Abstract
Severe early childhood caries (ECC), while strongly associated with Streptococcus mutans using selective detection (culture, PCR), has also been associated with a widely diverse microbiota using molecular cloning approaches. The aim of this study was to evaluate the microbiota of severe ECC using anaerobic culture. The microbial composition of dental plaque from 42 severe ECC children was compared with that of 40 caries-free children. Bacterial samples were cultured anaerobically on blood and acid (pH 5) agars. Isolates were purified, and partial sequences for the 16S rRNA gene were obtained from 5,608 isolates. Sequence-based analysis of the 16S rRNA isolate libraries from blood and acid agars of severe ECC and caries-free children had >90% population coverage, with greater diversity occurring in the blood isolate library. Isolate sequences were compared with taxon sequences in the Human Oral Microbiome Database (HOMD), and 198 HOMD taxa were identified, including 45 previously uncultivated taxa, 29 extended HOMD taxa, and 45 potential novel groups. The major species associated with severe ECC included Streptococcus mutans, Scardovia wiggsiae, Veillonella parvula, Streptococcus cristatus, and Actinomyces gerensceriae. S. wiggsiae was significantly associated with severe ECC children in the presence and absence of S. mutans detection. We conclude that anaerobic culture detected as wide a diversity of species in ECC as that observed using cloning approaches. Culture coupled with 16S rRNA identification identified over 74 isolates for human oral taxa without previously cultivated representatives. The major caries-associated species were S. mutans and S. wiggsiae, the latter of which is a candidate as a newly recognized caries pathogen.
INTRODUCTION
Early childhood caries (ECC), dental caries of the primary dentition, also known as nursing (bottle) caries, is the most common chronic infectious disease of childhood in the United States, affecting 28% of the population (6). Advanced forms of this disease, severe ECC, can destroy the primary dentition and is the major reason for hospital visits for young children (42). Severe ECC disproportionately affects disadvantaged ethnic and socioeconomic groups and can affect over 50% of the children in these groups (2, 19, 23, 38, 55).
Dental caries is caused by an interaction between acidogenic bacteria, a carbohydrate substrate which is frequently sucrose, and host susceptibility (51). The acidogenic and acid-tolerant bacterial species Streptococcus mutans is recognized to be the primary pathogen in early childhood caries (4, 8, 31, 50). S. mutans is detected in caries-free populations but is not detected in all cases of childhood caries (1, 27), suggesting that other species may be cariogenic pathogens.
Studies of severe ECC using culture have historically focused on selected bacterial groups, particularly S. mutans and other Streptococcus species and Lactobacillus, Actinomyces, and Veillonella species (27, 31, 33, 52). Isolates were generally identified phenotypically, sometimes only to the genus level, and thus, their relationships to currently defined human oral taxa on the basis of a 16S rRNA-defined taxonomy (11) are unclear. Culture studies demonstrated a strong association of S. mutans with ECC and severe ECC and also reported significant associations with selected Actinomyces and Lactobacillus species. Primary isolation on acid media has been used to select for acid-tolerant species that would be present in active carious lesions. Total counts were higher on acid agar from children with initial caries (45) and severe ECC (21) than from caries-free children. Acid broth enrichment was found to select for Streptococcus oralis, S. mutans, Actinomyces israelii, and Actinomyces naeslundii in severe ECC (31).
PCR of the 16S rRNA gene with cloning and subsequent sequencing has been used to evaluate the diversity of the microbiota of early childhood caries, and combined with use of species/taxon-specific probes to the 16S rRNA gene to evaluate disease associations between children with and without carious lesions (1, 3, 9, 18), these molecular methods were found to detect a wider diversity of species than previous culture-based studies, including unnamed taxa not previously recognized by culture. Species associated with samples from carious lesions included Gram-negative anaerobic species of Prevotella, Porphyromonas, and Selenomonas and an unnamed Bifidobacterium species which was associated with caries deep into dentine (3). Anaerobic culture, using 16S rRNA gene sequences for isolate identification, of carious dentine in adults detected a wide diversity of species that was comparable to the diversity found by cloning and sequencing analyses of the same samples (35). Further, the latter study indicated that increased proportions of actinobacteria were detected by culture than by clonal analysis (35), indicating the ability of anaerobic culture to detect a component of oral microbial diversity that was underrepresented by molecular methods.
The primary purpose of this study was to evaluate the microbiota of severe ECC to determine whether there were species, other than S. mutans, that were significantly associated with caries and that could be pathogens for this infection. The study hypothesized that the microbiota would differ between plaque from children with carious lesions and plaque from caries-free children and that any new acidogenic caries-associated taxa would be candidates as caries pathogens. By using enriched and acid isolation media, strict anaerobiosis for sample handling, and strain identification by 16S rRNA sequence analysis and by sampling a sufficient number of children to adequately differentiate disease categories, we anticipated that several isolates would belong to previously uncultured oral taxa previously known only from molecular cloning studies.
MATERIALS AND METHODS
Study population.
Medically healthy 2- to 6-year-old children were recruited from the dental clinics at the Departments of Pediatric Dentistry at Boston University Goldman School of Dentistry and Tufts University and Cambridge Health Alliance, Cambridge, MA. The children in this study were from among severe ECC and caries-free children for whom the diet, the microbiota by selective culture, and clonal analysis have been described (21, 24, 36). Children with severe ECC (14) had extensive caries in the primary dentition that affected over 36% of tooth surfaces with an average of 4 pulpally involved teeth (21) and were scheduled for restorative treatment under general anesthesia. Caries-free children, determined by visual and radiographic examination, had no cavities or enamel white spot lesions, which can represent early stages of tooth enamel demineralization. Inclusion criteria were that the child was medically healthy and had not used antibiotics within the last 3 months and the parent or guardian was willing to consent to the child's clinical examination and microbial sampling. The study design was explained to the child's parent or guardian, from whom informed consent was obtained if they were willing to participate with their child. The study design, protocol, and informed consent were approved by the Institutional Review Boards of Boston University, Tufts University, Cambridge Health Alliance, and the Forsyth Institute.
Clinical examination, sociodemographic information, and bacterial sampling.
Parents or guardians of the children completed a survey that included child demographic information and a 24-h food survey of a typical day (36). For the severe ECC children, clinical investigators recorded clinical measurements and took plaque samples while the child was under general anesthesia during the visit for dental treatment. For the caries-free children, clinical measurements and sampling were performed in the dental clinic. Clinical measurements comprised enumerating the teeth and extent of caries by tooth surface (mesial, buccal, lingual, labial, and occlusal), plaque and gingival indexes, and gingival bleeding (24).
Plaque samples were taken with sterile wooden toothpicks (32) from the buccal and interproximal surfaces of molars to include plaque from carious lesions in the severe ECC children. Each plaque sample was put in a 5-ml gastight vial containing 2 ml prereduced anaerobically sterilized (PRAS) Ringer's solution (49) with 3-mm glass beads (44) and placed in insulated bags with frozen freezer blocks. Samples were transported to the microbiology laboratory and processed within 4 h of sampling.
