Abstract
We evaluated the analytical, work flow, and clinical performance of the Versant CT/GC DNA 1.0 assay (Versant CT/GC assay, where “CT” represents Chlamydia trachomatis and “GC” represents Neisseria gonorrhoeae). The assay simultaneously detects Chlamydia trachomatis and Neisseria gonorrhoeae in swab and first-catch urine (FCU) specimens. The limit of detection (LoD) was determined to be 342 copies/ml for C. trachomatis and 137 copies/ml for GC. The Versant CT/GC assay detected 15 C. trachomatis serovars and 46 GC strains. The Versant CT/GC assay demonstrated no cross-reactivity with 136 potentially cross-reacting organisms. Clinical concordance of the Versant CT/GC assay to the Aptima Combo 2 (AC2) assay from Gen-Probe was demonstrated using 1,129 patient specimens, including 589 urine and 540 swab specimens. Discrepant specimens were subjected to DNA sequencing to identify the presence of amplified targets and to identify false-positive and false-negative results. Overall percent agreement was greater than 98%. Positive and negative percent agreements for detection of C. trachomatis were 94.4% and 99.1%, respectively, in urine specimens and 95.8% and 99.8%, respectively, in swab specimens. Positive percent agreement for the detection of N. gonorrhoeae was 100% in both urine and swab specimens, and negative percent agreements were 99.6% and 99% in urine and swab specimens, respectively. In conclusion, the performance of the Versant CT/GC assay was comparable to that of the AC2 assay. The Versant CT/GC assay can be recommended for the detection of C. trachomatis and N. gonorrhoeae in swab and urine specimens of symptomatic and asymptomatic individuals.
INTRODUCTION
Chlamydia trachomatis and Neisseria gonorrhoeae infections are the two most common sexually transmitted diseases (STDs) in the United States, with annual direct and indirect costs exceeding $2.4 billion and $1 billion, respectively (12). In 2008, 1,210,523 chlamydia cases (401.3 per 100,000 population) and 336,742 gonorrhea cases (111.6 per 100,000 population) were reported to the Centers for Disease Control and Prevention (CDC) (3). Most women and men with chlamydial infection and many women with gonorrheal infection do not have symptoms that would prompt them to seek medical care (11, 15). Therefore, most infections remain undiagnosed and untreated, increasing the risk of substantial morbidity and economic cost (2, 8, 17, 18). Thus, to identify and treat infected individuals and their partners and to reduce this high disease incidence and economic burden, a rapid, accurate, reliable, and cost-effective diagnostic test for both infections is needed (14).
Diagnostic test manufacturers have developed new-generation nucleic acid amplification tests (NAATs) that have been shown to give superior performance to culture, direct fluorescent-antibody assay, and enzyme immunoassays (4, 7, 9, 10, 16). In this study, we describe the analytical, work flow, and clinical performance of the new Versant CT/GC DNA 1.0 assay (Versant CT/GC assay, where “CT” represents Chlamydia trachomatis and “GC” represents Neisseria gonorrhoeae) on the fully integrated Versant kinetic PCR (kPCR) molecular system. The Versant CT/GC DNA 1.0 assay (kPCR) and Versant urine transport kit (UTK) are CE marked and are not commercially available in the United States. The clinical performance of the Versant CT/GC assay is compared to that of the Aptima Combo 2 (AC2) assay from Gen-Probe for the detection of C. trachomatis and N. gonorrhoeae infections in both female and male first-catch urine (FCU) and swab specimens.
MATERIALS AND METHODS
The Versant kPCR molecular system consists of a sample preparation (SP) module designed for automated sample preparation and an amplification/detection (AD) module designed for real-time (or kinetic) PCR and detection. The Versant CT/GC assay is run on the Versant kPCR molecular system.
Sample preparation.
