Abstract
Clostridium perfringens is a ubiquitous and versatile pathogenic bacterium and is implicated in the etiology of the poultry diseases necrotic enteritis (NE) and poultry gangrene (PG). In this study, multilocus sequence typing was used to investigate genotypic relationships among 139 C. perfringens isolates from 74 flocks. These isolates had multiple disease, host, and environmental origins. The results indicated a polymorphic yet highly clonal population, with 79.6% of all isolates partitioning into one of six clonal complexes or two dominant sequence types, ST-9 and ST-31. The most prolific clonal complex, CC-1, contained 27.3% of all isolates and was not clearly associated with one particular disease. The subtypes CC-4 and ST-31 were highly associated with NE and represented 9.4% and 7.2% of the total isolates, respectively. No PG-associated and NE-associated C. perfringens isolates shared the same sequence type or clonal complex. NE-associated subtypes were more clonal and appeared more evolutionarily divergent than PG-associated subtypes, which tended to cluster in the more ancestral lineages alongside isolates from asymptomatic chickens and turkeys. Toxin gene screening identified cpb2 throughout these isolates and correlated the presence of netB with NE pathology. Previous investigations into the genetic basis of C. perfringens pathogenicity have focused on toxins and other variable genetic elements. This study presents the first sequence-based comparison of C. perfringens isolates recovered in clinical cases of PG and NE and demonstrates that niche specialization is observable in the core genomes of poultry-associated C. perfringens isolates, a concept with both epidemiological and evolutionary significance.
INTRODUCTION
Clostridium perfringens is a Gram-positive spore-forming anaerobe considered to be one of the most common pathogenic bacteria in nature (36). C. perfringens resides primarily in soil, sediments, the alimentary tracts of animals, and feces and is recognized for its array of potent exotoxins, including food-borne emetic toxin, and the ability to initiate or exacerbate necrotizing soft tissue infections, such as gas gangrene (11). C. perfringens-mediated diseases are a significant cause of economic loss to livestock industries (1, 40, 43). In poultry, C. perfringens is directly implicated in the etiology of necrotic enteritis (NE) and is often associated with poultry gangrene (PG), a progressive necrosis of soft tissues (23). Phylogenetic studies to date have revealed considerable genetic diversity within C. perfringens, as well as evidence of niche partitioning correlated with human disease; the most striking example is an association between specific strains and the incidence of food-borne enterotoxemia (24, 28, 31, 32). Phylogenetic relationships and niche partitioning among isolates associated with the poultry diseases NE and PG have yet to be fully investigated.
NE can occur in both subclinical and acute forms in poultry, resulting in decreased production efficiency parameters and, in some outbreaks, mortality as high as 50% within a flock (4, 23). Key risk factors include the prerequisite disturbance of the intestinal mucosa, often via coccidial infection by Eimeria spp., diets with high levels of protein and energy, and high-viscosity diets, particularly those including wheat or barley (4, 23). Enteric mucosal disturbance with high dietary viscosity and nutrient availability creates optimal conditions for the proliferation of endogenous C. perfringens, which is accompanied by toxin production and subsequent NE pathology.
Poultry gangrene infections commonly involve C. perfringens and other members of the histotoxic clostridia, primarily Clostridium septicum, although some nonclostridial species have also been implicated (12, 21, 39). Poultry gangrene can be initiated via environmental contamination of an external wound; however, recent evidence supports an endogenous, likely gastrointestinal, route of infection in cases of subcutaneous gas gangrene (26). Despite its presence in infected tissues, the contribution of C. perfringens to the development of PG remains unclear.
Genetic relationships between C. perfringens strains of disparate origins, including human and livestock host species, have been investigated (3, 10, 16, 32, 34). Such phylogenetic studies have elucidated distinct lineages associated with specific disease syndromes attributed to this pathogen. The most obvious association is a clear partitioning of strains capable of inducing food-borne illness into lineages distinct from those of strains isolated from other niches, e.g., gangrenous muscle and healthy gastrointestinal tracts (GITs) (24, 28). Investigations into the genetic relationships among C. perfringens strains recovered from poultry with NE and isolates obtained from asymptomatic poultry tissues, alimentary tracts, and the production environment have identified distinct subtypes associated with NE that are capable of inducing disease in experimental NE models, whereas isolates from other lineages do not reproduce the disease (5, 20, 40). Recently, the presence of a novel toxin, NetB, was shown to be highly correlated with C. perfringens isolates implicated in NE disease, suggesting that certain poultry strains of C. perfringens have acquired specialized pathogenic capabilities (17, 18, 20). Toxin production by C. perfringens with subsequent detection in affected tissues is often considered diagnostic for specific diseases caused by these bacteria and forms the basis for a typing scheme based on the presence of the four major toxins—α, β, ε, and ι—produced by certain strains of the species (11, 36).
In this study, multilocus sequence typing (MLST) was utilized to investigate phylogenetic relationships and disease or niche partitioning in a diverse collection of C. perfringens isolates from broiler chickens and turkeys. The MLST technique offers unambiguous, high-resolution results, in the form of sequence data and subsequent allelic profile data, useful for analyses of population phylogeny and clonal characteristics, e.g., BURST (based upon related sequence types) analysis (14, 16). MLST has proven useful in linking specific C. perfringens subtypes to distinct pathotypes, e.g., type C subtypes in porcine enteritis and human food poisoning subtypes (16, 28). The typing scheme employed here is based largely on those previously published for this species (2, 16). Additional loci were included to allow for a population-level interspecies comparison between this collection of C. perfringens and a C. septicum population of poultry origin from a previous study published by our group (27). This investigation is, to our knowledge, the first to specifically compare C. perfringens isolates recovered from both NE and PG, and it provides insight into the phylogenetic and etiological characteristics of the isolates responsible for each disease.
MATERIALS AND METHODS
Clostridium perfringens isolates and demographics.
In order to examine a broad genotypic range of isolates from poultry gangrene, necrotic enteritis, and asymptomatic conditions, target isolates were selected for MLST partially on the basis of unique RAPD (random amplification of polymorphic DNA) PCR profiles. RAPD profiles were generated with Ready-To-Go RAPD analysis beads and Primer 2, provided with the kit (GE Healthcare, Piscataway, NJ) (44). The isolates included were collected from 74 poultry flocks in 8 states of the United States (Alabama, Delaware, Georgia, Minnesota, Missouri, Pennsylvania, Virginia, and Wisconsin) between 2004 and 2009.
A total of 65 PG-associated and 20 poultry NE-associated C. perfringens isolates were included in the analysis. They are listed in full in Table 1 and are summarized by origin in Table 2. PG-associated isolates were cultured from 39 birds displaying symptoms of clostridial myonecrosis. NE-associated isolates were cultured from the GITs of 20 birds with typical necrotic enteritis GIT pathology. Fifty-four isolates with no strict disease association were also included for comparison. These isolates were derived from 12 asymptomatic flockmates of the birds with PG, 8 birds from farms where PG was not endemic in production systems with histories of PG, 15 birds from production systems with recent histories of NE, and 8 birds in production systems where NE is uncommon, as well as from environmental sources, including litter and Alphitobius diaperinus beetles from a production system with recent NE and 8 litter samples from a turkey production system with a history of PG disease. PG-associated isolates were representative of both turkeys (n = 28) and broiler chickens (n = 37), while NE-associated isolates were all of broiler chicken origin (n = 20). Collected isolates with no strict disease association were representative of turkey (n = 18) and broiler chicken (n = 36) production settings. In total, of the 139 isolates (excluding reference strains), 79 were of GIT origin, 41 were obtained from infected muscle, 6 were from liver, 1 was from blood, 1 was from spleen, and the remainder were derived from environmental sources.
