Abstract
The incidence of simian virus 40 (SV40) infections in rhesus macaques infected with simian-human immunodeficiency viruses (SHIV) and in uninfected animals was determined using PCR. Rates varied from 5% in peripheral blood mononuclear cells of uninfected monkeys to 19.6% in SHIV-infected macaques. Much higher detection rates, up to 75%, were found in lymph nodes and spleen samples of SHIV-infected animals. Sequence analysis of PCR amplicons revealed that they form two genetic clusters, one containing the majority of known SV40 strains and the other formed by variants with 7% genetic difference. Based on this difference, we propose two SV40 types: “type 1” or “classical type” for the majority of SV40 strains and “type 2” for the novel SV40 variants. The genome of one variant, SV40-Ri257, was completely sequenced and analyzed. The agnogene of SV40-Ri257 extends into the VP2 open reading frame and encodes a typical agnoprotein fused to a C-terminal hydrophobic region. The transcriptional control region (TCR) of SV40-Ri257 is the least conserved region compared to type 1 viruses. Particularly, the 3′ end of the TCR, containing the early promoter and enhancer region, exhibits considerable variation. Further analysis of SHIV-infected macaques with type-specific PCRs revealed that the TCR of type 1 was completely conserved, whereas this region in type 2 varied considerably within the early enhancer region. We provide evidence here for the existence of a novel SV40 type in rhesus macaques and show that double infections with both types frequently occur.
INTRODUCTION
Simian virus 40 (SV40) is the best-studied polyomavirus and one of the best-studied viruses. Discovered in 1960 as a contaminant virus in poliovirus cultures for vaccine production (27), it was the first animal virus whose genome was completely sequenced (9). SV40 has been the subject of numerous studies since it was discovered that this small DNA virus could induce tumors when injected in rodents (7, 8), while it also was capable of in vitro transformation of cells (13, 15).
The natural host of SV40 is the rhesus macaque (Macaca mulatta), while other macaque species harbor closely related viruses (14). In macaques, depending on age and origin, the infection rate can be as high as 95% (30), but infection of healthy animals usually does not result in disease symptoms. In immunocompromised macaques, however, SV40 can cause disease symptoms that are similar to progressive multifocal leukoencephalopathy (PML), a rare and fatal disease in humans caused by the related JC polyomavirus (JCV) (11).
Despite great interest in the biology of SV40, knowledge of the natural history of SV40 is still relatively limited. Several viruses have been isolated from different macaque species, such as rhesus (12, 16, 23, 25) and cynomolgus (3, 29) macaques. Genetic analysis revealed that the majority of SV40 that had not been cultured in vitro possess a transcriptional control region (TCR) with only one 72-bp “repeat,” indicating that the duplication previously reported in other SV40 strains had been obtained during cell culture passage (5, 17, 24). Several authors have studied SV40 in immunocompromised macaques (12, 16, 23) and have shown that infection of macaques with simian immunodeficiency virus (SIV) results in a significantly higher ability to detect SV40. In addition, immunocompromised macaques have shown been used to study meningoencephalitis due to SV40 infection (2, 6, 26).
We describe here the results of a comprehensive screening of DNA samples isolated from blood of healthy macaques and from various tissues of animals infected with chimeric simian-human immunodeficiency viruses (SHIV) to get further insight into the incidence of infection, tissue tropism, and genetic variation of SV40 in healthy rhesus macaques and in monkeys with various levels of immunodeficiency due to experimental SHIV infection. Our data point to lymph nodes and spleens of SHIV-infected macaques as organs with a high prevalence of SV40 but, more importantly, reveal that a significant percentage of monkeys were infected with a new type of SV40, with some animals showing evidence for double infections with the novel, and the classical SV40 type.
MATERIALS AND METHODS
Tissue samples and DNA extraction.
Rhesus macaques (Macaca mulatta) were housed at the Biomedical Primate Research Centre in Rijswijk, Netherlands. Animals were of Chinese or Indian origin, and these groups were separately housed. Healthy animals were from the breeding colony, and blood was collected during routine health surveillance. Other macaques were used in AIDS vaccine trials, and had been experimentally infected with simian-human immunodeficiency virus chimeras (SHIVs). From this group, blood and tissue (spleen and lymph node) samples were taken at the time of euthanasia and stored frozen at −20°C.
Nucleic acids were extracted from peripheral blood mononuclear cells (PBMC), spleen tissue, and lymph nodes by using the Puregene blood kit (Gentra Systems, Minneapolis, MN).
SV40 diagnostic PCR.