Microbiological analysis of plaque samples.
Samples were dispersed anaerobically by vortexing the anaerobic gastight sample vials for 30 s. Serial 10-fold dilutions of each sample were performed in PRAS Ringer's solution under oxygen-free argon (49). Appropriate dilutions were plated in duplicate on acid agar, comprising Trypticase soy agar (20 g/liter), brain heart infusion agar (26 g/liter), yeast extract (10 g/liter), and hemin (5 mg/liter), with the pH being adjusted to pH 5.0 with HCl before the agar was autoclaved. Samples were also plated on blood agar (BA), comprising Trypticase soy agar (20 g/liter), brain heart infusion agar (26 g/liter), yeast extract (10 g/liter), hemin (5 mg/liter), menadione (0.5 mg/liter), N-acetyl-muramic acid (10 mg/liter), and defibrinated sheep blood (50 ml/liter), and on fastidious anaerobe agar (FFA; Acumedia Manufacturers, Inc., Lansing, MI) with hemin (5 mg/liter) and defibrinated sheep blood (50 ml/liter). The primary isolation plates were incubated in an anaerobic chamber in an atmosphere of 80% N2, 10% H2, and 10% CO2 (49). After 8 to 10 days of anaerobic incubation, colonies from dilutions yielding about 30 to 100 colonies were counted. From the acid agar, 30 to 50 colonies of each sample were subcultured to acid and to blood agar. From the blood agar, 15 to 20 colonies from a BA plate and 15 to 20 colonies from an FFA plate were subcultured to a BA plate to obtain a total of 30 to 40 isolates from blood agar for each sample. Subcultures were reincubated anaerobically, and new colonies were examined for purity by evaluation of colony morphology. Mixed cultures were replated and cultured until there was colony homogeneity.
16S rRNA sequencing of isolates.
Strains were identified by harvesting cells from blood agar, even if primary isolation was on the acid agar. For the initial 10 samples, bacterial cells were harvested in 100 μl 50 mM Tris-EDTA buffer, pH 7.6, and DNA was purified using a DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA). For subsequent samples, colony PCR was performed by touching a sterile pipette tip to a young colony (3 to 4 days) and putting cells directly in the PCR mix. The 16S rRNA gene was amplified using universal bacterial primers F24 and F25 for 16S rRNA (16). PCRs were performed in 50 μl mix (NEB 10× ThermoPol reaction buffer, 1 U NEB Taq DNA polymerase, 10 mM Roche PCR nucleotide mix) under cycling conditions of 94°C for 15 min; 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 90 s plus 1 s per cycle; 72°C for 15 min; and a 4°C hold. For purified DNA, the initial denaturing step was decreased from 15 min to 5 min. The PCR amplification product (1,500 bases) was examined using 1% agarose gel electrophoresis.
The PCR product was treated with exonuclease I (USB Corporation, Cleveland, OH) and shrimp alkaline phosphatase (USB Corporation, Cleveland, OH), following the manufacturer's instructions, under conditions of 37°C for 15 min, 80°C for 15 min, and a 4°C hold to remove primers and deoxynucleoside triphosphates. Purified amplicons in the 16S rRNA gene were sequenced using an ABI Prism cycle sequencing kit on an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, CA). The sequencing primer was F15 (positions 519 to 533, reverse, 5′-TTA CCG CGG CTG CTG-3′) as previously described (24, 37).
Isolates that failed PCR with the F24 and F25 universal primers or in sequencing were treated as follows. DNA from a bacterial colony was isolated using 15 μl GeneReleaser reagent (Bioventures, Inc., Murfreesboro, TN), following the manufacturer's protocol, using the following conditions: 65°C for 30 s, 8°C for 30 s, 65°C for 90 s, 97°C for 180 s, 8°C for 60 s, 65°C for 180 s, 97°C for 60 s, 65°C for 60 s, and an 80°C hold. This was followed by PCR using the same reagents used for the initial PCR (with primers F24 and F25) but with primers amplifying 800 bp (primer 8FPL [forward, 5′-AGA GTT TGA TCC TGG CTC AG-3′] and primer 806R [reverse, 5′-GGA CTA CCA GGG TAT CTA AT-3′]) (40). Cycling conditions were 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min; 72°C for 5 min; and a 4°C hold. Electrophoresis on a 1% agarose gel was used to verify that amplicons were of 800 bases in size. Positive amplicons were sent to Genewiz, Inc. (South Plainfield, NJ), for cleanup and sequencing with the 806R primer.
Taxon identification using HOMD.
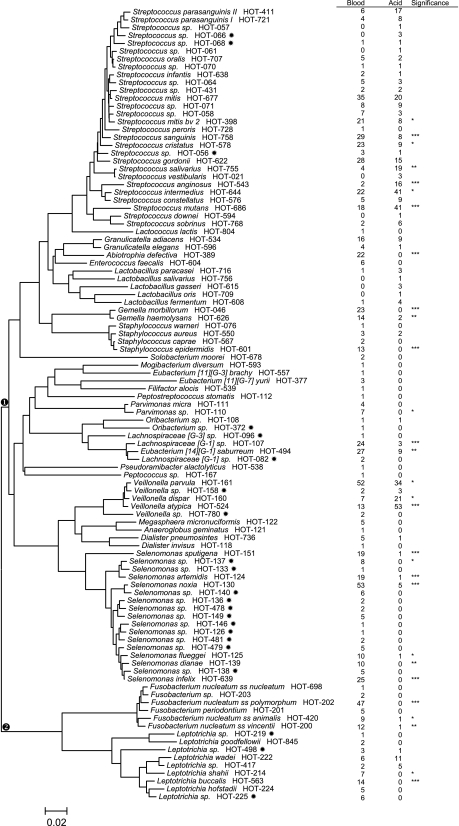
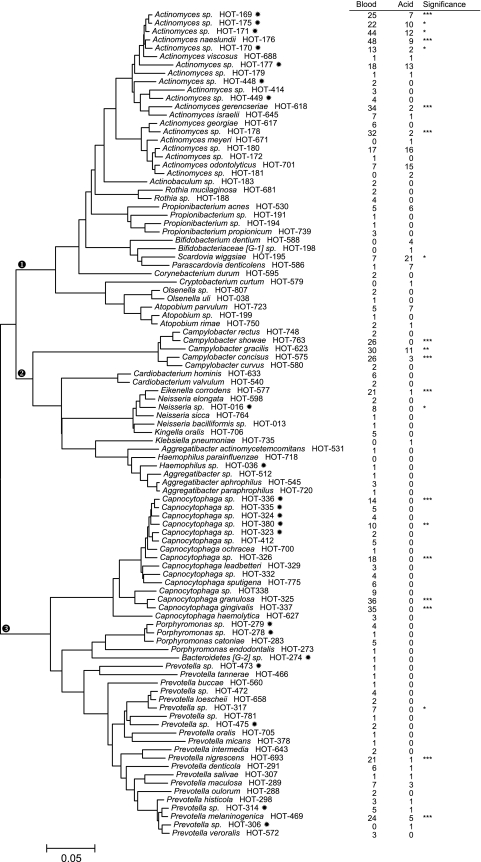
Isolate sequences approximately 400 to 800 bases long were examined by BLAST analysis against a 16S rRNA reference sequence set at the Human Oral Microbiome Database (HOMD) (10, 11). The Human Oral Microbiome Database (version 10) contains 619 species and phylotypes based on 98.5% similarity cutoffs of full 1,540-base 16S rRNA sequences. Each oral species or phylotype in the database has been given a Human Oral Taxon (HOT) number. Isolate identifications required a single read of approximately 350 to 500 bp. The threshold for BLAST identification of 500-base partial sequences was a 98% match (an approximately 8-base mismatch for partial sequences). The majority of partial sequences could be identified to species or human oral taxons, and the few for which some ambiguity existed were place in the taxa with the highest similarity match. Strains which did not match the HOMD references were clustered into extended taxa as previously described (11). The phylogenetic trees for the species found in this study were created from the full-length reference sequences in HOMD using the neighbor-joining method (Fig. 1 and 2).