Table 1.
Characterization of isolates by sequence type, disease association, and toxin genes
| Isolate | Geography | Host | Source | Disease association | Sequence type | Toxin gene |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| plc | cpb | etx | iA | cpb2 | cpe | netB | ||||||
| AGTP01-0001 | WI | Turkey | Muscle | PG | ST-1 | + | − | − | − | + | − | − |
| AGTP01-0002 | MO | Turkey | Muscle | PG | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0003 | MO | Turkey | Muscle | PG | ST-3 | + | − | − | − | + | − | − |
| AGTP01-0004 | DE | Broiler | GIT | ASd | ST-4 | + | − | − | − | − | − | − |
| AGTP01-0005 | WI | Turkey | Muscle | PG | ST-5 | + | − | − | − | − | − | − |
| AGTP01-0006 | WI | Turkey | Muscle | PG | ST-6 | + | − | − | − | + | − | − |
| AGTP01-0007 | WI | Turkey | Muscle | PG | ST-3 | + | − | − | − | − | − | − |
| AGTP01-0008 | WI | Turkey | Muscle | PG | ST-7 | + | − | − | − | + | − | − |
| AGTP01-0009 | WI | Turkey | Muscle | PG | ST-6 | + | − | − | − | + | − | − |
| AGTP01-0010 | MO | Turkey | Muscle | PG | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0011 | WI | Turkey | Muscle | PG | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0012 | DE | Broiler | GIT | AS | ST-9 | + | − | − | − | − | − | − |
| AGTP01-0013 | DE | Broiler | GIT | AS | ST-10 | + | − | − | − | + | − | − |
| AGTP01-0014 | DE | Broiler | Blood | AS | ST-9 | + | − | − | − | − | − | − |
| AGTP01-0015 | DE | Broiler | Liver | AS | ST-10 | + | − | − | − | − | − | − |
| AGTP01-0016 | DE | Broiler | Muscle | PG | ST-11 | + | − | − | − | − | − | − |
| AGTP01-0017 | DE | Broiler | GIT | AS | ST-9 | + | − | − | − | − | − | − |
| AGTP01-0018 | DE | Broiler | Muscle | PG | ST-12 | + | − | − | − | − | − | − |
| AGTP01-0019 | WI | Turkey | Muscle | PG | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0020 | MO | Turkey | Muscle | PG | ST-3 | + | − | − | − | + | − | − |
| AGTP01-0021 | MO | Turkey | Muscle | PG | ST-13 | + | − | − | − | − | − | − |
| AGTP01-0022 | WI | Turkey | Muscle | PG | ST-14 | + | − | − | − | − | − | − |
| AGTP01-0023 | MN | Turkey | Muscle | PG | ST-15 | + | − | − | − | + | − | − |
| AGTP01-0024 | MN | Turkey | Muscle | PG | ST-15 | + | − | − | − | + | − | − |
| AGTP01-0025 | DE | Broiler | GIT | PG | ST-3 | + | − | − | − | + | − | − |
| AGTP01-0026 | DE | Broiler | Muscle | PG | ST-16 | + | − | − | − | − | − | − |
| AGTP01-0027 | DE | Broiler | Muscle | PG | ST-10 | + | − | − | − | + | − | − |
| AGTP01-0028 | DE | Broiler | Muscle | PG | ST-11 | + | − | − | − | − | − | − |
| AGTP01-0029 | DE | Broiler | Muscle | PG | ST-15 | + | − | − | − | − | − | − |
| AGTP01-0030 | DE | Broiler | Liver | PG | ST-10 | + | − | − | − | + | − | − |
| AGTP01-0031 | DE | Broiler | Muscle | PG | ST-17 | + | − | − | − | + | − | − |
| AGTP01-0032 | DE | Broiler | Muscle | PG | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0033 | DE | Broiler | Muscle | PG | ST-17 | + | − | − | − | − | − | − |
| AGTP01-0034 | DE | Broiler | Liver | PG | ST-18 | + | − | − | − | − | − | − |
| AGTP01-0035 | DE | Broiler | Muscle | PG | ST-19 | + | − | − | − | + | − | − |
| AGTP01-0036 | DE | Broiler | Muscle | PG | ST-20 | + | − | − | − | + | − | − |
| AGTP01-0037 | DE | Broiler | Muscle | PG | ST-21 | + | − | − | − | + | − | − |
| AGTP01-0038 | DE | Broiler | Muscle | PG | ST-18 | + | − | − | − | + | − | − |
| AGTP01-0039 | DE | Broiler | Muscle | PG | ST-18 | + | − | − | − | + | − | − |
| AGTP01-0040 | DE | Broiler | Muscle | PG | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0041 | MN | Turkey | Muscle | PG | ST-3 | + | − | − | − | + | − | − |
| AGTP01-0042 | DE | Broiler | Muscle | PG | ST-17 | + | − | − | − | + | − | − |
| AGTP01-0043 | MO | Turkey | Muscle | PG | ST-22 | + | − | − | − | + | − | − |
| AGTP01-0044 | VA | Turkey | Muscle | PG | ST-23 | + | − | − | − | + | − | − |
| AGTP01-0045 | VA | Turkey | Muscle | PG | ST-24 | + | − | − | − | + | − | − |
| AGTP01-0046 | VA | Turkey | Muscle | PG | ST-14 | + | − | − | − | − | − | − |
| AGTP01-0047 | MN | Turkey | Muscle | PG | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0048 | DE | Broiler | GIT | AS | ST-4 | + | − | − | − | + | − | − |
| AGTP01-0049 | DE | Broiler | GIT | PG | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0050 | GA | Broiler | GIT | PG | ST-25 | + | − | − | − | + | − | − |
| AGTP01-0051 | GA | Broiler | GIT | AS | ST-9 | + | − | − | − | + | − | − |
| AGTP01-0052 | GA | Broiler | GIT | PG | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0053 | GA | Broiler | GIT | PG | ST-9 | + | − | − | − | + | − | − |
| AGTP01-0054 | DE | Broiler | GIT | PG | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0055 | DE | Broiler | GIT | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0056 | DE | Broiler | GIT | PG | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0057 | MN | Turkey | GIT | PG | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0058 | MN | Turkey | GIT | PG | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0059 | MN | Turkey | GIT | AS | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0060 | MN | Turkey | GIT | AS | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0061 | DE | Broiler | GIT | PG | ST-4 | + | − | − | − | + | − | − |
| AGTP01-0062 | MN | Turkey | GIT | PG | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0063 | MN | Turkey | GIT | AS | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0064 | MN | Turkey | GIT | PG | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0065 | MN | Turkey | GIT | AS | ST-27 | + | − | − | − | + | − | − |
| AGTP01-0066 | MN | Turkey | GIT | AS | ST-9 | + | − | − | − | − | − | − |
| AGTP01-0067 | MN | Turkey | GIT | AS | ST-28 | + | − | − | − | + | − | − |
| AGTP01-0068 | MN | Turkey | GIT | AS | ST-29 | + | − | − | − | + | − | − |
| AGTP01-0069 | DE | Broiler | GIT | AS | ST-12 | + | − | − | − | + | − | − |
| AGTP01-0070 | MN | Turkey | GIT | PG | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0071 | MN | Turkey | GIT | AS | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0072 | MN | Turkey | GIT | PG | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0073 | MN | Turkey | GIT | AS | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0074 | MN | Turkey | GIT | AS | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0075 | DE | Broiler | Muscle | PG | ST-17 | + | − | − | − | + | − | − |