For the SV40 screening of DNA samples, we made use of a published seminested PCR assay targeted to the early region encoding the small and large T antigens (21). We improved the sensitivity of the assay by modifying the test to a fully nested-PCR using the primers SV40-for2 and SV40-rev as the outer set of primers and SV40-forin and PYV-rev as inner primer set (Table 1). The outer PCR was performed in a 50-μl volume using 1 μg of DNA, 2 U of Maxima Hot Start Taq DNA polymerase (Fermentas GMBH, St. Leon-Rot, Germany), 5 μl of 10× Hot Start PCR buffer, 1 pmol of each primer, 2.5 mM MgCl2, and 200 μM concentrations of each deoxynucleoside triphosphate. Amplification was performed with an enzyme activation step of 4 min at 96°C, followed by 40 amplification cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s. In the second amplification reaction, 2 μl of the PCR product of the outer PCR was used as a template. The conditions for the second amplification were identical to those for the outer PCR, except that 2.25 mM MgCl2 was used.
Table 1.
PCR primers sequences used in this study
| PCR | Primer | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| SV40 diagnostic | SV40-for2 | CTTTGGAGGCTTCTGGGATGCAACT | 574 |
| SV40 rev | GCATGACTCAAAAAACTTAGCAATTCTG | ||
| SV40-forin | CTGGTGTTGATGCAATATACTGC | 493 | |
| PYV-rev | GAAAGTCTTTAGGGTCTTCTACC | ||
| Type specific | SV40 ctrl out-F | ATATGCCTTTCTCATMAGAGG | 791 (classical strain 776) |
| SV40 ctrl out-R | CAGACACAGTAGCAATTAGGTC | 703 (type 2 strain Ri257) | |
| SV40 classic-F | CTAGAAGGTCCATTAGCTGCAAA | 666 | |
| SV40 classic-R | AAACTTACCAGTTAACTTTCTGG | ||
| SV40 type 2-F | AAAAGGTCCATTAATTGCAGGG | 575 | |
| SV40 type 2-R | ACCTTACCTATTTGCCAGTGTTG | ||
| Late region | Late-Fout | GCAAATGCAGTTAGTAGACATAC | 2,350 |
| Late-Rout | TGGTCCTATAGTTAGCTAGCAC | ||
| Late-Fin | GCCTCCAAAATCAGGTTGATG | 2,010 | |
| Late-Rin | GGAATTCTGGCCACACTGTAG | ||
| Early region | Early-Fout | GCAGTTTACTGATGACTCTCCAG | 3,450 |
| Early-Rout | CTATTCTAGAAGTAGTGAGGAGGC | ||
| Early-Fin | AGAATTCCATTGCCTAATTTA | 3,274 | |
| Early-Rin | ATATTCCTCTTATGAGAAAGGC |
Amplification and sequencing of SV40 genome.
The genome of the new SV40 type was amplified in two overlapping fragments, more or less equivalent to the early and late gene regions. Primers were designed using the published sequence of SV40-YNDQ38 and are given in Table 1. The late region was amplified as a 2,010 bp-long fragment using 5 U of Maxima Hot Start Taq DNA polymerase. Both outer and inner PCR were performed with 1.5 mM MgCl2 for 40 cycles with a 2-min elongation time at 72°C. For the amplification of the 3,274-bp early fragment we made use of the Expand High Fidelity PCR System (Roche Diagnostics Nederland BV, Almere, Netherlands). The outer and inner PCRs were carried out in a 50-μl volume, using 1.5 mM MgCl2 and 5.25 U of Expand High Fidelity enzyme mix. Both amplification reactions consisted of an initial denaturation step of 4 min at 95°C, followed by 40 cycles of 15 s at 94°C and 30 s at 55°C and a 2-min extension at 72°C.
The early and late fragments were cloned in the pJET1.2 vector (CloneJET PCR cloning kit; Fermentas GmbH, St. Leon-Rot, Germany) and sequenced by using a primer-walking strategy.
Type-specific PCR.
To distinguish between the different SV40 types, we designed a nested-PCR assay with a SV40-specific outer primer set (SV40 ctrl out -F and SV40 ctrl out-R) and two type-specific inner primer sets (SV40 classic-F and -R; SV40 type 2-F and -R) (Table 1). The reaction mixtures were identical to the diagnostic PCR, but the outer PCR was supplemented with 3.5 mM MgCl2, whereas the inner reaction was performed with 2.5 mM MgCl2. In both reactions, amplification was performed with an enzyme activation step of 4 min at 96°C, followed by 40 amplification cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 45 s.