Fig. 1.
Phylogenetic tree of Firmicutes and Fusobacteria species and phylotypes detected. Partial 16S rRNA sequences of isolates were compared with sequences in the Human Oral Microbiome Database. The trees were created from the full 16S rRNA reference sequence. HOT numbers are the reference taxon numbers in the Human Oral Microbiome Database. The star after the HOT numbers indicates a taxon previously recognized only from clone sequences. Numbers of children with taxa detected by primary isolation on either blood or acid agar are in the table to the right of the tree. Species or phylotypes that differed in detection frequencies are noted: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (chi-square test). The scale indicates the phylogenetic distance.
Statistical analyses.
Children were divided into the severe ECC and caries-free categories. Mean differences between severe ECC and caries-free children in age, plaque, gingival indices, and percentage of gingival sites bleeding were evaluated by t test. Contingency tables were used to evaluate associations between each disease category and gender and race-ethnicity and were compared by chi-square or Fisher's exact test. Counts from blood and acid isolation media were compared by the Kruskall-Wallis test (Mann-Whitney U test).
For sequence-based comparison, we combined isolate 16S rRNA gene sequences into three meta-libraries and assigned them into operational taxonomic units (OTUs). One library included all sequences from blood agar, the second included all sequences from acid agar, and the third included all blood and acid isolates combined. These meta-libraries were aligned to the 16S ribosomal SILVA reference database (39), and analyses were performed using MOTHUR software (41), as we previously described for a cloning and sequencing analysis of this population (24). A distance matrix was created, and sequences were assigned to phylotypes on the basis of the OTUs calculated from the genetic distance between sequences. Phylotype richness, coverage from Good's coverage estimation, (17, 43), and library diversity from the Shannon-Weaver diversity index (41) were calculated at evolutionary distances of 0.02, 0.03, 0.04, and 0.05 (22) for all three libraries. Using hypothesis-based comparison between the libraries, we also tested whether the blood and acid libraries have the same structure using the Cramer-von Mises test with ∫-LIBSHUFF (41).
For HOMD-derived taxa, phylotype or species detection frequencies were calculated by child for blood and acid isolation media and in severe ECC and caries-free disease categories and were compared by chi-square or Fisher's exact test. This analysis was performed for all children and separately for children in whom S. mutans was not detected. Children were divided into those in whom neither, one, or both of the species Streptococcus mutans and Scardovia wiggsiae were detected, and detection frequencies in these groups were compared by severe ECC and caries-free categories by chi-square analysis.
Statistical analyses were performed using SPSS software (SPSS Inc., Chicago, IL).
RESULTS
The severe ECC and caries-free (control) groups were balanced by age (3.65 and 3.91 years, respectively), gender, race, and ethnicity (Table 1). The racial classification of the children was 35% white, 34% black, 28% Asian, and 2.4% other races, with 24% of the children being Hispanic. Severe ECC children had more plaque and gingivitis than caries-free children (Table 1). Median acid counts for caries-free and severe ECC children were 1.23 × 107 and 2.98 × 107, respectively, and blood counts were 1.93 ×108 and 5.94 ×108, respectively. Counts were higher for severe ECC than caries-free children only for blood agar (P < 0.05). A total of 5,608 isolates were characterized: from caries-free children, 1,334 and 1,300 with acid and blood isolation, respectively, and from severe ECC children, 1,404 and 1,570 with acid and blood isolation, respectively.
Table 1.
Demographic and clinical characteristics of study population
| Characteristic | Caries free (n = 40) | Severe ECC (n = 42) | P value |
|---|---|---|---|
| Mean age (yr) ± SEM | 3.68 ± 0.16 | 3.97 ± 0.12 | 0.160a |
| No. (%) male | 21 (52.5) | 24 (57.1) | 0.61b |
| No. (%) by race: | |||
| White | 13 (32.5) | 16 (38.1) | |
| Black | 14 (35) | 14 (33.3) | |
| Asian | 12 (30) | 11 (26.2) | |
| Other/mixed | 1 (2.5) | 1 (2.4) | 0.959b |
| No. (%) Hispanic ethnicity | 10 (25) | 10 (25) | 1.0b |
| Clinical characteristics | |||
| Mean % carious surfaces | 0 | 33 ± 2.7 | |
| Mean plaque index ± SEM | 0.59 ± 0.07 | 1.54 ± 0.12 | <0.0001a |
| Mean gingival index ± SEM | 0.22 ± 0.07 | 1.74 ± 0.14 | <0.0001b |
| Mean percent sites bleeding ± SEM | 1.9 ± 1.0 | 52.1 ± 5.8 | <0.0001b |
Student's t test.
Chi-square test.
OTU data.
At a 0.02 evolutionary distance (98% similarity), 220 phylotypes were observed from acid agar, 420 from blood agar, and 563 from combined acid and blood agar (Table 2). The number of phylotypes from acid and blood media (563 phylotypes) was less than the sum of phylotypes on both media (640 phylotypes), reflecting detection of some taxa on both isolation media. The sum of acid and blood agar taxa was more than the total number of blood agar taxa, indicating that the acid agar taxa were not just a subset of the blood agar taxa. At the lower similarities, the number of phylotypes decreased, with the combined acid and blood agar consistently yielding higher numbers of phylotypes than either isolation medium by itself. Using Good's coverage estimation, higher coverage was observed from the acid agar (94.57%) than from the blood agar (91.38%) (Table 2). There was greater diversity, calculated using the Shannon-Weaver diversity index, from the blood isolation than the selective acid isolation. The blood and acid libraries were significantly different from each other using ∫-LIBSHUFF (P < 0.001).
Table 2.