| AGTP01-0076 | DE | Broiler | Muscle | PG | ST-12 | + | − | − | − | + | − | − |
| AGTP01-0077 | DE | Broiler | Muscle | PG | ST-12 | + | − | − | − | − | − | − |
| AGTP01-0078 | DE | Broiler | Liver | PG | ST-15 | + | − | − | − | − | − | − |
| AGTP01-0079 | DE | Broiler | Liver | PG | ST-17 | + | − | − | − | − | − | − |
| AGTP01-0080 | DE | Broiler | Spleen | PG | ST-17 | + | − | − | − | + | − | − |
| AGTP01-0081 | DE | Broiler | GIT | PG | ST-15 | + | − | − | − | − | − | − |
| AGTP01-0082 | MN | Turkey | Litter | AS | ST-2 | + | − | − | − | + | − | − |
| AGTP01-0083 | MN | Turkey | Litter | AS | ST-9 | + | − | − | − | − | − | − |
| AGTP01-0084 | DE | Broiler | Liver | PG | ST-10 | + | − | − | − | − | − | − |
| AGTP01-0085 | DE | Broiler | GIT | PG | ST-4 | + | − | − | − | − | − | − |
| AGTP01-0086 | MN | Turkey | Litter | AS | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0087 | DE | Broiler | GIT | PG | ST-15 | + | − | − | − | + | − | − |
| AGTP01-0088 | DE | Broiler | GIT | PG | ST-12 | + | − | − | − | + | − | − |
| AGTP01-0089 | MN | Turkey | Litter | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0090 | MN | Turkey | Litter | AS | ST-6 | + | − | − | − | + | − | − |
| AGTP01-0091 | MN | Turkey | Litter | AS | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0092 | MN | Turkey | Litter | AS | ST-30 | + | − | − | − | + | − | − |
| AGTP01-0093 | MN | Turkey | Litter | AS | ST-26 | + | − | − | − | + | − | − |
| AGTP01-0101 | MN | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0102 | MN | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0103 | MN | Broiler | GIT | NE | ST-39 | + | − | − | − | + | − | + |
| AGTP01-0104 | MN | Broiler | GIT | NE | ST-32 | + | − | − | − | + | − | + |
| AGTP01-0105 | MN | Broiler | GIT | NE | ST-32 | + | − | − | − | + | − | + |
| AGTP01-0106 | MN | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0107 | MN | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0108 | MN | Broiler | GIT | NE | ST-39 | + | − | − | − | + | − | + |
| AGTP01-0109 | MN | Broiler | GIT | NE | ST-39 | + | − | − | − | + | − | + |
| AGTP01-0110 | MN | Broiler | GIT | NE | ST-32 | + | − | − | − | + | − | + |
| AGTP01-0111 | PA | Broiler | GIT | NE | ST-33 | + | − | − | − | + | − | − |
| AGTP01-0112 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | − |
| AGTP01-0113 | PA | Broiler | GIT | AS | ST-10 | + | − | − | − | + | − | + |
| AGTP01-0114 | PA | Broiler | GIT | AS | ST-34 | + | − | − | − | + | − | + |
| AGTP01-0116 | PA | Broiler | GIT | AS | ST-35 | + | − | − | − | − | − | − |
| AGTP01-0117 | PA | Broiler | GIT | AS | ST-36 | + | − | − | − | − | − | + |
| AGTP01-0118 | PA | Broiler | GIT | AS | ST-34 | + | − | − | − | + | − | + |
| AGTP01-0119 | PA | Broiler | GIT | AS | ST-37 | + | − | − | − | + | − | − |
| AGTP01-0121 | PA | Broiler | GIT | AS | ST-37 | + | − | − | − | + | − | − |
| AGTP01-0122 | PA | Broiler | GIT | AS | ST-34 | + | − | − | − | − | − | + |
| AGTP01-0123 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0124 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0125 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0126 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0127 | PA | Broiler | GIT | NE | ST-31 | + | − | − | − | + | − | + |
| AGTP01-0128 | PA | Broiler | GIT | NE | ST-38 | + | − | − | − | + | − | + |
| AGTP01-0129 | AL | Broiler | GIT | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0130 | AL | Broiler | GIT | AS | ST-33 | + | − | − | − | + | − | + |
| AGTP01-0131 | AL | Broiler | GIT | AS | ST-30 | + | − | − | − | + | − | − |
| AGTP01-0132 | AL | Broiler | GIT | AS | ST-33 | + | − | − | − | + | − | + |
| AGTP01-0133 | AL | Broiler | GIT | AS | ST-40 | + | − | − | − | + | − | − |
| AGTP01-0134 | AL | Broiler | GIT | AS | ST-33 | + | − | − | − | + | − | + |
| AGTP01-0135 | AL | Broiler | GIT | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0136 | AL | Broiler | GIT | AS | ST-33 | + | − | − | − | + | − | + |
| AGTP01-0137 | AL | Broiler | GIT | AS | ST-4 | + | − | − | − | + | − | − |
| AGTP01-0138 | AL | Broiler | GIT | AS | ST-41 | + | − | − | − | + | − | − |
| AGTP01-0139 | AL | Broiler | GIT | AS | ST-4 | + | − | − | − | + | − | − |
| AGTP01-0140 | AL | Broiler | GIT | AS | ST-14 | + | − | − | − | + | − | − |
| AGTP01-0141 | AL | Broiler | GIT | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0142 | AL | Broiler | GIT | AS | ST-17 | + | − | − | − | + | − | − |
| AGTP01-0143 | GA | Broiler | Beetle (larvae)a | AS | ST-4 | + | − | − | − | + | − | − |
| AGTP01-0144 | GA | Broiler | Beetle (adult)a | AS | ST-39 | + | − | − | − | + | − | − |
| AGTP01-0145 | GA | Broiler | GITb | NE | ST-34 | + | − | − | − | + | − | − |
| AGTP01-0146 | GA | Broiler | Litter | AS | ST-8 | + | − | − | − | + | − | − |
| AGTP01-0147 | GA | Broiler | GITb | AS | ST-9 | + | − | − | − | − | − | + |
| AGTP01-0148 | GA | Broiler | GITb | NE | ST-39 | + | − | − | − | + | − | + |
| ATCC 13124c | Gas gangrene | ST-42 | + | − | − | − | − | − | − | |||
| SM101c | Food poisoning | ST-43 | + | − | − | − | − | + | − | |||
| Strain 13c | Soil | ST-44 | + | − | − | − | + | − | − | |||
Table 2.
Origins of isolates from poultry or poultry environments examined in this study
| Disease and source | No. of isolates |
||
|---|---|---|---|
| By host |
Total | ||
| Broiler | Turkey | ||
| NEa | |||
| GIT | 20 | 0 | 20 |
| Subtotal | 20 | 0 | 20 |
| PGb | |||
| Muscle | 19 | 22 | 41 |
| GITc | 12 | 6 | 18 |
| Liverc | 5 | 0 | 5 |
| Spleenc | 1 | 0 | 1 |
| Subtotal | 37 | 28 | 65 |
| Noned | |||
| GIT | 31 | 10 | 41 |
| Liver | 1 | 0 | 1 |
| Blood | 1 | 0 | 1 |
| Litter | 1 | 8 | 9 |
| Beetles | 2 | 0 | 2 |
| Subtotal | 36 | 18 | 54 |
| Total | 93 | 46 | 139 |
Isolates were recovered from 20 birds with symptoms of necrotic enteritis confirmed by typical intestinal gross pathology.
Isolates were recovered from 39 birds with symptoms of poultry gangrene confirmed by typical gross pathology of clostridial myonecrosis.
Isolates were recovered from birds with characteristic symptoms of poultry gangrene but were derived from tissues other than the gangrenous muscle lesions themselves.