DNA sequencing and genome analysis.
Gel-purified DNA from PCR or column-purified plasmid was sent to Baseclear BV (Leiden, Netherlands) for sequencing, and the raw sequence data were analyzed using SeqMan Pro version 8.1 (Lasergene Software; DNASTAR, Madison, WI). MacVector version 11.0.4 (MacVector, Inc., Cary, NC) was used for analysis of the completed nucleotide sequences. Structural analysis of the agnoprotein was performed using the PSIpred, TMpred, and HMMTOP v2.0 programs as found on the ExPASy proteomics server (http://www.expasy.ch/tools/).
Phylogenetic analysis.
Phylogenetic analysis was based on large T-antigen gene sequences and was performed by using the neighbor-joining method as implemented in MEGA version 4 (28). The EMBL database accession numbers of the SV40 strains used in our analyses were as follows: AF038616 (SV40 K661), AF156107 (SV40 VA45-54), AF316139 (SV40 776), and DQ218418 (SV40 YND38).
Nucleotide sequence accession number.
The sequence of the SV40-Ri257 strain has been deposited in the EMBL sequence database under accession number FN812745. Large T gene and TCR sequences were deposited under accession numbers FN824622 to FN824658 and FN812742 to FN812744, respectively.
RESULTS
Incidence of SV40 infections in healthy and SHIV-infected macaques.
We analyzed 246 DNA samples obtained from 56 rhesus macaques. DNA was extracted from various tissues of these animals that had been experimentally infected with SHIV and suffer various levels of immunodeficiency. The tissues included 56 spleen samples, 51 samples from PBMC, 53 mesenteric lymph node (LN) samples, 30 inguinal LN samples, 44 axillary LN samples, and 8 samples isolated from mixed axillary and inguinal lymph nodes. In all, 45 animals were found to be SV40 positive in at least one tissue (83.3%). Detection rates varied between 75% in the mixed LN DNA samples to a 19.6% detection rate determined in PBMC of SHIV-infected macaques (Table 2). Likewise, we also performed the diagnostic SV40 PCR on 221 PBMC DNA samples from healthy breeding colony animals. In sharp contrast to the rates in the immunocompromised animals, only 11 of 221 animals (5%) tested positive in the diagnostic SV40 PCR.
Table 2.
PCR detection of SV40 in tissues of SHIV-infected rhesus macaques
| Tissuea | No. of samples tested | No. of SV40-positive samples | % Positive |
|---|---|---|---|
| Spleen | 56 | 19 | 33.9 |
| PBMC | 51 | 10 | 19.6 |
| Mesenteric LN | 53 | 14 | 26.4 |
| Axillary LN | 44 | 21 | 47.7 |
| Inguinal LN | 30 | 10 | 33.3 |
| Mixed axillary LN/inguinal LN | 8 | 6 | 75 |
LN, lymph nodes.
Detection of a new SV40 type in rhesus macaques.
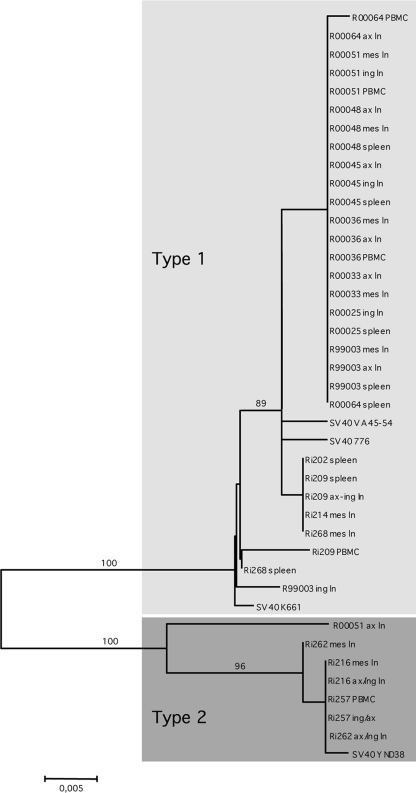
To determine which SV40 strain(s) infected the macaques in our colony and whether any differences could be found between viruses infecting different tissues from the same animal, we analyzed tissue DNA samples from 19 SHIV-infected macaques. We obtained 37 sequences by direct sequence analysis of the amplicons that were produced in the diagnostic PCR. The genetic variation is shown in Fig. 1. In the neighbor-joining tree, two clusters of viral sequences can be distinguished. The largest cluster, which includes a few subclusters formed by minor variants, consists of sequences that are 99% identical to the majority of SV40 large T gene sequences that have been deposited in the GenBank database. We tentatively designated it the “classical” or “type 1” SV40.