A new nucleic acid extraction and purification procedure was developed using Siemens' proprietary, highly uniform magnetic silica beads. The sample preparation step is fully automated on the Versant sample preparation module. For sample preparation, proteinase K and a chaotropic lysis buffer were added to 0.25 ml of swab transport medium or urine specimen to inactivate DNases and to lyse bacterial particles. Following bacterial lysis, magnetic silica beads and the internal control (IC) were added to the mixture, and released C. trachomatis/N. gonorrhoeae DNAs and other nucleic acids were captured by the magnetic silica beads. The mixture was incubated at 62°C for 15 min. The magnetic silica beads with captured DNA were washed by a series of wash steps (850 μl in wash I, 450 μl in wash II, and 450 μl in wash III) to remove any remaining sample components. Purified DNA was eluted in 100 μl of elution buffer and incubated at 74°C for 10 min. Finally, 25 μl of purified DNA was automatically eluted and added to a 96-well PCR plate containing 25 μl of the CT/GC primer-probe mix and enzyme mix provided in the Versant CT/GC DNA 1.0 assay (kPCR).
Real-time PCR amplification and detection.
Chlamydia trachomatis-specific PCR primers and probe were designed to target the GenBank nucleic acid sequence of the 7.5-kb C. trachomatis cryptic plasmid. The C. trachomatis probe was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and Black Hole Quencher at the 3′ end; this primer-probe set also detects the new variant Chlamydia trachomatis that has a 377-bp deletion in the cryptic plasmid. Neisseria gonorrhoeae-specific PCR primers and probe were designed to target the nucleic acid sequence of the 7 to 8 copies of pivNG gene loci of N. gonorrhoeae. The N. gonorrhoeae probe was labeled with Cy5 at the 5′ end and Black Hole Quencher at the 3′ end. The IC fragment is a 195-bp double-stranded DNA (dsDNA) fragment derived from Methanobacterium thermoautrophicum; the IC probe was labeled with hexachloro-6-carboxy-fluorescein (HEX) at the 5′ end and Black Hole Quencher at the 3′ end. The Versant amplification and detection module automatically performs the amplification and detection steps in a single, sealed reaction well using a master mix containing 4.5 units of Taq DNA polymerase, 60 to 90 nM oligonucleotide primers and probes, 10× PCR buffer, 0.4 to 0.8 mM deoxynucleoside triphosphates (dNTPs), reference dye (carboxy-X-rhodamine [ROX]), MgCl2, and 0.75 units of AmpErase uracil N-glycosylase (UNG). The PCR thermocycling condition is as follows: after 10 min at 50°C for UNG activation and 15 min at 95°C for Taq activation, 40 PCR cycles of 15 s at 95°C and 1 min at 62°C.
Assay amplification profiles and reference standard curves.
C. trachomatis and N. gonorrhoeae reference standards, consisting of a 1.6-kb C. trachomatis dsDNA fragment and a 1.0-kb N. gonorrhoeae dsDNA fragment, were prepared using plasmid clones containing a region of sequences targeted by this assay. Each C. trachomatis and N. gonorrhoeae reference standard stock was evaluated by two independent, validated analytical methods: an assay for the A260 and phosphate assay. The nominal concentration of the reference standard was determined from its phosphate content using U.S. National Institute of Standards and Technology phosphate standard reference materials. Diluted C. trachomatis and N. gonorrhoeae reference standards were processed and tested using the same extraction and Versant CT/GC assay methods used for the test samples. A linear regression (mean threshold cycle [CT] values versus the nominal concentration of the standards) supplied the slope and intercept values for quantification of samples based on CT values. Sample preparation (SP) recovery was also accessed by testing both C. trachomatis and N. gonorrhoeae reference standard curves with (and without) the SP step.
Analytical sensitivity. (i) LoD.