Isolates were obtained from the poultry environment and from birds with no apparent signs of disease.
Collection of NE samples.
Live chickens with outward signs of necrotic enteritis (e.g., depression and/or diarrhea) during outbreaks with a characteristic mortality spike were euthanized by cervical dislocation and were dissected in order to obtain the portion of the GIT from the duodenum to the ileocecal junction. GIT tissues were shipped overnight to the lab on ice, but not frozen. Upon arrival, sections of the duodenum, jejunum, and ileum were excised, flushed internally with 10 ml of sterile 0.1% peptone water (Becton, Dickinson and Company, Sparks, MD) to expel the luminal contents, and sliced longitudinally to expose the mucosa. The identities of tracts with characteristic gross NE pathology (thin, friable, diphtheritic membranes and “Turkish towel” appearance, etc.) were recorded for subsequent bacterial (sub)type association (4).
Collection of PG samples.
Samples from PG-associated birds and poultry environments were collected and processed as described previously (27). Birds displaying signs of clostridial myonecrosis (i.e., depression, ataxia, crepitant patches of skin with underlying edema) were identified, and tissue samples (gangrenous muscle lesions, GIT, liver, and occasionally spleen and/or blood) were collected by trained personnel and were shipped overnight on ice for microbiological analysis. GIT sections were processed like NE samples, while muscle and organ samples were rinsed externally with 0.1% peptone water (Becton, Dickinson and Company). Blood samples were diluted in sterile 0.1% peptone water (Becton, Dickinson and Company) and were plated directly onto the appropriate selective medium as detailed below.
Collection of samples from asymptomatic poultry and environmental sources.
Samples from asymptomatic poultry were collected and processed in a manner similar to that described for NE and PG samples. Litter samples were collected from multiple areas within a sampled poultry house. Eleven grams of litter was diluted with 99 ml of sterile 0.1% peptone water (Becton, Dickinson and Company), and bacteria were extracted with a stomacher (IUL, Barcelona, Spain) operating at 5 strokes/s for 60 s prior to plating as described below. Litter beetles (Alphitobius diaperinus) were ground in a sterile 1.5-ml microcentrifuge tube with a plastic pestle prior to being streaked onto agar plates for bacterial isolation.
Processing of GIT and tissue samples.
The excised sections of duodenum, jejunum, and ileum described above were combined by bird; 99 ml of sterile 0.1% peptone water (Becton, Dickinson and Company) was added; and the mucosa was extracted with a stomacher (IUL, Barcelona, Spain) operating at 5 strokes/s for 60 s. Other tissues (gangrenous muscle lesions, GIT, and liver, etc.) were combined with 99 ml of sterile 0.1% peptone water (Becton, Dickinson and Company) and were extracted as described for GITs unless indicated otherwise above (i.e., blood and beetle samples). Samples were plated on Perfringens Agar Base (tryptose sulfite cycloserine [TSC] agar [Oxoid Ltd., Cambridge, United Kingdom] supplemented with 400 mg/liter d-cycloserine and 5% egg yolk emulsion) using an Autoplate 4000 spiral plater (Spiral Biotech, Norwood, MA). After approximately 15 min, plates were overlaid with Perfringens Agar Base, prepared as described above but without egg yolk emulsion. Plates were incubated anaerobically (AnaeroPack system; Mitsubishi Gas Chemical America, Inc., New York, NY) for 48 h at 37°C, after which presumptive C. perfringens isolates were identified by hydrogen sulfide production (black colony color) and lecithinase production (outer halo of egg yolk hydrolysis). Discrete morphology-positive colonies were used to inoculate Bacto brain heart infusion broth (Becton, Dickinson, and Company) supplemented with 5 g/liter yeast extract (Fisher BioReagents, Fair Lawn, NJ) and 0.5 g/liter l-cysteine (Fisher BioReagents), and cultures were grown for 24 h at 37°C under anaerobic conditions. Total genomic DNA was isolated from the broth cultures with the DNeasy 96 blood and tissue kit (Qiagen Inc., Valencia, CA). Toxin types were established by PCR toxinotyping (45).
Toxin gene screening.
All isolates used in the MLST scheme were toxinotyped (Table 1) using PCR assays for the α, β, ε, ι, and β2 toxin genes and the C. perfringens enterotoxin (CPE) gene (9, 22, 44). Isolates were also assayed for the presence of netB by using a previously published PCR assay (17). Suitable reference isolates for toxin types were included as positive controls in the toxinotyping scheme.
Sequencing of housekeeping genes.
DNA was isolated from broth cultures of confirmed C. perfringens strains using one of two methods: either a High Pure PCR template preparation kit (Roche, Mannheim, Germany) or a DNeasy 96 blood and tissue kit (Qiagen Inc., Valencia, CA). PCR amplification and sequencing of each of the 11 chromosomally located housekeeping gene segments were performed using the primer sets and annealing temperatures listed in Table 3. The primers developed for this study were designed using Primer 3 (Integrated DNA Technologies) (29). PCR mixtures contained 5 μl 10× PCR buffer, 1.5 μl 50 mM MgCl2, 1 μl 10 mM deoxynucleoside triphosphates (dNTPs), 10 pmol each primer, 2.5 U Platinum Taq (Invitrogen, Carlsbad, CA), 1 μl template DNA (not quantified), and sterile distilled H2O to a final volume of 50 μl. Reactions were performed with initial denaturation for 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at the appropriate annealing temperature, and 1 min/kb of expected product at 72°C, with a final extension of 2 min at 72°C. Agarose gel electrophoresis (1% agarose, Tris-borate-EDTA [TBE]) was performed to confirm the presence and correct size of the PCR amplicon. PCR products were purified (PureLink PCR purification kit; Invitrogen) and sequenced in both directions (Eurofins MWG Biotech, Huntsville, AL).
Table 3.
Primers for C. perfringens housekeeping genes
| Gene | Primera | Sequence (5′ → 3′) | Amplicon size (bp) | Annealing temp (°C) | Source or reference |
|---|---|---|---|---|---|
| ddl | F | AGTGGTAATTCTGTAGTTCATGCCT | 700 | 55 | This study |
| R | CCCTTACTAGCATGTCTACTCTAGC | This study | |||
| dnaK | F | CTCAGAAGGTGCTAGAACAACTCCA | 710 | 58 | This study |
| R | TTGGACCAGTTGCATCAGCAGT | This study | |||
| glpK | F | TCCTAGAGAAGGATGGGTTGAGCA | 619 | 57 | This study |
| R | CCCAGCTATTCCAGCTATAGGAAC | This study | |||
| recA | F | GCTTTAGGAATTGGTGGAGTACCA | 734 | 57 | This study |
| R | CCATATGAGAACCAAGCTCCACTC | This study | |||
| gyrA | F | CAGTTTATGATGCATTAGTTAGAATGGCAC | 751 | 57 | This study |
| R | GCAAGCATTATAACTCCAAATGTGTCTTGC | This study | |||
| groEL | F | AGCAAGAGAAATTGAACTTGAAGATGC | 750 | 58 | This study |
| R | CCGCCTACTTCATCAGATATAACAACGCC | This study | |||
| tpi | F | ACTCCAATAATCGCAGGAAACTGG | 711 | 57 | This study |
| R | TGCAACTAAGCTAGCTCCACCAAC | This study | |||
| plc | F | AGCTTATTCTATACCTGACACAGGG | 630 | 57 | This study |
| R | CCTGGGTTGTCCATTTCCCATTCT | This study | |||
| dut | F | TTAAGTATTTTGATAACGCAAC | 441 | 50 | 16 |
| R | CTGTAGTACCAAATCCACCACG | 16 | |||
| gmk | F | TAAGGGAACTATTTGTAAAGCC | 475 | 50 | 16 |
| R | TACTGCATCTTCTACATTATCG | 16 | |||
| sod | F | GATGCTTTAGAGCCATCAATAG | 475 | 50 | 16 |
| R | AATAATAAGCATGTTCCCAAAC | 16 |
F, forward primer; R, reverse primer.