Fig. 1.
Neighbor-joining phylogeny based on a 470-nt PCR fragment amplified from the early region of SV40. Gray boxes indicate the proposed SV40 types. Phylogenetic analysis was performed by the neighbor-joining method using the ML matrix model as implemented in MEGA version 4 (28). Bootstrap values (as a percentage of 1,000 resamplings) are indicated. The bar indicates the number of nucleotide residue replacements per site.
The second, smaller, cluster is formed by seven sequences that were obtained from tissues of four rhesus macaques. These variants cluster strongly together (100% bootstrap) and show an average 7% nucleotide sequence difference with the classical SV40 sequences. Because of the clear distinction from the other SV40 strains, we tentatively labeled the viruses from the small cluster “type 2.” Remarkably, only one SV40 sequence found in GenBank, derived from the SV40 strain YNQD38 (GenBank accession no. DQ218418 [unpublished data]), showed 99% identity with our sequences.
Genome sequencing and molecular characterization.
A representative genome of the new SV40 type 2 (strain SV40-Ri257) was amplified in two overlapping fragments from DNA isolated from PBMC of macaque Ri257. Using a primer-walking strategy, the fragments were sequenced, and the complete circular polyomavirus genome sequence was obtained by joining both sequences. The resultant genome of SV40-Ri257 was 5.125 nucleotides (nt) in length and was aligned to the SV40 reference strain 776 (SV40-776; GenBank accession no. AF316139) for further molecular characterization. The DNA alignment had 26 gaps over a total length of 5,221 nt, and the overall nucleotide identity was 88%. The alignments of the individual early (small T, large T) and late genes (VP1, VP2/3) are shown in the Fig. S1A to D in the supplemental material. Nucleotide similarities were 92% (small T), 87% (large T), 90% (VP1), and 91% (VP2/VP3), respectively. The protein identities/similarities of the viral genes are shown in Table 3. A slightly higher similarity was seen between the protein sequences compared to the nucleotide comparisons.
Table 3.
Comparison of proteins from SV40 types 1 and 2
| Protein | Aligned length (aa)a | No. (%) |
No. of gaps in alignment | |
|---|---|---|---|---|
| Identities | Similarities | |||
| Small T antigen | 174 | 169 (97) | 3 (1) | 0 |
| Large T antigen | 708 | 651 (91) | 32 (4) | 5 |
| VP2/VP3 | 353 | 332 (94) | 16 (4) | 2 |
| VP1 | 364 | 353 (96) | 6 (1) | 0 |
aa, amino acids.
Curiously, the recognized agnogene of SV40-Ri257 extends into the VP2 open reading frame and is 270 nt in size, in contrast to the SV40-776 agnogene that is only 186 nt in length. The N-terminal 61 amino acid residues of the agnoprotein encoded by SV40-Ri257 are highly similar to type 1 agnoproteins (83%), but most similar to the type 2 agnoprotein of SV40-YNDQ38 (96%). The C-terminal 29 amino acids, however, are unique to this SV40 strain and are also missing in the agnoprotein of YNDQ38. A BLAST search in the protein databases did not reveal any similarities with other known proteins or polypeptides. This region is hydrophobic in nature and has an isoelectric point of 7.00, which is in sharp contrast with the N-terminal part of the protein which has a pI of 11.03. Structural analysis of this region using PSIpred, TMpred, and HMMTOP v2.0 programs (ExPASy proteomics server [http://www.expasy.ch/tools/]) points to the presence of a transmembrane domain in the C-terminal extension of the agnoprotein.
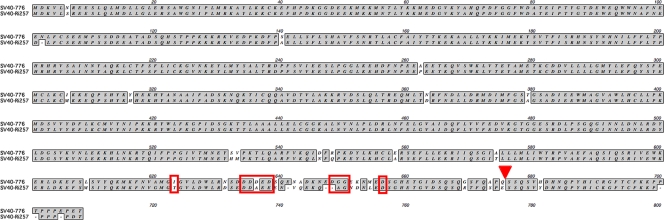
Because of polymorphisms found in the C-terminal region of the large T antigen, this region, starting with the Gly residue at position 622, has been used to for strain assignment of type 1 SV40 (25). Alignment of the large T antigens of SV40-776 (type 1) and SV40-Ri257 revealed a high number of polymorphisms distributed over the whole protein sequence but which were most dense at the C terminus (Fig. 2). The polymorphisms seen in that region differ considerably from those that are used for type 1 strain designation. In contrast, the same region is 100% identical between SV40-Ri257 and SV40-YNQD38. This emphasizes the divergence of those strains but also implies that the C terminus of the large T antigen of type 2 viruses may be more conserved than in the classical SV40 strains.