The limit of detection (LoD) of the assay was determined by testing assay sensitivity panels prepared using heated-inactivated C. trachomatis (lymphogranuloma venereum II, strain 434) and N. gonorrhoeae (ATCC 19424) cells diluted in Micro Test M4RT (Remel, Lenexa, KS) or a negative urine pool containing preservative. Initial cell stock concentrations were determined using C. trachomatis or N. gonorrhoeae reference standards. The assay LoD was determined to be the lowest concentration of analyte that was detected greater than or equal to 95% of the time, using a statistical logistic regression model, on the basis of the hit rate at each level of the panel. The panels were tested using three different lots of reagents on three Versant kPCR molecular systems.
(ii) C. trachomatis serovar and N. gonorrhoeae isolate detection.
C. trachomatis serovar panels were used to assess the ability of the assay to detect the most prevalent C. trachomatis serotypes. These panels were prepared by diluting 15 heat-inactivated C. trachomatis serovars to concentrations ranging from 0.0067 to 0.67 inclusion-forming units (IFU) per assay in tryptic soy broth (TSB) buffer. N. gonorrhoeae isolate panels were prepared by diluting extracted genomic DNAs of 46 different N. gonorrhoeae strains into Tris-EDTA (TE) buffer to 50, 25, 10, and 1 copy per PCR reaction. The assay sensitivity for each serovar or isolate was determined on the basis of a 100% hit rate in triplicate at the lowest level tested.
Analytical specificity.
The Versant CT/GC assay was also challenged with 137 potentially cross-reactive organisms in M4RT (or TE buffer) at 1.0 × 106 cells per assay or 2.0 × 105 copies of genomic DNA per assay for all organisms except Chlamydia pneumoniae and Chlamydia psittaci, both of which were tested at 1.0 × 105 IFU per assay. The organisms listed in Table 1 are typical of organisms potentially found in urogenital sites that C. trachomatis/N. gonorrhoeae NAAT manufacturers use to challenge NAATs for cross-reactivity and to determine analytical specificity.
Table 1.
Organisms that are potentially cross-reactive with C. trachomatis or N. gonorrhoeae
| Microorganism or virus |
|---|
| Achromobacter xerosis |
| Acinetobacter calcoaceticus |
| Acinetobacter lwoffii |
| Aerococcus viridans |
| Aeromonas hydrophila |
| Alcaligenes faecalis |
| Arcanobacterium pyogenes |
| Bacillus subtilis |
| Bacillus thuringiensis |
| Bacteroides thetaiotaomicrona |
| Brevibacterium linens |
| Candida albicans |
| Candida glabrata |
| Candida parapsilosis |
| Candida tropicalis |
| Chlamydia pneumoniae (4)b |
| Chlamydia psittaci (6BC)c |
| Chromobacterium violaceum |
| Citrobacter freundii (C. braakii) |
| Corynebacterium genitalium |
| Corynebacterium renale |
| Corynebacterium xerosis |
| Deinococcus radiodurans |
| Derxia gummosa |
| Edwardsiella tarda |
| Enterobacter aerogenes |
| Enterobacter cloacae |
| Enterococcus avium |
| Enterococcus faecalis |
| Enterococcus faecium |
| Epstein-Barr virus (EBV) |
| Erysipelothrix rhusiopathiae |
| Escherichia coli |
| Escherichia intermedia |
| Flavobacterium odoratum (Myroides odoratus) |
| Gardnerella vaginalis |
| Gemella haemolysans |
| Haemophilus influenzae |
| Haemophilus parainfluenzae |
| Helicobacter pylori |
| Hepatitis A virus (HAV) |
| Hepatitis B virus |
| Herpes simplex virus type 1 (HSV-1) |
| Herpes simplex virus type 1 (HSV-2) |
| Human immunodeficiency virus type 1 |
| Human papillomavirus type 16 |
| Human papillomavirus type 18 |
| Kingella kingae |
| Klebsiella oxytoca |
| Klebsiella pneumoniae |
| Lactobacillus acidophilus |
| Lactobacillus brevis |
| Lactobacillus casei |
| Lactobacillus delbrueckii lactis |