Multilocus sequence typing and phylogenetic analysis.
Bidirectional sequencing reads for each of the 11 housekeeping gene segments were aligned and trimmed to a uniform length (Bionumerics, version 6.0; Applied Maths, Inc., Austin, TX). Ambiguities were resolved when appropriate with the CAP3 Sequence Assembly Program (13) (http://pbil.univ-lyon1.fr/cap3.php) and by examination of the raw electropherograms in Bionumerics. Nucleotide sequence data for three Clostridium perfringens reference strains, ATCC 13124, SM101, and strain 13, were obtained from the NCBI repository and were included in subsequent analyses for purposes of comparison (24, 33). Once imported, aligned, and trimmed, the sequences from all 142 isolates were compared first by allele and then by allelic profile using Bionumerics, at which point the data were used to assign each isolate to a sequence type (ST). After initial MLST analysis in Bionumerics, the START2 software package (available at http://pubmlst.org/software/analysis/start2/) was used to assemble and analyze concatenated sequence information for each ST (14). Based on a representative of each ST, the Maynard-Smith index of association (IA) was calculated to test for recombination, and the ratio of synonymous to nonsynonymous mutations (dN/dS) was computed by the Nei-Gojobori method as a measure of selection (25, 35). The eBURST tool for MLST analysis (available at http://eburst.mlst.net/default.asp) was used to identify clonal complexes (CCs) (defined by 10 or more identical loci), singleton STs, and possible ancestral genotypes (7, 41). Both STs and CCs were considered to be C. perfringens MLST subtypes.
Concatenated sequence data for a representative of each distinct ST were imported into the MEGA 4.0 software package (http://www.megasoftware.net/) in order to examine isolate and ST relatedness at sequence-level resolution. After complete deletion of alignment gaps, a total of 5,758 positions were used in each concatenated sequence as a data set for phylogeny calculations. An evolutionary phylogeny was constructed in MEGA 4 using the neighbor-joining method and maximum composite likelihood (MCL) to estimate evolutionary distances, and topology was validated by bootstrapping (1,000 replicates) (8, 30, 37, 38). To establish evolutionary relevance, the reference strain SM101 (ST-43) was used to root the tree. The optimum tree generated was condensed where bootstrap support for the clustering of taxa was less than 50% of the replicates (37).
Statistical analysis.
All statistical comparisons were performed in GraphPad Prism, version 5.0 (GraphPad Software, Inc., La Jolla, CA). Fisher's exact test was used to test the correlation of toxin gene detection with disease status.
Nucleotide sequence accession numbers.
The nucleotide sequences of the loci analyzed in this study were submitted to GenBank under accession numbers HM624505 to HM624643 (ddl), HM624366 to HM624504 (dnaK), HM624088 to HM624226 (glpK), HM624922 to HM625060 (recA), HM625200 to HM625338 (gyrA), HM625339 to HM625477 (groEL), HM624644 to HM624782 (tpi), HM625061 to HM625199 (plc), HM624227 to HM624365 (dut), HM625478 to HM625616 (gmk), and HM624783 to HM624921 (sod).
RESULTS
Allele sequencing and gene characteristics.
The first identified allele of the dnaK gene contained an internal 18-bp deletion, and allele 11 of the dut gene contained an internal 3-bp insertion. The total number of alleles for all loci analyzed averaged 12.2, with the most polymorphic gene, glpK, possessing 16 alleles and the least polymorphic, gmk, possessing 5 alleles (Table 4). The average p-distance (number of nucleotide differences/total nucleotides) was used as an indicator of polymorphism within a set of alleles. The greatest average p-distance was observed for the sod gene, at 0.028 difference/nucleotide, and the lowest for the gmk gene, with 0.011 difference/nucleotide (Table 4). These cursory indices of polymorphism were fully supported by examination of the percentage of polymorphic sites per allele; the maximum percentage of polymorphism was found, again, in the sod gene, in which 7.65% of sites were polymorphic, and the minimum in the gmk gene, in which 1.07% of sites were polymorphic (Table 4). All allele sequences examined were coding sequences; thus, the ratio of nonsynonymous to synonymous mutations was used as a measure of selective pressure on each allele. Based on this analysis, all genes possessed dN/dS ratios of <1, indicating purifying selection (Table 4). Two genes, recA and sod, possessed dN/dS values of zero; all mutations present in those genes were identified as synonymous. Significant linkage disequilibrium was detected between the genes examined, as determined by a Maynard-Smith IA (35) value of 2.635 (P, < 0.001, based on one representative of each ST).
Table 4.
Allele information
| Gene | Function | Sequence (bp)a | No. of alleles | No. (%) of polymorphic sites | Avg p-distanceb | dN/dSc |
|---|---|---|---|---|---|---|
| ddl | d-Ala-d-Ala ligase | 588 | 15 | 33 (5.61) | 0.019 | 0.1155 |
| dnaK | Chaperone protein | 708 | 14 | 37 (5.23) | 0.013 | 0.0598 |
| dut | dUTP nucleotidohydrolase | 340 | 13 | 21 (6.18) | 0.017 | 0.817 |
| glpK | Glycerol kinase | 522 | 16 | 20 (3.83) | 0.015 | 0.0552 |
| gmk | Deoxyguanylate kinase | 373 | 5 | 4 (1.07) | 0.011 | 0.5211 |
| groEL | Chaperonin | 597 | 14 | 28 (4.69) | 0.016 | 0.0356 |
| gyrA | DNA topoisomerase | 693 | 10 | 20 (2.89) | 0.014 | 0.0779 |
| plc | Phospholipase C (alpha toxin) | 440 | 15 | 26 (5.91) | 0.020 | 0.0918 |
| recA | DNA repair | 564 | 8 | 27 (4.79) | 0.020 | 0.0000 |
| sod | Superoxide dismutase | 366 | 13 | 28 (7.65) | 0.028 | 0.0000 |
| tpiA | Triosephosphate isomerase | 588 | 11 | 14 (2.38) | 0.012 | 0.0157 |
Sequence types and eBURST analysis.
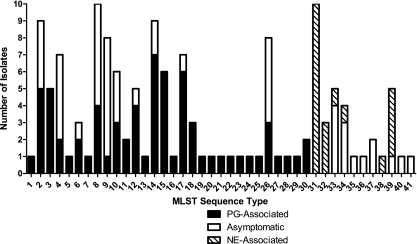
From the 139 isolates typed by MLST, 41 unique sequence types (STs) were identified (Fig. 1). The C. perfringens reference strains ATCC 13124, strain 13, and SM101 segregated into individual sequence types containing a single isolate each, bringing the total number of STs to 44. In five of the eight cases where multiple isolates were obtained from an individual bird with PG, multiple STs were identified. On average, each ST contained 3.2 isolates, with the most prolific STs, ST-8 and ST-31, containing 10 isolates each, and 23 STs containing 1 isolate each. Of STs containing PG-associated isolates, 16 of 26 (61.5%) contained only PG-associated isolates, while the remainder (n = 10) contained both PG- and asymptomatic-bird-associated isolates. Three of six (50.0%) STs containing NE-associated isolates contained exclusively NE-associated isolates, while the remaining STs contained both asymptomatic-bird- and NE-associated isolates. No ST contained isolates of both PG-associated and NE-associated origins.