Phylotype richness, coverage, and diversity of acid and blood isolate libraries
| Library | Evolutionary distance (% similarity) | No. of phylotypes | Good's coverage (%) | Shannon-Weaver diversity index |
|---|---|---|---|---|
| Acid isolates | 0.02 (98) | 220 | 94.57 | 3.81 ± 0.05 |
| 0.03 (97) | 172 | 96.13 | 3.51 ± 0.07 | |
| 0.04 (96) | 135 | 97.52 | 3.33 ± 0.06 | |
| 0.05 (95) | 117 | 98.11 | 3.26 ± 0.06 | |
| Blood isolates | 0.02 (98) | 420 | 91.38 | 5.19 ± 0.05 |
| 0.03 (97) | 322 | 94.27 | 4.93 ± 0.05 | |
| 0.04 (96) | 277 | 95.29 | 4.73 ± 0.05 | |
| 0.05 (95) | 221 | 96.67 | 4.52 ± 0.05 | |
| Acid and blood | 0.02 (98) | 563 | 93.67 | 4.98 ± 0.05 |
| isolates | 0.03 (97) | 425 | 95.75 | 4.67 ± 0.05 |
| 0.04 (96) | 346 | 96.88 | 4.44 ± 0.05 | |
| 0.05 (95) | 280 | 97.67 | 4.30 ± 0.04 |
HOMD species and phylotypes.
Of the 5,608 isolates characterized, 5,112 (92.2%) were found to have >98% sequence similarity identity to 198 HOMD groups (11), 337 (6.0%) isolates to 52 extended groups, and 139 (2.8%) isolates to 45 potentially novel groups. Taxa for which no isolates had previously been cultured were 45 HOMD taxa, 29 extended HOMD taxa, and 45 potentially novel groups. There was increased diversity from blood agar, with identification of 185 HOMD taxa, whereas 88 HOMD taxa were identified from the acid agar. The species richness or diversity between disease groups varied between isolation media and comprised 69 taxa from caries-free children and 60 taxa from severe ECC children from acid agar, but 144 taxa from caries-free children and 160 taxa from severe ECC children from blood agar.
The species and phylotypes identified to HOMD groups are shown in two phylogenic trees (Fig. 1 and 2), with the numbers of children with taxa detected from acid and blood agars presented in a table alongside each tree. Figure 1 illustrates the taxa detected in the phyla Firmicutes and Fusobacteria. The majority of phylotypes detected were in the Firmicutes (Fig. 1), including 28 Streptococcus species or phylotypes, 5 Lactobacillus species, 5 Veillonella species or phylotypes, and 17 Selenomonas species or phylotypes. Streptococcus species that were detected more frequently from blood agar than acid agar were Streptococcus mitis 2 and Streptococcus sanguinis and also (at P < 0.05) Streptococcus mitis 1 and Streptococcus gordonii. Streptococcus salivarius, Streptococcus cristatus, Streptococcus intermedius, Streptococcus anginosus, and S. mutans were detected more frequently on acid agar. Lactobacillus species were detected more frequently from acid agar than blood agar, although detection frequencies were too low for significant differences. Among the Veillonellaceae, Veillonella parvula was detected more frequently from blood agar, whereas Veillonella dispar and Veillonella atypica were detected more frequently from acid agar isolation. Selenomonas species were preferentially isolated from blood agar. Isolates for 13 previously recognized, but not yet cultivated, members of the Veillonellaceae were identified. In the phylum Fusobacteria, Fusobacterium and Leptotrichia species were preferentially isolated from blood agar (Fig. 1).
Figure 2 illustrates the species and phylotypes in the phyla Actinobacteria, Proteobacteria, and Bacteroidetes. Actinomyces taxa (17) dominated the Actinobacteria and included seven newly cultivated taxa. Most Actinomyces were preferentially isolated on blood agar, although several were also isolated on acid agar, suggesting a degree of acid tolerance. Bifidobacterium, Scardovia, and Parascardovia species were preferentially isolated on acid agar. Taxa in the Proteobacteria and Bacteroidetes were preferentially isolated on blood agar, with only Campylobacter gracilis and Prevotella melaninogenica being detected on acid agar. Isolates for 12 previously uncultured taxa were identified in the phylum Bacteroidetes.
Fig. 2.
Phylogenetic tree of taxa detected in Actinobacteria, Proteobacteria, and Bacteroidetes. Methods for isolate identification and tree preparation and definitions of the symbols are as described in the legend to Fig. 1. Species or phylotypes that differed in detection frequencies are noted: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (chi-square test).
Disease associations.
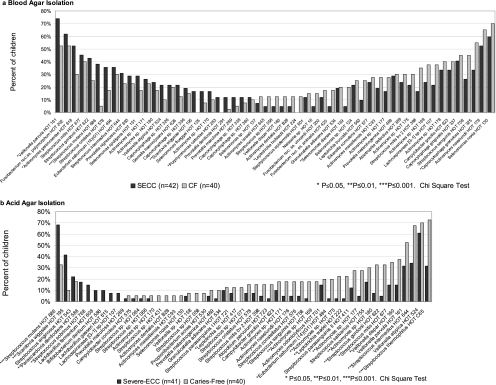
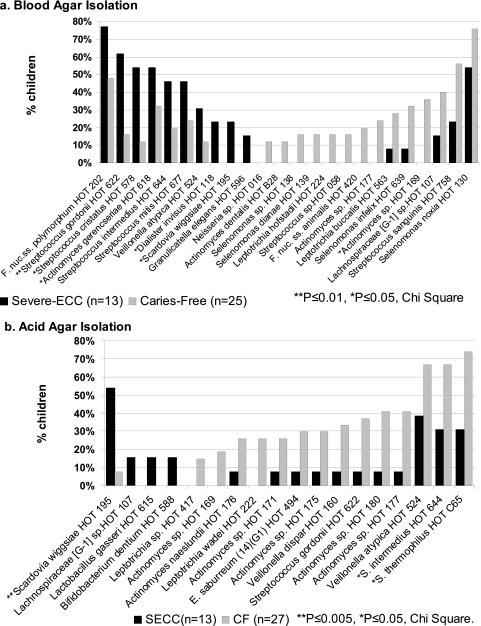
Species and phylotype associations with severe ECC and caries-free children are illustrated in Fig. 3 to 5. The major species from blood agar isolation detected more frequently from severe ECC than caries-free children (Fig. 3a) were S. mutans and Scardovia wiggsiae. Other species detected more frequently in severe ECC children from blood agar, but not significantly, were V. parvula, Actinomyces gerensceriae, and Porphyromonas catoniae. Species detected more frequently in caries-free children from blood agar, but not significantly, included Capnocytophaga granulosa, Leptotrichia hofstadii, and Selenomonas dianae. The major severe ECC-associated species from acid agar (Fig. 3b) were S. mutans, S. wiggsiae, Parascardovia denticolens, and Streptococcus sobrinus. Streptococcus mutans and S. wiggsiae detection remained significant after adjustment for multiple comparisons. Parascardovia denticolens, Streptococcus sobrinus, Lactobacillus fermentum, Bifidobacterium dentium, Lactobacillus gasseri, and Lachnospiraceae sp. HOT 107 were detected only from severe ECC children on acid agar. From acid agar, caries-free children had higher detection frequencies of Streptococcus thermophilus, S. intermedius, Streptococcus gordonii, Leptotrichia wadei, and unnamed Actinomyces sp. taxa HOT 180, HOT 177, HOT 175, and HOT 169.