| Lactobacillus jensenii |
| Lactobacillus plantarum |
| Leuconostoc mesenteroides |
| Listeria monocytogenes |
| Micrococcus luteus |
| Moraxella (Branhamella) catarrhalis |
| Moraxella lacunata |
| Moraxella osloensis |
| Morganella morganii |
| Mycoplasma genitaliuma |
| Mycoplasma hominisa |
| Neisseria (Bergeriella) denitrificans |
| Neisseria cinerea (5) |
| Neisseria elongata (3) |
| Neisseria flava |
| Neisseria flavescens (3) |
| Neisseria lactamica (4) |
| Neisseria meningitides Aa |
| Neisseria meningitides Ba |
| Neisseria meningitides C (3)a |
| Neisseria meningitides Da |
| Neisseria meningitides Xa |
| Neisseria meningitides Ya |
| Neisseria meningitides Za |
| Neisseria mucosa (2) |
| Neisseria mucosa var. heidelbergeneisis |
| Neisseria perflava (3) |
| Neisseria polysacchareae |
| Neisseria sicca (3) |
| Neisseria subflava (2) |
| Paracoccus denitrificans |
| Pasteurella multocida |
| Peptostreptococcus (Ruminococcus) productusa |
| Propionibacterium acnes |
| Proteus mirabilis |
| Proteus vulgaris |
| Providencia stuartii |
| Pseudomonas aeruginosa |
| Pseudomonas fluorescens |
| Pseudomonas putida |
| Rahnella aquatilis |
| Saccharomyces cerevisiae |
| Salmonella enterica subsp. enterica |
| Salmonella enterica subsp. enterica serovar Minnesota |
| Salmonella enterica subsp. enterica serovar Typhimurium |
| Serratia marcescens |
| Shigella sonnei |
| Staphylococcus aureus |
| Staphylococcus epidermidis |
| Staphylococcus faecalis (S. haemolyticus) |
| Staphylococcus saprophyticus |
| Streptococcus agalactiae (2) |
| Streptococcus bovis |
| Streptococcus mitis |
| Streptococcus mutans |
| Streptococcus pneumoniae |
| Streptococcus pyogenes (group A) |
| Streptococcus salivarius |
| Streptococcus sanguinis |
| Trichomonas vaginalisa |
| Ureaplasma urealyticuma |
| Vibrio choleraea |
| Yersinia enterocolitica |
Extracted genomic DNA rather than whole organisms were tested.
Values in parentheses represent the number of different strains tested in this study.
The strain of Chlamydia psittaci used in this study.
Assay work flow study.
An assay work flow study was conducted using a batch size of 96 (94 samples plus 2 external controls) on the Versant kPCR molecular system. Operator hands-on time as well as time for completion of SP and AD modules was measured.
Clinical performance with prospectively collected specimens and comparison with AC2 assay.
Male FCU (n = 312), female FCU (n = 277), male urethral swab (n = 280), and female endocervical swab (n = 260) specimens were prospectively collected from symptomatic and asymptomatic patients attending two Los Angeles County Department of Public Health STD clinics in Los Angeles, CA. Specimens were collected by use of the Siemens Versant UTK and the M4RT combo collection kit. The Siemens UTK contains 400 μl of urine preservative buffer in order to stabilize the DNA in urine. Also, urine and swab samples from the same individual were collected using Gen-Probe Aptima urine and swab collection kits, according to their respective manufacturer's package insert instructions.
All specimens were tested for C. trachomatis and N. gonorrhoeae with the AC2 assay at the Los Angeles County Department of Public Health Laboratory and with the Versant CT/GC assay at the Siemens Healthcare Diagnostics facility in Berkeley, CA. The AC2 assay was used as a comparative method because of its high sensitivity and specificity as well as its ability to simultaneously detect both C. trachomatis and N. gonorrhoeae in a single specimen. Test results from the two assays were compared. Percent concordance of the Versant CT/GC assay compared to the AC2 assay was determined.