Fig. 2.
Alignment of the large T proteins of SV40-Ri257 and SV40-776. Red boxes indicate regions of amino acid polymorphisms used in strain designation for classical type SV40. The red arrowhead indicates a deletion-insertion site also used in strain designation.
The major structural protein VP1 of SV40-Ri257 and -YNQD38 is identical, except for position 32, where SV40-Ri257 possesses a Glu residue and SV40-YNQD38 VP1 contains a Lys at that position. Both SV40 type 2 viruses differ at four sites in VP1 from the type 1 reference strain 776. Two mutations (Ala→Thr at position 6 and Cys→Ala at position 127) are located outside the recognized VP1 surface loops (19). The other amino acid differences are found in a neutralization epitope in the EF-loop (22). Especially the amino acid residue at position 178 (Gln in strain 776 and Leu in type 2 viruses) is likely involved in virus neutralization (22). The other amino acid (Val→Ser at position 187) has not been the subject of neutralization studies.
TCR.
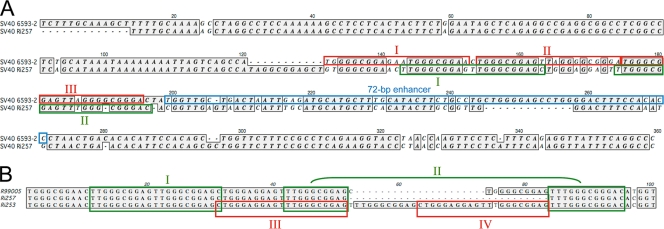
The TCR, also referred to as noncoding control region, is located between open reading frames of the T antigens and the agnoprotein and directs the transcription of the early genes toward the left and of the late genes toward the right (i.e., in opposite directions). It contains promoter/enhancer sequences and also contains the origin of DNA replication (for a review, see reference 31). This region is the most variable part of the genome, and its structure is influenced by its culture on cell lines (5, 17, 24). The prototype SV40 strain 776 is a cell culture-adapted laboratory strain, which TCR contains three 21-bp repeated sequences and a duplicated 72-bp enhancer element. The latter has probably been acquired during passage in culture (nonarchetypal TCR). If SV40 is isolated directly from its natural host, most type 1 SV40 TCRs are characterized by a single 72-bp enhancer, in addition to the viral ori and multiple 21-bp repeats (archetypal TCR). In Fig. 3A the TCR from the new type 2 has been aligned with the corresponding region from strain 6593-2, an SV40 characterized directly from the brain of an immunocompromised rhesus macaque (16). The TCR of SV40-Ri257 is more divergent than the coding sequences on the genome. The 5′ end of the TCR, which includes the viral ori and the AT-rich tract (nt 1 to 120), is highly similar in both viruses, except that the TCR of strain 6593-2 has an additional stretch of bases at its 5′ end, which is probably due to a duplication. The 3′ end of the SV40 TCR contains a 21-bp repeat region that is part of the early promoter and an enhancer region consisting of one or more 72-bp repeated areas. In SV40 6593-2, the TCR contains two 21-bp repeats (II and III), an additional sequence that is very similar to the 21-bp repeat (I), and a single 72-bp enhancer sequence. In the TCR of SV40-Ri257, three 10-bp repeated sequences (TTGGGCGGAG; nt 143 to 152, 153 to 162, and 174 to 183) can be distinguished, in addition to one imperfect 10-bp repeat (TTGGGCGGGA; nt 185 to 194). These repeats can form two, not fully duplicated, 21-bp repeats (I and II; Fig. 3A). The supposed enhancer region of SV40-Ri257 has limited homology to the 72-bp enhancer of strain 6593-2 (nt 199 to 271).
Fig. 3.
(A) Alignment of TCR of SV40 6593-2 and SV40 Ri257. The 21-bp repeats are indicated by boxes (6593-2 in red, Ri257 in green). Separate 21-bp repeats are indicated by Roman numerals. The 72-bp enhancer region of 6593-2 is indicated by a blue box. (B) Alignment of variant TCR of SV40 type 2 detected in tissue DNA samples from three different rhesus macaques. Green boxed sequences represent the putative 21-bp repeats of SV40 Ri257 (I and II). Red boxes show alternative 21-bp sequences in the TCR of SV40 Ri253 (III and IV).