Fig. 1.
Sequence type disease partitioning.
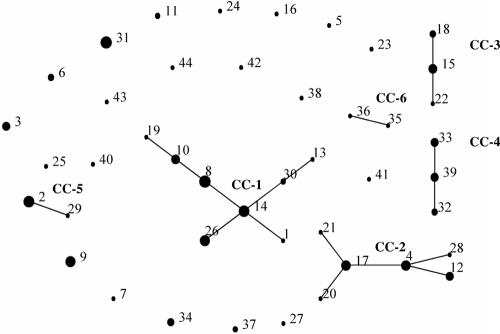
BURST (eBURST implementation) analysis defined a clonal complex (CC) as comprising isolates for which 10 of 11 alleles were identical (Fig. 2). An alternate CC definition, 9 of 11 alleles identical, resulted in the addition of only a single isolate to each of the first two CCs, so the more stringent definition was used. Overall, six CC subtypes, containing 113 of the 142 total isolates, were identified; the largest (CC-1) comprised a total of 38 isolates. Twenty STs were identified as singletons, with no observed CC associations. CC-1, CC-2, CC-3, and CC-5 all contained PG-associated isolates, with CC-5 containing only PG-associated isolates (n = 10). CC-4 contained both NE-associated and asymptomatic-bird-associated isolates but was the only CC that contained NE-associated isolates. CC-6 contained two single-isolate STs, ST-35 and ST-36, neither of which was associated with disease. All other STs clustered singly in the eBURST analysis. Based on ratios of single- and multiple-locus variants, eBURST analysis is able to identify probable founding or ancestral genotypes for CCs with more than two members. eBURST identified ST-14 and ST-15 as the probable ancestral genotypes for CC-1 and CC-3, respectively, whereas multiple candidates for the ancestral genotype were identified for CC-2 and CC-4, and no ancestral genotype was predicted for CC-5 or CC-6.
Fig. 2.
eBURST representation of relationships between sequence types and clonal complexes. Clonal complexes are designated CC-1 to CC-6. Separate dots represent sequence types, which are designated by sequence type number. The size of each dot correlates with the number of isolates in that sequence type.
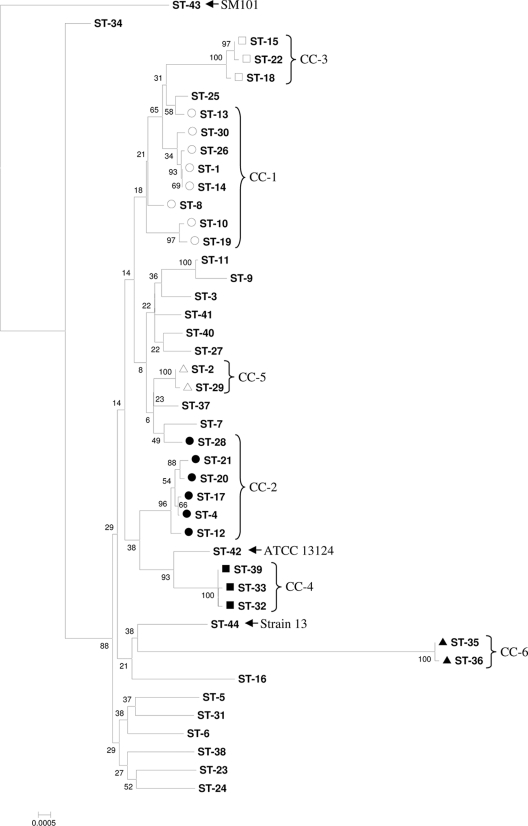
Phylogeny.
The optimum inferred phylogenetic tree was generated by the neighbor-joining and maximum composite likelihood (MCL) methods with the sum of branch length equal to 0.082 base substitution per site. Clonal complexes, as identified by ST profile-based eBURST analysis, generally, but not exclusively, clustered together in the tree (Fig. 3) and were often interspersed with singleton STs. Generally, STs containing PG-associated isolates were scattered throughout the phylogenetic tree and belonged to a variety of clusters, whereas STs containing NE-associated isolates were most commonly restricted to a small number of distantly related subtypes. Much of the dendrogram was found to be dominated by a large clade that contained CC-1 and CC-3 as well as a substantial number of closely related STs; however, it is noteworthy that no STs containing NE-associated isolates were observed to be closely related to this group. CC-4 contained NE-associated isolates with the highest interisolate sequence homology and clearly represented a phylogenetically distinct subtype. The C. perfringens reference strain ATCC 13124 had the ST most closely related to CC-4, while C. perfringens strain 13 clustered loosely with CC-6. The C. perfringens reference strain SM101, used to root the tree, clustered nearest the NE-associated ST-34 but diverged from it with a high number of substitutions per base. These two STs (ST-34 and ST-43 [SM101]) diverged appreciably from each other and also diverged considerably from the STs in the bulk of the tree.
Fig. 3.
Neighbor-joining phylogeny of C. perfringens isolates from the current study. The tree was generated on the basis of a maximum composite likelihood model and was condensed where bootstrap support was <50% of replicates. Clonal complexes are designated CC-1 (○), CC-2 (●), CC-3 (□), CC-4 (■), CC-5 (▵), and CC-6 (▴).
Toxin gene screening.
All isolates toxinotyped (n = 142) were identified as toxinotype A (45), meaning that all isolates possessed the α toxin gene but none of the genes for additional major toxins (Table 1). Thus, the β, ε, and ι toxins were not found in any of the environmental isolates or reference strains (ATCC 13124, SM101, strain 13) tested, but the β2 toxin was found in 111 of 142 (78.2%) total isolates screened (Table 5). The cpe enterotoxin also was not found in any of the poultry isolates examined here but was present in the reference isolate C. perfringens SM101. The netB toxin was identified in 17 of 20 (85.0%) NE-associated isolates, no PG-associated isolates, and 10 of 54 (18.5%) isolates not associated with disease (Table 5).
Table 5.
Prevalence of C. perfringens toxin genes according to disease association
| Disease association | Prevalence (no. of isolates) |
||
|---|---|---|---|
| cpa | cpb2 | netB | |
| PG | 65 | 47 | 0 |
| NE | 20 | 20 | 17 |
| None (asymptomatic birds) | 54 | 43 | 10 |
| Total | 139 | 110 | 27 |
DISCUSSION
The MLST schemes that formed the methodological basis for this work have enabled increasingly specific comparisons of C. perfringens populations, first by examination of isolates of diverse host and disease origins (16, 28) and then by the application of a more highly discriminatory scheme to disease-associated isolates of poultry origin (2). The method utilized in this study was adapted from a previously published MLST schema (16) with the addition of three loci, dnaK, groEL, and gyrA, in order to allow for direct comparison between the results obtained from C. perfringens poultry isolates from this study and those from previous work performed on poultry-associated C. septicum strains (27). These additional loci are popular targets for phylogenetic reconstruction from molecular markers, and all three were observed in this study to possess 10 or more alleles. The addition of the dnaK and gyrA loci to the scheme increased the number of STs identified from 34 to 44; however, the addition of the groEL locus did not affect the number of STs identified.