Discrepant specimen resolution.
Specimens that tested positive by the Versant CT/GC assay and the AC2 assay were considered consensus positive for C. trachomatis and/or N. gonorrhoeae. Similarly, specimens that tested negative by both assays were considered consensus negative. Positive percent agreement (PPA) and negative percent agreement (NPA) of the two assays, along with their 95% confidence intervals (CIs), were then calculated (5, 6).
Specimens that showed discrepant results compared to the AC2 assay results were retested by the Versant assay. If the retested Versant assay results became concordant with the AC2 assay results, the specimen was not sequenced. Clinical samples that remained discrepant were purified using a Qiagen QIAamp MinElute kit. Purified DNA from these discrepant samples was then subjected to touchdown PCR with primers flanking the target amplification region of the C. trachomatis cryptic plasmid. PCR products from samples that produced no band were subjected to a second round of PCR. Conditions for touchdown PCR were as follows: initial 95°C for 10 min, a touchdown phase of 10 cycles of 94°C for 30 s and 67°C for 45 s with a 0.5°C decrease in temperature per cycle, followed by 72°C for 1 min and a normal amplification phase of 39 cycles of 94°C for 30 s and 62°C for 45 s and then 72°C for 1 min and a final extension step of 72°C for 10 min. PCR products and those that produced a band were purified and subjected to sequencing analysis with a primer specific to the C. trachomatis cryptic plasmid.
RESULTS
Amplification profiles and reference standard curves.
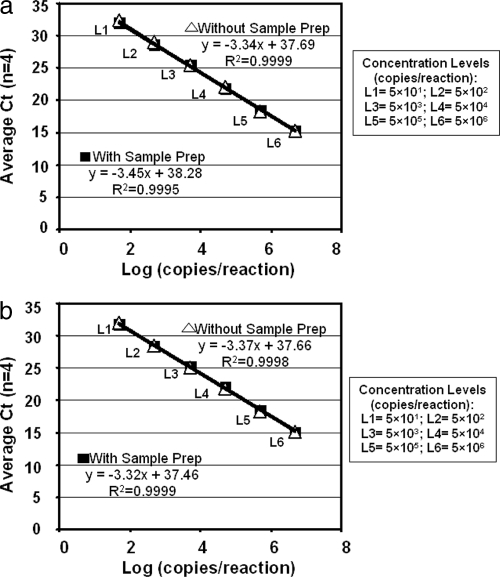
The slopes of the standard curves (range, −3.315 to −3.455) were nearly identical to the expected slope (−3.3) for 100% efficiency of PCR amplification (Fig. 1). Comparable sample preparation recovery rates were 80% to 100% for both 1.6-kb C. trachomatis and 1.0-kb N. gonorrhoeae dsDNA (data not shown).
Fig. 1.
Amplification of C. trachomatis and N. gonorrhoeae standard curves. Diluted C. trachomatis (a) and N. gonorrhoeae (b) reference standards were either tested directly in the AD step of the Versant CT/GC assay (denoted Without Sample Prep) or processed and tested by both SP and AD steps of Versant CT/GC assay (denoted With Sample Prep).
Analytical sensitivity.
Using logistical regression, LoDs were determined to be 342 copies/ml for C. trachomatis and 137 copies/ml for N. gonorrhoeae. Assay sensitivity for all 15 C. trachomatis serovars ranged from 0.067 to 1 IFU per assay. Assay sensitivity for genomic DNA of all 46 N. gonorrhoeae isolates ranged from 1 to 50 copies per assay.
Analytical specificity.