Infections of SV40 types in rhesus macaques: evidence for double infections with both types.
To evaluate the incidence of infection in rhesus macaques with either the classical or the new type, we set up type-specific PCR assays targeted to the different TCRs. To prevent cross-contamination, separate rooms were used to isolate DNA, to prepare the PCR master mixes, to add template DNA, and to analyze the PCR products. In addition, the different PCR assays were performed on separate days. Forty-five animals that had previously been tested positive in our SV40 LTag-PCR were retested using the type-specific PCR assay. Depending on the availability of the DNA, several tissues from individual animals were tested. Thirty-seven macaques were confirmed to be SV40 positive using the new assay. The classical type was detected in 53 samples from 33 animals (89% incidence). The infection rate of SV40 type 2 was lower: 17 positive tissues from 13 animals (35%). Interestingly, in nine animals we were able to amplify the TCR of both types, indicating that double infections with both SV40 types occur.
Sequence analysis of amplicons revealed a complete conservation of the TCR region in the classical type 1 viruses. Most type 2 TCRs were identical to the Ri257 sequence, but two variant TCR sequences were detected (Fig. 3B). In both variants the 5′ ends of the TCR were completely conserved, as was the sequence believed to contain the homologue to the 72-bp enhancer of the classical type. Both variant TCRs are likely the result of incomplete duplication events within the early promoter region. SV40 detected in the PBMC of animal R99005 had an 11-bp insertion within the early promoter region and, in the axillary and inguinal lymph nodes of macaque Ri253, a variant TCR was amplified that contained a 32-bp insert. The latter resulted in the formation of two complete 21-bp perfectly repeated sequences (III and IV) in place of the repeated sequences I and II.
DISCUSSION
The two best-characterized human polyomaviruses, JCV and BKV, can both cause serious and, in the case of JCV and PML, fatal disease in immunocompromised persons (20). Similarly, SV40 can cause PML-like symptoms in macaques with an impaired immune system (10–12, 16), making the macaque a potential model to study polyomavirus-induced pathology. To our knowledge, we are the first to perform a relatively large-scale SV40 PCR-screening of healthy and immunocompromised macaques. Our findings strengthen the validity of this animal model since, as in humans, the detection rate of this virus increases considerably in immunocompromised individuals. The data also indicate that PCR screening of blood for polyomaviruses can lead to a severe underestimation of the number of viremic macaques, because in our analyses the lymph nodes (particularly the axillary nodes), and the spleen showed a significantly higher SV40 infection rate than was found in blood. In previous studies, SV40 was detected in brain, kidney, and lung tissue (2, 18, 26, 29). Together, these data show that the tissue tropism of SV40 is much like that of human polyomaviruses in infected humans (1, 4, 20).
The initial analysis of large T antigen gene PCR fragments indicated that four monkeys were infected with a genetic variant of SV40. BLAST analysis with the genome of one of the variant viruses, SV40-Ri257, revealed that it differed by 12% from the majority of SV40 genomes. Only one SV40 sequence that was deposited in GenBank, YNDQ38, had 99% identity with SV40-Ri257. This led us to conclude that SV40-Ri257 and also YNDQ38 are representatives of a novel type of SV40 (type 2). Further screening of SV40-positive animals with type-specific PCR assays showed that this SV40 variant was relatively common in our colony animals, since it was found in at least 35% of the SV40-positive animals.
It comes as a surprise that almost 50 years after the first description of SV40 (27) a new SV40 type was described which, at least in our colony, is frequently found. The only closely related SV40 found in GenBank, YNDQ38, has been isolated from a Chinese rhesus macaque, but in our study only 12 of 37 SV40-positive animals were also Chinese-origin rhesus macaques (as determined by mitochondrial DNA analysis [data not shown]), while the remaining 25 monkeys were Biomedical Primate Research Centre (BPRC)-bred Indian macaques. These data suggest that SV40 type 2 is not restricted to animals from a specific geographical region (China) but is native to all rhesus macaques. However, it is difficult to comprehend why the new virus type has not been detected before in other primate centers, especially those in North America. One reason could be that these centers have imported animals from regions where type 1 viruses are the only circulating SV40. A more pragmatic explanation could be that SV40 type 2 viruses are of Chinese origin but have accidentally spread between animals from different geographical regions that were housed at the BPRC. Genetic typing nowadays excludes mixing of animals with a different genetic background, and improved housing conditions and diagnostics eliminate accidental spread of viruses. However, in past these were not standard procedures, and nonpathogenic viruses, like SV40, could have been introduced in other breeding groups without being noticed. Obviously, the analysis of samples collected from wild-living macaques is the only method to unravel the natural history of SV40.