Based on the allelic polymorphism of the housekeeping genes examined here, considerable genetic diversity exists in the core genome of this collection of Clostridium perfringens strains. The loci examined here averaged 12.2 alleles and identified 44 STs among 139 poultry-associated C. perfringens type A isolates and 3 reference strains. In comparison, Jost et al. (16) identified 24.4 average alleles per locus and 80 STs over eight genes in 132 isolates; however, the C. perfringens isolates studied by Jost et al. were obtained from a much more diverse set of host species and toxinotypes (16). A study by Chalmers et al. (2) identified 5.9 average alleles per loci and 22 STs over nine loci in 61 isolates from NE-affected and healthy poultry. In contrast, a congruent MLST analysis performed previously by our group with a collection of 108 poultry gangrene-associated Clostridium septicum isolates identified an average of 2.7 alleles per loci and 10 total STs (27). C. septicum provides a useful comparison, since it is often considered the more primary etiological agent in the pathology of poultry gangrene (21, 26, 27, 39). The levels of allelic and ST diversity in the core genomes of the isolates analyzed here are high given the demographics of this collection of C. perfringens strains in comparison to those of similar studies and contrast strongly with the very low diversity observed in orthologous genes of C. septicum strains of similar host and environmental origins (27). An explanation for the comparative differences in core genome locus diversity may be that C. septicum exists primarily as a highly specialized opportunistic pathogen, whereas C. perfringens is more plastic in its ability to occupy a much wider range of ecological niches (11, 36).
Although considerable polymorphism existed in the loci analyzed, the sequence data characteristics measured in this study also suggested a highly clonal population. Significant linkage disequilibrium was observed between all 11 loci (IA, 2.635), indicating low recombination rates in the core genomes of the C. perfringens isolates examined (35). This hypothesis is substantiated by the observation that 113 of the 142 isolates (79.6%) partitioned into six clonal complexes, stringently defined as groups of isolates sharing 10 or more of the 11 loci examined, and two abundant sequence types, ST-9 and ST-31, containing 8 and 10 isolates, respectively. Further, the concatenated-sequence-based phylogeny constructed here also supports the clustering of isolates into clonal complexes as identified by eBURST analysis while further detailing the phylogenetic relationships between clades and singleton STs. Of the dominant subtypes observed, clonal complex 1 (CC-1) is of particular interest, because it was clearly the most prolific subtype identified in the study, representing 27.3% of the total poultry isolates examined. CC-1 was recovered from all geographic areas sampled, was present in both turkeys and broilers, and was not conclusively associated with disease. The exact mechanisms responsible for the unique abundance and distribution of CC-1 are not entirely clear from the results presented here, although eBURST analysis suggests that this clonal complex of isolates is genotypically ancestral to the poultry-associated strains examined. The possibility of a genetic determinant responsible for the relative frequency of this subtype justifies further investigation focusing on the genomics and phenotypic characteristics of the strains included, which may provide valuable insight into biomarkers of ecological fitness in bacteria from the poultry environment. Also noteworthy was the observation of two unique subtypes, CC-4 and ST-31, both of which were highly associated with necrotic enteritis, representing 25% and 50% of NE cases as well as 9.4% and 7.2% of all poultry isolates examined, respectively. It is likely that the abundance of these disease-associated subtypes is a direct result of selection due to their apparent advantage in the initiation of NE infection under appropriate environmental conditions. Both subtypes were found only in broilers but were recovered from disparate geographic areas and harbored the netB toxin gene in 87.0% of isolates. Thus, particular core genetic elements correspond here to the incidence of ecological dominance, which is evident both in the general case of relative representation in a collection of poultry strains and in the specific case of niche association in the incidence of poultry NE disease.
The PCR-based survey of C. perfringens toxin genes is an important complement to the phylogenetic characterization of this population. Recent genotypic studies have provided mixed support for the importance of cpb2 in poultry enteric disease (15, 42). Overall, cpb2 was present in 78.2% of all isolates surveyed; however, the presence of this gene was significantly correlated with C. perfringens isolates recovered in cases of NE (P, 0.007) but not PG (P, 0.093). Although netB was also highly correlated with NE-associated isolates (P, <0.001) and was absent in PG-associated isolates, there was no significant correlation between the presence of cpb2 and netB (P, 0.195). These correlations extended to phylogenetic clades as well; cpb2 was present in all clonal complexes except CC-6 and was ubiquitous in the NE-associated subtypes CC-4 and ST-31. NE-associated subtypes contained comparatively high numbers of netB-positive isolates; netB was absent in all PG-associated subtypes and present in 18.5% of non-disease-associated isolates. Further investigation of cpb2 and netB toxin expression may support phylogenetic disease associations in vivo (15, 19).
Clostridium perfringens isolates recovered from the affected individuals of a flock suffering from an NE outbreak are often genotypically clonal, suggesting that specific genetic elements may be linked to the pathogenesis of this disease (4–6, 10, 20). The results of this study support this view, since isolates recovered in cases of NE were observed to be highly clonal and more evolutionarily removed from their most recent common ancestors than most non-NE strains, based on analysis of core genome loci. In addition, NE-associated isolates were not found in the dominant PG-associated clades of the phylogenetic tree, suggesting that these strains are genetically distinct. Overall, no PG-associated and NE-associated isolates shared the same ST or clonal complex. Interestingly, two unique STs residing in the dominant PG clades harbor the NetB gene, although its phenotypic expression was not investigated here. The fact that these two STs were not isolated from diseased birds does not preclude the possibility that these isolates could potentially instigate NE pathology. In contrast to these findings for NE isolates, the PG-associated isolates were more genetically polymorphic and in many instances were closely related to isolates from asymptomatic poultry. The exact role of C. perfringens in PG remains unclear, particularly since C. septicum has been implicated as the primary etiologic agent in most recent outbreaks (21, 39), which may explain why clear lineages of C. perfringens associated with PG were generally not observed. A conserved ability of most C. perfringens type A strains to contribute to gangrene under the appropriate conditions might also explain the intermingling of isolates from healthy and diseased specimens. Despite the general trends discussed above, one clonal complex, CC-3, was found in both turkeys and broilers and was observed to be strictly associated with PG. Phylogenetic analysis indicated that CC-3 has clearly diverged from the remainder of PG-associated isolates. Additional investigation of isolates localized to CC-3 would be beneficial to establish its validity as a virulent subtype.
In order to directly compare sequence information from a prior study (2) with the data described here, the sequence orientation, length, and region were standardized between the two studies. Some discriminatory data were lost when the two schemes were merged, in that 22 STs were condensed into 18 STs in the prior study and 44 STs into 34 STs in the current study. Nevertheless, these condensed STs retained all the disease and demographic associations of the separate schemes. A strong association emerged between the lone NE-associated clonal complex from the current study (CC-4) and the two most abundant NE-associated STs (Chalmers ST-1 and Chalmers ST-10) from the prior study (Fig. 4). Combined, these two STs from the prior study contained 26 isolates, of which 22 were both NE associated and netB positive (2). Further, two additional NE-associated and netB-positive STs from the prior study were closely related to ST-31 from the current study, a particularly dominant NE-associated subtype (Fig. 4). The demographic association between the two studies also extended to non-disease-associated STs: several non-disease-associated STs from the prior study clustered with the non-disease-associated subtypes CC-2 and ST-9 of the current study (Fig. 4). Notably, the prior study did not observe as high a prevalence of cpb2 among the 61 isolates it examined as was observed in the current study, further confounding the significance of this toxin in poultry isolates of C. perfringens.
Fig. 4.
Neighbor-joining phylogenetic comparison of C. perfringens isolates from the current study (STs followed by “AGTP”) with those from a prior study (STs followed by “Chalmers”) (2). The tree was generated on the basis of a maximum composite likelihood model and was condensed where bootstrap support was <50% of replicates. ST numbers are equivalent for Fig. 3 and 4 but have been condensed in Fig. 4 due to the merging of the typing schemes. Clonal complexes are indicated by symbols: ○, CC-1; ●, CC-2; □, CC-3; ■, CC-4; ▵, CC-5; ▴, CC-6.