Fig. 3.
Major species detected in severe ECC and caries-free children. (a) Blood agar isolation. Taxa detected in over 10% of either severe ECC or caries-free children are ordered from most frequent in severe ECC children from the left axis and most frequent in caries-free children from the right axis. Fusobacterium nuc. ss., Fusobacterium nucleatum subspecies. Streptococcus mutans and Scardovia wiggsiae showed the highest associations with severe ECC children. Species detected more frequently in caries-free children were Leptotrichia hofstadii, Selenomonas dianae, and Capnocytophaga granulosa. Several taxa previously recognized only from clone sequences, identified to HOT numbers, were detected at higher proportions from caries-free children, although differences were not statistically significant. (b) Acid (pH 5) agar isolation. Taxa detected in at least 5% of either severe ECC or caries-free children are ordered from most frequent in severe ECC children from the left axis and most frequent in caries-free children from the right axis. S. mutans, S. wiggsiae, Parascardovia denticolens, and S. sobrinus showed significant associations with severe ECC children. Caries-free children had higher detection frequencies of Streptococcus thermophilus, S. intermedius, Streptococcus gordonii, Leptotrichia wadei, and several unnamed Actinomyces species.
For the extended HOMD groups (11), Actinomyces dentalis HOT B29 was detected on blood agar in 2.5% of severe ECC and caries-free children, Actinomyces massiliensis HOT C93 was detected most frequently from blood agar (P < 0.0001) in 27% of the caries-free children and 19% in the severe ECC children, and S. thermophilus HOT C65 was significantly associated with acid agar (P < 0.0001) and caries-free children (Fig. 3b). Detection of additional provisional HOMD groups did not differ between acid and blood isolation. Comparisons between isolates in novel taxa were not performed, as only partial sequence data were available and the groups need verification.
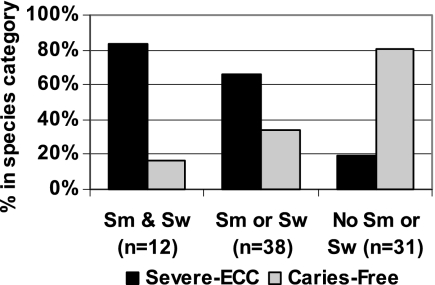
From both blood and acid agars, S. mutans and S. wiggsiae were the principal species associated with severe ECC, and the combination of detection of these species and disease category was examined (Fig. 4). Of the children with both species, over 80% were in the severe ECC disease category, whereas almost 80% of the children in whom neither species was detected were caries free.
Fig. 4.
S. mutans (Sm) and S. wiggsiae (Sw) in severe ECC and caries-free children. Eighty percent of children with both S. wiggsiae and S. mutans had severe ECC, whereas almost 80% of children with neither species were caries free, which was significantly different (P < 0.0001, chi-square).
S. mutans was detected in none of the severe ECC children. A preliminary analysis examined the caries-associated microbiota in the absence of S. mutans (Fig. 5). S. mutans was considered positive in a child if it was detected on either blood or acid agar for primary isolation. From blood agar (Fig. 5a), the major species associated with severe ECC in the absence of S. mutans were S. gordonii, S. cristatus, A. gerensceriae, Dialister invisus, and S. wiggsiae. From acid primary isolation (Fig. 5b), S. wiggsiae was associated with severe ECC, being detected in over 50% of the children. S. thermophilus and S. intermedius were detected more frequently in the S. mutans-free caries-free children (Fig. 3b).
Fig. 5.
Major species detected in the absence of S. mutans. (a) Major species detected from blood agar. Species were detected in >10% of either severe ECC or caries-free children. Species detected more frequently in severe ECC children in the absence of S. mutans were S. gordonii, S. cristatus, S. intermedius, A. gerensceriae, Dialister invisus, and S. wiggsiae. (b) Species detected from acid agar. Species were detected in >10% of either severe ECC or caries-free children. S. wiggsiae was associated with severe ECC children, being detected in over 50% of the children. Species detected more frequently in the S. mutans-free caries-free children were S. thermophilus and S. intermedius.
DISCUSSION
This study demonstrated that a wide diversity of bacterial species could be detected from dental plaque samples from young children by using anaerobic culture using enriched blood agar and acid agar for primary isolation. A novel finding of this study was that a newly named species, Scardovia wiggsiae, was significantly associated with severe early childhood caries, including in those children in whom the primary pathogen of dental caries, S. mutans, was not detected. A previous report in which molecular methods were used associated Scardovia wiggsiae (previously classified to be an unidentified Bifidobacterium species) with advanced dentinal caries in young children (3). The present study represents one of the largest culture-based studies of the microbiota of early childhood caries in which identifications from over 5,600 isolates from blood and acid agars were analyzed. The size of the subject population, at least 40 children in each clinical category, was dictated by the numbers of children needed to show statistically significant differences between groups.
Microbial diversity.
Molecular cloning and sequencing analysis of dental caries in children has revealed a widely diverse microbiota (1, 3, 18). When 16S rRNA clones could not be identified from searches in existing databases, it was assumed that the clones represented uncultured or uncultivable species. In the current study, isolation on the enriched blood agar medium detected a greater diversity of species than isolation on acid agar, indicating that the nutrients in blood and a neutral pH favor growth of many fastidious species in dental plaques. Anaerobic culture frequently detects higher counts than aerobic incubation for oral and clinical biofilms and many environmental samples because only the outer bacterial layer is exposed to environmental oxygen, and thus, most bacteria survive anaerobic conditions (29). The diversity by culture was greater than that by clonal analysis of samples from children in the same population, in part because of limitations of the primers used for the clonal analysis (24). Some taxa were identified only by clonal analysis, indicating that the cultural and molecular approaches were complementary. Similar to Wade and coworkers, we found that the microbiota of dental caries was mainly cultivable compared with taxa detected using clonal analysis (35). The majority of previously unrecognized taxa (new species) and taxa recognized only from clone sequences were detected by primary isolation on blood agar incubated anaerobically. Most of the unnamed/previously uncultivated taxa were in the genera Streptococcus, Selenomonas, Actinomyces, and Capnocytophaga. Using phenotype-based identifications, many would likely have been misclassified to named species.