The Versant CT/GC assay demonstrated no cross-reactivity with the 136 potentially cross-reacting organisms that were tested (Table 1). However, five N. cinerea strains have been tested, including one ATCC strain (ATCC 14685), three Siemens in-house strains, and one Quality Control for Molecular Diagnostics (QCMD; 2010, Glasgow, Scotland) strain (QCMD NG10-02). Only one N. cinerea strain from QCMD presented a positive result, at 1e5 cells/ml. All of the other four N. cinerea strains gave negative results when they were tested at 1e7 cells/ml.
Work flow.
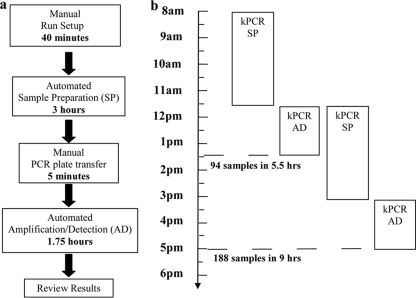
The work flow of the Versant CT/GC assay is shown in Fig. 2. The total time to obtain results was about 5.5 h for a batch size of 94 urine specimens. On average, a run required 40 min of hands-on reagent setup and specimen loading, followed by 3 h of automated SP processing time in the SP module. After the amplification plate was sealed, the plate was transferred to the AD module (5 min). This was followed by 1.75 h for a fully automated amplification and detection step. As shown in Fig. 2b, while a first batch of specimens was run in the AD module, a second batch of specimens was started on the Versant SP module, thus enabling a total of 188 specimens (94 specimens per run) to be analyzed and reported in a 9-hour shift. In addition, a third batch of specimens was left in the Versant AD module at the end of a shift, and postassay data review was completed on the following day. The total time to obtain results was less than 6 h for a batch size of 94 M4RT swab specimens (work flow details not shown).
Fig. 2.
Work flow of Versant CT/GC DNA 1.0 assay (kPCR). Distinct steps in the work flow of a Versant CT/GC assay run are identified. The amount of time specified for each step was calculated on the basis of a full Versant CT/GC assay run with 94 urine specimens and 2 controls.
Clinical comparison with AC2 assay.
A total of 589 FCU and 540 swab specimens were prospectively collected and tested with the Versant CT/GC assay and the AC2 assay. The clinical presentation of the study patients, which includes their symptomatic status by specimen type and their test results from the Versant CT/GC assay, is presented in Table 2. The prevalence rates of C. trachomatis in FCU specimens of symptomatic and asymptomatic patients were 7.3% and 10.3%, respectively, and those in swab specimens were 6.6% and 10.9%, respectively (Table 2). The prevalence rates of N. gonorrhoeae of symptomatic and asymptomatic patients were 7.3% and 0.9%, respectively, in FCU specimens and 8.1% and 0.9%, respectively, in swab specimens (Table 2). On the basis of initial results from both assays, the Versant CT/GC assay was comparable to the AC2 assay with over 98% overall agreement. Among the 86 Versant CT/GC assay-positive FCU specimens, 56 were positive for C. trachomatis and 30 were positive for N. gonorrhoeae, and among the 82 AC2 assay-positive urine specimens, 54 were positive for C. trachomatis and 28 were positive for N. gonorrhoeae (Table 3). Positive and negative percent agreements between the Versant CT/GC assay and the AC2 assay for detection of C. trachomatis in FCU specimens were 94.4% (95% CI, 84.9% to 98.1%) and 99.1% (95% CI, 97.8% to 99.6%), respectively, with an overall agreement of 98.6% (Table 3). For the detection of N. gonorrhoeae in FCU specimens, positive and negative percent agreements between the two assays were 100% (95% CI, 87.9% to 100%) and 99.6% (95% CI, 98.7% to 99.9%), respectively, with an overall agreement of 99.7% (Table 3).
Table 2.