In conclusion, MLST was used to successfully subtype 139 poultry-derived C. perfringens isolates by using DNA sequence and allelic profile data. MLST and sequence-based phylogenetic inference identified a polymorphic yet predominantly clonal population, with a phylogeny characterized by dominant clonal complexes and sequence types, and the remaining isolates existing as low-abundance STs. Disease-associated C. perfringens isolates segregated distinctly based on ST, clonal complex, and the concatenated-sequence-based phylogeny. Notably, NE-associated isolates were not found in the dominant PG-associated clades of the sequence-based phylogeny, nor were PG-associated isolates found in the dominant NE-associated subtypes CC-4 and ST-31. PG-associated isolates were often closely related to isolates from asymptomatic poultry and generally clustered in clades predicted to be evolutionarily ancestral. These results indicate that certain subtypes have evolved the specialized ability to cause NE disease in poultry and that this specialization is evident in the core genome. Here we have shown for the first time that isolates from poultry NE and PG disease are distinct. Detection of disease niche partitioning in the core genome has epidemiological relevance, because these genes are largely conserved and stably inherited, whereas many C. perfringens virulence factors associated with specific disease can be lost or acquired depending on environmental conditions and association with other bacteria (11, 20, 36). Further phylogenetic analyses focused on disease-associated isolates are necessary in order to fully understand the correlation between the core housekeeping genome and the more-variable virulence factors in this species. The application of such information may allow the identification of conserved genetic biomarkers for strains implicated in disease etiology or of prominent ecological fitness determinants in both general and disease-specific niches.
ACKNOWLEDGMENTS
Sincere thanks to Gabhan Chalmers and Patrick Boerlin of the University of Guelph for providing MLST data for comparison (2).
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1. Baker A. A., Davis E., Rehberger T., Rosener D. 2010. Prevalence and diversity of toxigenic Clostridium perfringens and Clostridium difficile among swine herds in the Midwest. Appl. Environ. Microbiol. 76:2961–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chalmers G., et al. 2008. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J. Clin. Microbiol. 46:3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chalmers G., Martin S., Prescott J., Boerlin P. 2008. Typing of Clostridium perfringens by multiple-locus variable number of tandem repeats analysis. Vet. Microbiol. 128:126–135 [DOI] [PubMed] [Google Scholar]
- 4. Cooper K., Songer J. 2009. Necrotic enteritis in chickens: a paradigm of enteric infection by Clostridium perfringens type A. Anaerobe 15:55–60 [DOI] [PubMed] [Google Scholar]
- 5. Cooper K. K., et al. 2010. Virulence for chickens of Clostridium perfringens isolated from poultry and other sources. Anaerobe 16:289–292 [DOI] [PubMed] [Google Scholar]
- 6. Engström B. E., et al. 2003. Molecular typing of isolates of Clostridium perfringens from healthy and diseased poultry. Vet. Microbiol. 94:225–235 [DOI] [PubMed] [Google Scholar]
- 7. Feil E. J., Li B. C., Aanensen D. M., Hanage W. P., Spratt B. G. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 9. Garmory H. S., et al. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gholamiandekhordi A., Ducatelle R., Heyndrickx M., Haesebrouck F., Van Immerseel F. 2006. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet. Microbiol. 113:143–152 [DOI] [PubMed] [Google Scholar]
- 11. Hatheway C. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hofacre C., French J., Page R., Fletcher O. 1986. Subcutaneous clostridial infection in broilers. Avian Dis. 30:620–622 [PubMed] [Google Scholar]
- 13. Huang X., Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolley K., Feil E., Chan M., Maiden M. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 15. Jost B. H., Billington S. J., Trinh H. T., Bueschel D. M., Songer J. G. 2005. Atypical cpb2 genes, encoding β2-toxin in Clostridium perfringens isolates of nonporcine origin. Infect. Immun. 73:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jost B., Trinh H., Songer J. 2006. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet. Microbiol. 116:158–165 [DOI] [PubMed] [Google Scholar]
- 17. Keyburn A., et al. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keyburn A., et al. 2006. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74:6496–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keyburn A., et al. 2010. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet. Res. 41:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lepp D., et al. 2010. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One 5:e10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G., et al. 2010. An outbreak of gangrenous dermatitis in commercial broiler chickens. Avian Pathol. 39:247–253 [DOI] [PubMed] [Google Scholar]
- 22. Meer R. R., Songer J. G., Park D. L. 1997. Human disease associated with Clostridium perfringens enterotoxin. Rev. Environ. Contam. Toxicol. 150:75–94 [DOI] [PubMed] [Google Scholar]
- 23. Miller D. 1998. Clostridial diseases, p. 61–68 In Swayne D. E., Glisson J. R., Jackwood M. W., Pearson J. E., Reed W. M. (ed.), A laboratory manual for the isolation and identification of avian pathogens, 4th ed. American Association of Avian Pathologists, Kennett Square, PA [Google Scholar]
- 24. Myers G., et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nei M., Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 26. Neumann A. P., Dunham S. M., Rehberger T. G., Siragusa G. R. 2010. Quantitative real-time PCR assay for Clostridium septicum in poultry gangrenous dermatitis associated samples. Mol. Cell. Probes 24:211–218 [DOI] [PubMed] [Google Scholar]
- 27. Neumann A. P., Rehberger T. 2009. MLST analysis reveals a highly conserved core genome among poultry isolates of Clostridium septicum. Anaerobe 15:99–106 [DOI] [PubMed] [Google Scholar]
- 28. Rooney A., Swezey J., Friedman R., Hecht D., Maddox C. 2006. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 172:2081–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rozen S., Skaletsky H. 2000. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 30. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 31. Sawires Y., Songer J. 2006. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe 12:23–43 [DOI] [PubMed] [Google Scholar]
- 32. Sawires Y., Songer J. 2005. Multiple-locus variable-number tandem repeat analysis for strain typing of Clostridium perfringens. Anaerobe 11:262–272 [DOI] [PubMed] [Google Scholar]
- 33. Shimizu T., et al. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siragusa G., Danyluk M., Hiett K., Wise M., Craven S. 2006. Molecular subtyping of poultry-associated type A Clostridium perfringens isolates by repetitive-element PCR. J. Clin. Microbiol. 44:1065–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith J. M., Smith N. H., O'Rourke M., Spratt B. G. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Songer J. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 38. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tellez G., Pumford N., Morgan M., Wolfenden A., Hargis B. 2009. Evidence for Clostridium septicum as a primary cause of cellulitis in commercial turkeys. J. Vet. Diagn. Invest. 21:374–377 [DOI] [PubMed] [Google Scholar]
- 40. Timbermont L., et al. 2009. Origin of Clostridium perfringens isolates determines the ability to induce necrotic enteritis in broilers. Comp. Immunol. Microbiol. Infect. Dis. 32:503–512 [DOI] [PubMed] [Google Scholar]
- 41. Turner K. M. E., Hanage W. P., Fraser C., Connor T. R., Spratt B. G. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Asten A. J., Nikolaou G. N., Gröne A. 2010. The occurrence of cpb2-toxigenic Clostridium perfringens and the possible role of the β2-toxin in enteric disease of domestic animals, wild animals and humans. Vet. J. 183:135–140 [DOI] [PubMed] [Google Scholar]
- 43. Wehnes C. A., et al. 2009. Benefits of supplementation of an electrolyte scour treatment with a Bacillus-based direct-fed microbial for calves. Probiotics Antimicrob. Proteins 1:36–44 [DOI] [PubMed] [Google Scholar]
- 44. Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoo H. S., Lee S. U., Park K. Y., Park Y. H. 1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 35:228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]