DNA methyltransferase (12), by the latter assay. The apparently enhanced analytic sensitivity did not translate into observed differences in clinical detection where agreement between the two methods (after resolving Roche N. gonorrhoeae-positive results with LC confirmatory testing) was excellent. However, it should be noted that we did not have any Abbott N. gonorrhoeae-positive female urine samples during the course of this study, likely because of the smaller number of female urine samples received coupled with the much lower prevalence of N. gonorrhoeae in our clinical practice. N. gonorrhoeae-positive female urine specimens may have been a specimen type where differences in N. gonorrhoeae sensitivity would have been more obvious. In support of this notion, the Roche assay was previously shown to have suboptimal sensitivity for the detection of N. gonorrhoeae in female urine samples and was not FDA approved for this indication (10). In contrast, the Roche and Abbott C. trachomatis assays both target the same multicopy cryptic plasmid present in approximately 5 copies per bacterium (15) and, as expected, had more similar analytic sensitivities.
Fifth, we also examined the effects of transport media on the analytical performance of the Abbott method. The use of M4-RT medium in place of the Abbott transport medium appeared to have little if any effect on performance characteristics. Therefore, M4-RT medium may serve as a useful alternative collection medium for some laboratories that wish to simplify the collection of specimens by the use of a single transport medium that will also allow the culturing of viruses and Chlamydia. The one caveat is that we did observe several Roche C. trachomatis-positive specimens becoming negative upon retesting by the Roche, Abbott, and AC2 assays (Roche C. trachomatis-positive, Abbott C. trachomatis-negative specimens in Table 2), which is potentially related to the degradation of C. trachomatis-positive swab specimens upon storage. It is possible that the guanidinium thiocyanate-containing Abbott transfer buffer would have stabilized the target in these specimens through the denaturation of nucleases in the sample.
Finally, it should be noted that the Abbott assay provides a benefit in terms of automation and throughput. On average, the Abbott assay required 1.9 min of hands-on technologist time per patient sample, versus 2.9 min for the Roche assay, assuming testing of the maximum number of patient samples per run. Furthermore, the Roche assay requires manual extraction and PCR setup. In contrast, these functions are automated on the Abbott platform. The total durations of a sample run for each instrument platform were 255 min for the Roche assay and 420 min for the Abbott assay. However, it should be noted that the latter includes time for both automated extraction and PCR setup. Furthermore, the Abbot platform tests up to 93 patient samples per run, while the Roche platform tests only up to 22 patient samples per run.
In summary, the Abbott RealTime CT/NG assay appears to be robust, automated, and accurate in comparison to the well-established Roche Cobas Amplicor CT/NG assay. It did not appear to suffer to an appreciable degree from the nonspecific cross-reactivity in the N. gonorrhoeae portion of the assay noted for some other assays. It also appears to show an enhanced ability to detect dual C. trachomatis and N. gonorrhoeae infections. As such, it is a welcome addition to the nucleic acid amplification testing options for C. trachomatis and N. gonorrhoeae detection.
ACKNOWLEDGMENTS
This study was supported in part by grant support received from Abbott Laboratories.