Analysis of OTUs on the basis of 16S rRNA sequence data has been developed for examining diversity and coverage of the microbiota in samples (41) and emerges as the current “gold standard” for examining the comprehensiveness of samples. Sequence-based data for isolate identifications also allowed analysis of OTUs from meta-libraries assembled from sequences from isolates from acid and blood agars. A greater diversity was observed for isolation on blood agar, reflecting the greater nutrient value of this medium, as noted above. In contrast, the coverage was higher on acid than blood agar. This indicates that, perhaps because of the lower overall diversity observed on acid, we captured a higher proportion of taxa that could be cultured on acid agar than would be expected to grow on blood agar. A combination of acid and blood agar libraries did not improve coverage over that by acid agar culture, suggesting that additional species could be detected on blood agar if more isolates in each sample had been selected for identification. The purpose of this study, however, was to associate the most frequently encountered species with clinical categories; hence, 40 children in each disease category were sampled.
Microbiota on blood agar compared with acid agar.
The goal of plating samples directly onto an acid (pH 5) agar was to identify the acid-tolerant species as they existed in vivo in the plaque samples associated with carious lesions. Pathogens for dental caries would be expected to be acid tolerant to survive the acid environment of active carious lesions (45). Separating oral taxa from childhood caries into those that were acid tolerant and acid sensitive has previously been performed, most recently using isolation in broth (31) and on agar (45). In the current study, several taxa were selected for by using acid pH 5 primary isolation medium compared with blood agar, particularly S. mutans, S. salivarius, S. intermedius, S. anginosus, S. thermophilus, S. wiggsiae, and V. atypica. Parascardovia denticolens, Bifidobacterium dentium, and Lactobacillus species were detected almost exclusively on the acid medium. Svensater et al. (45) also reported that Bifidobacterium species and several oral streptococci, in addition to S. mutans, including S. anginosus, S. oralis, S. intermedius, S. oralis, and S. mitis, were isolated on acid agar using samples from children with initial caries. In similarity to the results of the current study, Marchant et al. (31) detected Streptococcus and Actinomyces israelii isolates from children with severe ECC from acid broth but not in a pH 7 broth. However, Marchant et al. reported A. naeslundii to be acid sensitive in their study, whereas in the current study isolates showed acid resistance. In contrast, Svensater et al. did not report isolation of acid-tolerant Actinomyces species (45). In the current study, using 16S rRNA gene sequence data for identifications, Actinomyces species were detected more frequently from the blood agar than acid agar, except for Actinomyces odontolyticus, although the difference was not significant. A. israelii was detected infrequently but was detected more frequently on blood. Differences between agar and broth isolation likely contributed to these differences since the current study and that of Svensater et al. (45) had similar findings when acid agar was used.
In contrast, other species were suppressed by acid isolation, including species in the acidogenic genera Selenomonas, Capnocytophaga, Prevotella, and Actinomyces. Without knowing acid tolerance, one might speculate that any acidogenic species detected in caries-associated plaque might be potential caries pathogens. It seems less likely that acid-sensitive species, however, would induce dental caries. Induced acid tolerance of strains has been examined for bacterial species in diverse oral genera (46). Species grouped into similar acid tolerance groups, whether from primary isolation, as in the current study, or in the laboratory testing of isolates (46). For example, both approaches categorized Lactobacillus species to be the most acid tolerant and Prevotella species to be in the more acid-sensitive group, whereas Actinomyces and most Streptococcus species showed intermediate levels of acid tolerance. The concordance between clinical sample and laboratory testing methods obtained when the acid tolerance of species was examined suggests that the approaches are complementary, and our data may provide insight into the inducible acid tolerance of oral bacteria. Knowing the acid tolerance characteristics of oral bacteria may provide additional insight into the ecology of species related to carious lesions.
Principal caries-associated microbiota.
Isolation of a wide diversity of species in samples from children with dental caries allowed us to seek new pathogens for this infection. Both blood and acid isolation media selected S. mutans and Scardovia wiggsiae to be the most disease associated. The association of S. mutans with severe early childhood caries has long been established in several culture-based studies using anaerobic culture on blood agar for isolation (27, 31, 33). The second mutans streptococcus, S. sobrinus, was detected infrequently: 5% from blood agar and 15% from acid agar and only in the presence of S. mutans. We also observed low frequencies of S. sobrinus detection in these children by selective culture for mutans streptococci (21) but higher detection frequencies (35%) for S. sobrinus by PCR (36). These differences based on microbiological methods suggest that species-specific PCR is a more sensitive approach than culture to detect S. sobrinus.
It has been recognized that species other than the mutans streptococci play an important role in dental caries (1, 3, 4, 54). Scardovia wiggsiae (HOT 195) is a newly named species (13) that was previously associated with advanced dentinal caries in young children using 16S rRNA gene probe analysis (3), having been detected as oral clone CX010 from a clonal analysis of plaque from periodontal pockets (37). Scardovia wiggsiae was detected in advanced dentinal caries of adults (35) and from occlusal carious lesions of children (30) as Scardovia genomospecies C1. The current study corroborates the finding of the study of Becker et al., which further clarifies its association with dentinal caries, although S. wiggsiae was also detected in early, white spot lesions (3). Bifidobacteriaceae are generally acidogenic and acid tolerant and have been significantly associated with caries in adults by culture (5, 34, 53) and in children (1, 30) and in our studies with severe ECC using PCR (36). In the current study, the combination of S. mutans and S. wiggsiae was highly associated with severe ECC. This disease-associated species combination has not previously been reported. Particularly intriguing was the preliminary observation of S. wiggsiae in association with severe ECC in the absence of detection of S. mutans, suggesting that S. wiggsiae may be independently associated with caries. An alternative interpretation would be that S. wiggsiae is a secondary invader and is involved with caries progression into dentine at later stages of the infection, when it has been observed that S. mutans is not the dominant species (18, 26). It is not clear why only one previous study associated S. wiggsiae with early childhood caries, but possible reasons include the different isolation media and length of anaerobic incubation used, the previous lack of 16S rRNA identification to the level of well-defined human oral taxa in culture-based studies, and poor amplification of Bifidobacteriaceae in clonal and sequencing analyses (24). Another possibility could be differences in the sites sampled for analysis.
Bifidobacteriaceae, Lactobacillus, and Actinomyces species.
Other named Bifidobacteriaceae species detected on the acid agar in severe ECC children were Parascardovia denticolens (18%) and Bifidobacterium dentium (10%) in the presence of S. mutans and B. dentium (15%) in the absence of S. mutans. B. dentium was the major species detected from childhood caries (30) using a selective medium for intestinal bifidobacteria modified for oral samples (5). Another unnamed Bifidobacterium species, HOT 198, was detected in one caries-free child (Fig. 2). Lactobacillus species were another group of aciduric, acidogenic anaerobic Gram-positive rod species detected and were mainly isolated from children with severe ECC, although rather infrequently and less frequently than from older children (18). In the current study, species detected were L. fermentum, Lactobacillus paracasei, and L. gasseri (in the presence and absence of S. mutans). In our study of ECC using DNA probes, L. fermentum and L. gasseri were detected in association with early carious, mainly white spot, lesions of ECC (25). Using selective isolation from children developing ECC (nursing caries), the major species detected were L. fermentum with Lactobacillus rhamnosus, Lactobacillus buchneri, and Lactobacillus casei, but Lactobacillus species comprised only 1.3% of total cultivable microbiota (31). In nursing caries, lactobacillus counts were higher at 10%, which was similar to the results of an earlier study (52), but the species of the strains were not determined (33). Lactobacillus species have been considered secondary invaders that flourish in the aciduric environment of active caries (28). Lactobacillus species frequently comprise low proportions of plaque bacteria and thus likely require targeted detection methods, for example, selective isolation (31) or quantitative PCR (7), to adequately evaluate their associations with early childhood caries.