Several groups also investigated the SV40 TCR in healthy and immunocompromised (SHIV- or SIV-infected) macaques (12, 16, 23). Viruses were obtained from kidney and brain tissue, blood, or urine. Most analyzed viruses had an archetypal regulatory region that contained a duplicated 21-bp region in the early promoter region and a single 72-bp enhancer sequence. Likewise, in our study all type 1 TCRs analyzed (n = 14; 10 animals) were 100% identical, and all had the archetypal structure. However, intriguingly, we did find some level of variation in the TCRs of the novel type. The most common variant was found in four of seven sequences, was identical to the TCR of the published SV40-YNDQ38 and to -Ri257, and had similarities with the protoarchetypal TCR of SV40-K661, which has a single 21-bp “repeat” in addition to a single 72-bp enhancer sequence. Two other variant regulatory regions had 11- and 32-bp insertions in the early promoter region. These duplication events resulted in the formation of new or alternative 21-bp repeated sequences, much like the archetypal TCR. The 32-bp insertion variant was found in the axillary and inguinal LNs of macaque Ri253, while the 11-bp variant was detected once in macaque R99005. Similarly to our findings, Lednicky et al. also detected minor TCR variants, mainly in PBMC (16), and concluded that nonarchetypal regulatory regions can arise de novo in individual macaques. Because of the limited number of animals and tissue types, this question cannot be properly addressed in the present study. The cause and consequence of the TCR rearrangements requires functional analysis, which is beyond the scope of the present study.
Screening of SV40-positive animals with type-specific PCRs showed that this type is relatively common in the animals tested and that several individuals were infected with both SV40 types. The 12% overall sequence difference can have considerable influence on the outcome of (diagnostic) PCR assays that rely on primers specific for the majority of SV40 isolates. Depending on the location of the primers, underestimation, or even lack of detection, may occur. Serological testing may also be influenced due to the sequence variation of the encoded proteins. Although the amino acid variation is less than DNA, mutations of single amino acid residues in an important epitope can have a major influence on antibody response patterns induced by such protein.
Furthermore, the sequence differences, especifically those in the TCR and agnoprotein, can significantly affect diverse biological properties, and it will be interesting to determine whether the SV40 types differ in tissue distribution, potential pathogenicity, and/or replication characteristics.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by European Community Research Infrastructures Program grant RII3-CT-2006-026155 from the European Primate Network: Specialized Infrastructures and Procedures for Biological and Biomedical Research (EUPRIM-NET).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Abend J. R., Jiang M., Imperiale M. J. 2009. BK virus and human cancer: innocent until proven guilty. Semin. Cancer Biol. 19:252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Axthelm M. K., et al. 2004. Meningoencephalitis and demyelination are pathologic manifestations of primary polyomavirus infection in immunosuppressed rhesus monkeys. J. Neuropathol. Exp. Neurol. 63:750–758 [DOI] [PubMed] [Google Scholar]
- 3. Bofill-Mas S., Albinana-Gimenez N., Pipkin P. A., Minor P. D., Girones R. 2004. Isolation of SV40 from the environment of a colony of cynomolgus monkeys naturally infected with the virus. Virology 330:1–7 [DOI] [PubMed] [Google Scholar]
- 4. Bofill-Mas S., Formiga-Cruz M., Clemente-Casares P., Calafell F., Girones R. 2001. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J. Virol. 75:10290–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang X., Axthelm M. K., Letvin N. L., Koralnik I. J. 2005. Rearrangement of simian virus 40 regulatory region is not required for induction of progressive multifocal leukoencephalopathy in immunosuppressed rhesus monkeys. J. Virol. 79:1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dang X., Wuthrich C., Axthelm M. K., Koralnik I. J. 2008. Productive simian virus 40 infection of neurons in immunosuppressed rhesus monkeys. J. Neuropathol. Exp. Neurol. 67:784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eddy B. E., Borman G. S., Berkeley W. H., Young R. D. 1961. Tumors induced in hamsters by injection of rhesus monkey kidney cell extracts. Proc. Soc. Exp. Biol. Med. 107:191–197 [DOI] [PubMed] [Google Scholar]