Evaluation of the real-time, N. gonorrhoeae confirmatory testing methods was performed as part of a verification of assays used in our clinical laboratory practice prior to and independent of the evaluation of the Abbott RealTime CT/NG method.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Abbot Molecular, Inc July 2008. Abbott RealTime CT/NG, in vitro test, ref. 8L07, 51-608201/R1, package insert. Abbott Molecular, Inc., Des Plaines, IL [Google Scholar]
- 2. Blackhawk Biosystems, Inc September 2006. AmpliTrol CT/GC, package insert. Blackhawk Biosystems, Inc., San Ramon, CA [Google Scholar]
- 3. Boel C. H., van Herk C. M., Berretty P. J., Onland G. H., van den Brule A. J. 2005. Evaluation of conventional and real-time PCR assays using two targets for confirmation of results of the COBAS AMPLICOR Chlamydia trachomatis/Neisseria gonorrhoeae test for detection of Neisseria gonorrhoeae in clinical samples. J. Clin. Microbiol. 43:2231–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI 2008. User protocol for evaluation of qualitative test performance; approved guideline, 2nd ed. CLSI document EP12-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Feavers I. M., Maiden M. C. 1998. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol. Microbiol. 30:647–656 [DOI] [PubMed] [Google Scholar]
- 6. Gaydos C. A., et al. 2010. Performance of the Abbott RealTime CT/NG for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 48:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaydos C. A., et al. 2003. Performance of the APTIMA Combo 2 assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in female urine and endocervical swab specimens. J. Clin. Microbiol. 41:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levett P. N., et al. 2008. Evaluation of three automated nucleic acid amplification systems for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in first-void urine specimens. J. Clin. Microbiol. 46:2109–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marshall R., et al. 2007. Characteristics of the m2000 automated sample preparation and multiplex real-time PCR system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 45:747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin D. H., et al. 2000. Multicenter evaluation of AMPLICOR and automated COBAS AMPLICOR CT/NG tests for Neisseria gonorrhoeae. J. Clin. Microbiol. 38:3544–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McAdam A. J. 2000. Discrepant analysis: how can we test a test? J. Clin. Microbiol. 38:2027–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyada C. G., Born T. L. 1991. A DNA sequence for the discrimination of Neisseria gonorrhoeae from other Neisseria species. Mol. Cell. Probes 5:327–335 [DOI] [PubMed] [Google Scholar]
- 13. Morre S. A., et al. 1998. Monitoring of Chlamydia trachomatis infections after antibiotic treatment using RNA detection by nucleic acid sequence based amplification. Mol. Pathol. 51:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmer H. M., Mallinson H., Wood R. L., Herring A. J. 2003. Evaluation of the specificities of five DNA amplification methods for the detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 41:835–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pickett M. A., Everson J. S., Pead P. J., Clarke I. N. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151:893–903 [DOI] [PubMed] [Google Scholar]
- 16. Shain E. B., Clemens J. M. 2008. A new method for robust quantitative and qualitative analysis of real-time PCR. Nucleic Acids Res. 36:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Unemo M., Norlen O., Fredlund H. 2005. The porA pseudogene of Neisseria gonorrhoeae–low level of genetic polymorphism and a few, mainly identical, inactivating mutations. APMIS 113:410–419 [DOI] [PubMed] [Google Scholar]
- 18. U.S. Preventive Services Task Force 2007. Screening for chlamydial infection: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 147:128–134 [DOI] [PubMed] [Google Scholar]
- 19. Van Der Pol B., et al. 2001. Enhancing the specificity of the COBAS AMPLICOR CT/NG test for Neisseria gonorrhoeae by retesting specimens with equivocal results. J. Clin. Microbiol. 39:3092–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whiley D. M., et al. 2006. Evidence that the gonococcal porA pseudogene is present in a broad range of Neisseria gonorrhoeae strains; suitability as a diagnostic target. Pathology 38:445–448 [DOI] [PubMed] [Google Scholar]
- 21. Whiley D. M., et al. 2004. A new confirmatory Neisseria gonorrhoeae real-time PCR assay targeting the porA pseudogene. Eur. J. Clin. Microbiol. Infect. Dis. 23:705–710 [DOI] [PubMed] [Google Scholar]
- 22. Whiley D. M., Tapsall J. W., Sloots T. P. 2006. Nucleic acid amplification testing for Neisseria gonorrhoeae: an ongoing challenge. J. Mol. Diagn. 8:3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Workowski K. A., Berman S. M. 2006. Sexually transmitted diseases treatment guidelines, 2006. MMWR Recommend. Rep. 55(RR-11):1–94 [PubMed] [Google Scholar]
- 24. Workowski K. A., Berman S. M. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recommend. Rep. 59(RR-12):1–110 [PubMed] [Google Scholar]