Twenty different species or phylotypes of Actinomyces were detected and were mainly detected from blood agar, as discussed above. A. gerensceriae was detected more frequently in children with severe ECC, whereas A. naeslundii and several unnamed Actinomyces phylotypes were detected in caries-free children, although associations were not significant if the multiple comparisons are considered. The lack of strong associations of Actinomyces species with ECC is as previously observed (3, 33, 47). Actinomyces sp. HOT 179 (previously known as clone B19SC), which was overabundant in children with active caries (9), was infrequently detected in the current study. A. israelii was associated with severe ECC (31), whereas in the current study it was associated with A. gerensceriae and caries. These differences may be related to different methods of strain identification, as these species are genetically similar. Like the lactobacilli, actinomycetes may be secondary invaders of dentin (20, 28).
Streptococcus species.
The principal caries-associated streptococci were discussed above. In caries-free children, the streptococci detected were S. sanguinis, S. parasanguinis, and S. mitis, which is generally in agreement with the findings of previous studies (31).
Gram-negative species.
Acidogenic and nonacidogenic Gram-negative species were detected in these children, consistent with descriptions of childhood caries from molecular cloning and sequencing and 16S rRNA probe analyses (1, 3, 9). Many of the Gram-negative species, however, were suppressed by the acid agar, exceptions being certain veillonellae, Leptotrichia wadeii, P. melaninogenica, and Campylobacter gracilis. Only Veillonella parvula was associated with severe ECC. The acid tolerance and association with caries-free children has not previously been reported for Leptotrichia wadeii, but its association with health is consistent with the species description of being isolated from saliva of a healthy individual (15). C. gracilis is resistant to inhibitory agents (dyes and indicators) (48), so acid tolerance is consistent with the general resistance of this species. Veillonella species are frequently associated with caries, and while they are not acidogenic, they may play a critical role in the caries biofilm by supporting growth or survival of cariogenic species, as has been described for the interactions between S. mutans and Veillonella species (33). Gram-negative species have been observed invading dentinal tubules, and it has been suggested that they may be involved in extension of lesions deep into dentine (28). All of the severe ECC children had deep dentinal caries, with over 25% of molars having pulp-invasive lesions (21). The presence of Gram-negative species may also reflect the gingival inflammation experienced by these children, particularly as interproximal plaque, adjacent to the gingival sulcus, was included in the sample sites assayed.
Study strengths and limitations.
A major strength of this study was use of the HOMD, a curated 16S rRNA sequence database, to characterize isolates. HOT phylotypes reflect a consensus of sequences rather than identifications to individual clones, for example, in the GenBank database. HOMD phylotypes are linked to a sequence available from NCBI or HOMD, providing a better method to communicate findings than using laboratory-specific identification schemes. Use of sequence-based data for strain identifications proved a more rapid method than our previous cultural studies that used combinations of phenotypic tests with protein profiles to identify isolates (49). Limitations of the study included use of partial 16S rRNA sequence data for strain characterization, which are not as robust as data from full gene sequences. For some species that are difficult to identify to the species level, more robust identifications could have been obtained by sequencing other genes, for example, housekeeping genes for oral streptococci in the S. mitis and S. oralis group (12). Further, we recognize that not all fastidious species or species in very low abundance were cultured.
Clinical relevance.
The principal clinical implication of this study was expanding our understanding of the cultural microbiota of severe ECC and identifying a new species, S. wiggsiae, as a candidate caries pathogen for early childhood caries. S. wiggsiae becomes a candidate, along with other aciduric, acidogenic species, for testing as a risk indicator for dental caries by examining its pathogenic potential in vitro and in model systems, associations with initial carious lesions, and ability to predict caries in longitudinal studies, either as a single species or in species combinations.
In conclusion, this study achieved its primary objective by associating a newly recognized species, Scardovia wiggsiae, with severe ECC. Since this species is acidogenic (13) and aciduric, as it was isolated on an agar of pH 5, S. wiggsiae is a candidate to be a new caries pathogen. Further, we isolated and identified as diverse a microbiota as had been described using cloning and sequencing analysis and have isolates of previously uncultivated oral taxa. Classification of oral bacteria into those with and without tolerance to acid isolation will provide new data for ecology studies examining relationships between species in dental caries.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants DE-014264 (to A.C.R.T.), DE-015847 (to A.C.R.T.), DE-016937 (to F.E.D.), DE-007151 (to E.K.), DE-007327 (to N.I.C. and E.K.) from National Institute of Dental and Craniofacial Research, an intern grant from the Harvard School of Dental Medicine (to J.H.), and a postdoctoral fellowship from the Kingdom of Saudi Arabia (to M.A.D.).
We acknowledge Rachel Montgomery, Benita Demirza, Caroline Young, David Okuji, and Sam Merabi for subject recruitment and clinical measurements and Shulin Charles Lu for assistance with sample processing. We thank William Wade and Bruce Paster for critical review of the manuscript.