- 8. Eddy B. E., Borman G. S., Grubbs G. E., Young R. D. 1962. Identification of the oncogenic substance in rhesus monkey kidney cell culture as simian virus 40. Virology 17:65–75 [DOI] [PubMed] [Google Scholar]
- 9. Fiers W., et al. 1978. Complete nucleotide sequence of SV40 DNA. Nature 273:113–120 [DOI] [PubMed] [Google Scholar]
- 10. Holmberg C. A., et al. 1977. Isolation of simian virus 40 from rhesus monkeys (Macaca mulatta) with spontaneous progressive multifocal leukoencephalopathy. J. Infect. Dis. 136:593–596 [DOI] [PubMed] [Google Scholar]
- 11. Horvath C. J., et al. 1992. Simian virus 40-induced disease in rhesus monkeys with simian acquired immunodeficiency syndrome. Am. J. Pathol. 140:1431–1440 [PMC free article] [PubMed] [Google Scholar]
- 12. Ilyinskii P. O., Daniel M. D., Horvath C. J., Desrosiers R. C. 1992. Genetic analysis of simian virus 40 from brains and kidneys of macaque monkeys. J. Virol. 66:6353–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen F., Koprowski H., Ponten J. A. 1963. Rapid transformation of human fibroblast cultures by simian virus. Proc. Natl. Acad. Sci. U. S. A. 50:343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koliaskina G. I. 1963. Virological and serological data on latent infection caused by Ob40 (Sv40) virus in Macaca rhesus monkeys. Vopr. Virusol. 29:450–452 (In Russian.) [PubMed] [Google Scholar]
- 15. Koprowski H., et al. 1963. Transformation of cultures of human tissue infected with simian virus SV40. Acta Unio Int. Contra Cancrum. 19:362–367 [PubMed] [Google Scholar]
- 16. Lednicky J. A., et al. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lednicky J. A., Butel J. S. 1997. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J. Gen. Virol. 78(Pt. 7):1697–1705 [DOI] [PubMed] [Google Scholar]
- 18. Lednicky J. A., Halvorson S. J., Butel J. S. 2002. PCR detection and DNA sequence analysis of the regulatory region of lymphotropic papovavirus in peripheral blood mononuclear cells of an immunocompromised rhesus macaque. J. Clin. Microbiol. 40:1056–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liddington R. C., et al. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354:278–284 [DOI] [PubMed] [Google Scholar]
- 20. Maginnis M. S., Atwood W. J. 2009. JC virus: an oncogenic virus in animals and humans? Semin. Cancer Biol. 19:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinelli M., et al. 2002. Simian virus 40 sequences and expression of the viral large T antigen oncoprotein in human pleomorphic adenomas of parotid glands. Am. J. Pathol. 161:1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murata H., Teferedegne B., Sheng L., Lewis A. M., Jr., Peden K. 2008. Identification of a neutralization epitope in the VP1 capsid protein of SV40. Virology 381:116–122 [DOI] [PubMed] [Google Scholar]
- 23. Newman J. S., Baskin G. B., Frisque R. J. 1998. Identification of SV40 in brain, kidney and urine of healthy and SIV-infected rhesus monkeys. J. Neurovirol. 4:394–406 [DOI] [PubMed] [Google Scholar]
- 24. O'Neill F. J., Greenlee J. E., Carney H. 2003. The archetype enhancer of simian virus 40 DNA is duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells. Virology 310:173–182 [DOI] [PubMed] [Google Scholar]
- 25. Peden K., et al. 2008. Recovery of strains of the polyomavirus SV40 from rhesus monkey kidney cells dating from the 1950s to the early 1960s. Virology 370:63–76 [DOI] [PubMed] [Google Scholar]
- 26. Simon M. A., et al. 1999. Association of simian virus 40 with a central nervous system lesion distinct from progressive multifocal leukoencephalopathy in macaques with AIDS. Am. J. Pathol. 154:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sweet B. H., Hilleman M. R. 1960. The vacuolating virus, SV40. Proc. Soc. Exp. Biol. Med. 105:420–427 [DOI] [PubMed] [Google Scholar]
- 28. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 29. van Gorder M. A., et al. 1999. Cynomolgus polyoma virus infection: a new member of the polyoma virus family causes interstitial nephritis, ureteritis, and enteritis in immunosuppressed cynomolgus monkeys. Am. J. Pathol. 154:1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verschoor E. J., et al. 2008. Seroprevalence of SV40-like polyomavirus infections in captive and free-ranging macaque species. J. Med. Primatol. 37:196–201 [DOI] [PubMed] [Google Scholar]
- 31. White M. K., Safak M., Khalili K. 2009. Regulation of gene expression in primate polyomaviruses. J. Virol. 83:10846–10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.