Clinical presentation of study patients by specimen type and symptomatic status
| Specimen type and symptomatic status | No. (%) of patients with indicated Versant CT/GC DNA 1.0 assay test result |
||||
|---|---|---|---|---|---|
| Total |
C. trachomatis |
N. gonorrhoeae |
|||
| Positive | Negative | Positive | Negative | ||
| Swab | |||||
| Symptomatic | 320 | 21 (6.6) | 299 (93.4) | 26 (8.1) | 294 (91.9) |
| Asymptomatic | 220 | 24 (10.9) | 196 (89.1) | 2 (0.9) | 218 (99.1) |
| Total for swab | 540 | 45 (8.3) | 495 (91.7) | 28 (5.2) | 512 (94.8) |
| FCU | |||||
| Symptomatic | 356 | 26 (7.3) | 330 (92.7) | 26 (7.3) | 330 (92.7) |
| Asymptomatic | 233 | 24 (10.3) | 209 (89.7) | 2 (0.9) | 231 (99.1) |
| Total for FCU | 589 | 50 (8.5) | 539 (91.5) | 28 (4.8) | 561 (95.2) |
Table 3.
Comparison of the Versant CT/GC DNA 1.0 assay (kPCR) and AC2 assaya
| Organism, specimen, and result | No. of specimens with the indicated result by AC2 assay |
% PPA (95% CI) | % NPA (95% CI) | % OPA | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| C. trachomatis | |||||
| Urine | 98.6 | ||||
| Positive | 51 | 5 | 94.4 (84.9–98.1) | ||
| Negative | 3 | 528 | 99.1 (97.8–99.6) | ||
| Swab | 99.4 | ||||
| Positive | 46 | 1 | 95.8 (86.0–98.9) | ||
| Negative | 2 | 489 | 99.8 (98.9–100) | ||
| N. gonorrhoeae | |||||
| Urine | 99.7 | ||||
| Positive | 28 | 2 | 100 (87.9–100) | ||
| Negative | 0 | 556 | 99.6 (98.7–99.9) | ||
| Swab | 99.1 | ||||
| Positive | 28 | 5 | 100 (87.9–100) | ||
| Negative | 0 | 506 | 99.0 (97.7–99.6) | ||
All positive and negative percent agreements were calculated using the Aptima assay as a reference. Indeterminate results (IND) from the Aptima Combo 2 assay were excluded from the analysis. Assay-positive and -negative results were calculated on the basis of the assay initial results. PPA, positive percent agreement; NPA, negative percent agreement; OPA, overall percent agreement.
Among the 80 Versant CT/GC assay-positive swab specimens, 47 were positive for C. trachomatis and 33 were positive for N. gonorrhoeae; among the 76 AC2 assay-positive swab specimens, 48 were positive for C. trachomatis and 28 were positive for N. gonorrhoeae (Table 3). For detection of C. trachomatis in swab specimens, positive and negative percent agreements between the two assays were 95.8% (95% CI, 86.0% to 98.9%) and 99.8% (95% CI, 98.9% to 100%), respectively, with an overall agreement of 99.4% (Table 3). For detection of N. gonorrhoeae in swab specimens, positive and negative percent agreements between the two assays were 100% (95% CI, 87.9% to 100%) and 99.0% (95% CI, 97.7% to 99.6%), respectively, with an overall agreement of 99.1% (Table 3).
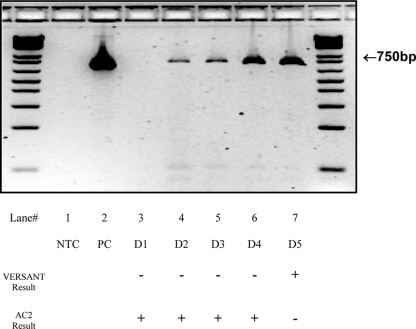
Table 3 indicates that there are 8 initial discrepant C. trachomatis urine results by adding two numbers diagonally. Similarly, there are 3 initial discrepant C. trachomatis swab results, 2 initial discrepant N. gonorrhoeae urine results, and 5 initial discrepant N. gonorrhoeae swab results. A total of five samples remained discrepant after they were retested; all five samples were discrepant in C. trachomatis detection. Touchdown PCR amplification results for these samples (D1 to D5) are shown in Fig. 3. PCR products from samples that produced no band (D1) when they were subjected to a second round of touchdown PCR testing remained negative (Fig. 3). When the PCR products of D2, D3, D4, and D5 were purified and subjected to sequencing analysis with the primer specific to the C. trachomatis cryptic plasmid, it was demonstrated that the sequence of these PCR products matched that of the C. trachomatis cryptic plasmid (data not shown).