Footnotes
Published ahead of print on 2 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Aas J. A., et al. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Azevedo T. D., Bezerra A. C., de Toledo O. A. 2005. Feeding habits and severe early childhood caries in Brazilian preschool children. Pediatr. Dent. 27:28–33 [PubMed] [Google Scholar]
- 3. Becker M. R., et al. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beighton D. 2005. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent. Oral Epidemiol. 33:248–255 [DOI] [PubMed] [Google Scholar]
- 5. Beighton D., et al. 2008. Isolation and identification of bifidobacteriaceae from human saliva. Appl. Environ. Microbiol. 74:6457–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beltran-Aguilar E. D., et al. 2005. Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis—United States, 1988-1994 and 1999-2002. MMWR Surveill. Summ. 54:1–43 [PubMed] [Google Scholar]
- 7. Byun R., et al. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42:3128–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caufield P. W., Li Y., Dasanayake A. 2005. Dental caries: an infectious and transmissible disease. Compend. Contin. Educ. Dent. 26:10–16 [PubMed] [Google Scholar]
- 9. Corby P. M., et al. 2005. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 43:5753–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewhirst F. E., et al. 2008. The Human Oral Microbiome Database. http://www.HOMD.org
- 11. Dewhirst F. E., et al. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Do T., et al. 2009. Population structure of Streptococcus oralis. Microbiology 155:2593–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Downes J., et al. 2011. Scardovia wiggsiae sp. nov., isolated from the human oral cavity and clinical material, and emended descriptions of the genus Scardovia and Scardovia inopinata. Int. J. Syst. Evol. Microbiol. 61:25–29 [DOI] [PubMed] [Google Scholar]
- 14. Drury T. F., et al. 1999. Diagnosing and reporting early childhood caries for research purposes. A report of a workshop sponsored by the National Institute of Dental and Craniofacial Research, the Health Resources and Services Administration, and the Health Care Financing Administration. J. Public Health Dent. 59:192–197 [DOI] [PubMed] [Google Scholar]
- 15. Eribe E. R., et al. 2004. Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp. nov., Leptotrichia hofstadii sp. nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov. Int. J. Syst. Evol. Microbiol. 54:583–592 [DOI] [PubMed] [Google Scholar]
- 16. Fox J. G., et al. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755–763 [DOI] [PubMed] [Google Scholar]
- 17. Good I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 18. Gross E. L., et al. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48:4121–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hardison J. D., et al. 2003. The 2001 Kentucky Childrens Oral Health Survey: findings for children ages 24 to 59 months and their caregivers. Pediatr. Dent. 25:365–372 [PubMed] [Google Scholar]
- 20. Hoshino E. 1985. Predominant obligate anaerobes in human carious dentin. J. Dent. Res. 64:1195–1198 [DOI] [PubMed] [Google Scholar]
- 21. Hughes C., et al. 2011. Aciduric microbiota and mutans streptococci in severe and recurrent severe early childhood caries. Pediatr. Dent. [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes J. B., Hellmann J. J., Ricketts T. H., Bohannan B. J. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin B. H., et al. 2003. Early childhood caries: prevalence and risk factors in Seoul, Korea. J. Public Health Dent. 63:183–188 [DOI] [PubMed] [Google Scholar]
- 24. Kanasi E., et al. 2010. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 44:485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanasi E., et al. 2010. Microbial risk markers for childhood caries in pediatricians' offices. J. Dent. Res. 89:378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loesche W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loesche W. J., Rowan J., Straffon L. H., Loos P. J. 1975. Association of Streptococcus mutans with human dental decay. Infect. Immun. 11:1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Love R. M., Jenkinson H. F. 2002. Invasion of dentinal tubules by oral bacteria. Crit. Rev. Oral Biol. Med. 13:171–183 [DOI] [PubMed] [Google Scholar]
- 29. Manganiello A. D., et al. 1977. Attempts to increase viable count recovery of human supragingival dental plaque. J. Periodontal Res. 12:107–119 [DOI] [PubMed] [Google Scholar]
- 30. Mantzourani M., et al. 2009. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 43:308–313 [DOI] [PubMed] [Google Scholar]
- 31. Marchant S., Brailsford S. R., Twomey A. C., Roberts G. J., Beighton D. 2001. The predominant microflora of nursing caries lesions. Caries Res. 35:397–406 [DOI] [PubMed] [Google Scholar]
- 32. Milgrom P., et al. 2000. Dental caries and its relationship to bacterial infection, hypoplasia, diet, and oral hygiene in 6- to 36-month-old children. Community Dent. Oral Epidemiol. 28:295–306 [DOI] [PubMed] [Google Scholar]
- 33. Milnes A. R., Bowden G. H. 1985. The microflora associated with developing lesions of nursing caries. Caries Res. 19:289–297 [DOI] [PubMed] [Google Scholar]
- 34. Modesto M., Biavati B., Mattarelli P. 2006. Occurrence of the family Bifidobacteriaceae in human dental caries and plaque. Caries Res. 40:271–276 [DOI] [PubMed] [Google Scholar]
- 35. Munson M. A., Banerjee A., Watson T. F., Wade W. G. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer C. A., et al. 2010. Diet and caries-associated bacteria in severe early childhood caries. J. Dent. Res. 89:1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paster B. J., et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Postma T. C., Ayo-Yusuf O. A., van Wyk P. J. 2008. Socio-demographic correlates of early childhood caries prevalence and severity in a developing country—South Africa. Int. Dent. J. 58:91–97 [DOI] [PubMed] [Google Scholar]
- 39. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Relman D. A., Schmidt T. M., MacDermott R. P., Falkow S. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293–301 [DOI] [PubMed] [Google Scholar]
- 41. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheller B., Williams B. J., Lombardi S. M. 1997. Diagnosis and treatment of dental caries-related emergencies in a children's hospital. Pediatr. Dent. 19:470–475 [PubMed] [Google Scholar]
- 43. Singleton D. R., Furlong M. A., Rathbun S. L., Whitman W. B. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Slots J., Rams T. E., Listgarten M. A. 1988. Yeasts, enteric rods and pseudomonads in the subgingival flora of severe adult periodontitis. Oral Microbiol. Immunol. 3:47–52 [DOI] [PubMed] [Google Scholar]
- 45. Svensater G., Borgstrom M., Bowden G. H., Edwardsson S. 2003. The acid-tolerant microbiota associated with plaque from initial caries and healthy tooth surfaces. Caries Res. 37:395–403 [DOI] [PubMed] [Google Scholar]
- 46. Svensater G., Larsson U. B., Greif E. C., Cvitkovitch D. G., Hamilton I. R. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12:266–273 [DOI] [PubMed] [Google Scholar]
- 47. Tang G., Samaranayake L. P., Yip H. K. 2004. Genotypic diversity of oral Actinomyces naeslundii genospecies 1 and 2 in caries-active preschool children. Oral Microbiol. Immunol. 19:371–378 [DOI] [PubMed] [Google Scholar]
- 48. Tanner A., Lai C. R. B. S. C. H., Listgarten M. A., Visconti R. A., Socransky S. S. 1981. Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and Description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int. J. Syst. Bacteriol. 31:432–445 [Google Scholar]
- 49. Tanner A. C. R., Maiden M. F., Macuch P. J., Murray L. L., Kent R. L., Jr 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85–98 [DOI] [PubMed] [Google Scholar]
- 50. Tanzer J. M., Livingston J., Thompson A. M. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028–1037 [PubMed] [Google Scholar]
- 51. Van Houte J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672–681 [DOI] [PubMed] [Google Scholar]
- 52. Van Houte J., Gibbs G., Butera C. 1982. Oral flora of children with “nursing bottle caries.” J. Dent. Res. 61:382–385 [DOI] [PubMed] [Google Scholar]
- 53. Van Houte J., Lopman J., Kent R. 1996. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 75:1008–1014 [DOI] [PubMed] [Google Scholar]
- 54. Van Houte J., Lopman J., Kent R. 1994. The predominant cultivable flora of sound and carious human root surfaces. J. Dent. Res. 73:1727–1734 [DOI] [PubMed] [Google Scholar]
- 55. Warren J. J., et al. 2009. A longitudinal study of dental caries risk among very young low SES children. Community Dent. Oral Epidemiol. 37:116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]