Fig. 3.
Touchdown PCR analysis of samples that yielded discrepant results in the Versant CT/GC assay and Gen-Probe AC2 assay. A total of five samples persistently yielded discrepant results in both the Versant CT/GC assay and Gen-Probe AC2 assay, even after they were retested. All five samples were discrepant in C. trachomatis detection. DNA from these clinical samples was extracted and subjected to touchdown PCR analysis with primers specific to the C. trachomatis cryptic plasmid. Lane 1, negative control (NTC); lane 2, C. trachomatis-positive control (PC); lanes 3 to 7, discrepant samples (D1 to D5).
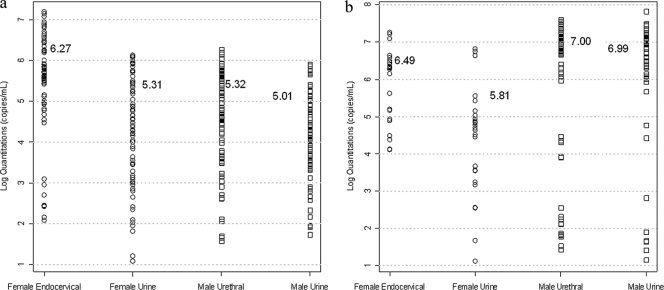
DNA copy numbers of the C. trachomatis and N. gonorrhoeae targets in clinical samples were determined with C. trachomatis and N. gonorrhoeae reference standards (Fig. 4). Geometric means are indicated by horizontal bars. The 95% quantiles of the DNA copy numbers in the C. trachomatis-positive clinical samples are 8.33 × 105 copies/ml for urine samples and 5.50 × 106 copies/ml for swab samples. The 95% quantiles of the DNA copy numbers in the N. gonorrhoeae-positive clinical samples are 2.78 × 107 copies/ml for urine samples and 2.99 × 107 copies/ml for swab samples.
Fig. 4.
Copy numbers of C. trachomatis and N. gonorrhoeae target DNA sequences in matched specimens from men and women. Copy numbers of C. trachomatis and N. gonorrhoeae target DNA sequences in FCU and swab samples from subjects who tested positive for C. trachomatis and N. gonorrhoeae infection were calculated and plotted on a logarithmic scale. These copy numbers were used to approximate the bacterial load in the individual clinical samples. (a) Copy numbers of C. trachomatis target DNA sequence in samples from C. trachomatis-positive subjects; (b) copy numbers of N. gonorrhoeae target DNA sequence in samples from N. gonorrhoeae-positive subjects.
DISCUSSION
Results of this study demonstrate that the clinical performance of the Versant CT/GC assay in detecting C. trachomatis and N. gonorrhoeae targets in a single endocervical or urethral swab specimen or in an FCU specimen has excellent agreement with that of the Gen-Probe AC2 assay. Overall agreements between the Versant CT/GC assay and AC2 assay for detection of C. trachomatis in urine and swab specimens were 98.6% and 99.4%, respectively; for detection of N. gonorrhoeae in urine and swab specimens, they were 99.7% and 99.1%, respectively. The rates of positive agreement between the two assays were over 94% and 100% for detection of C. trachomatis and N. gonorrhoeae, respectively, with the rate of negative agreement being over 99% for both C. trachomatis and N. gonorrhoeae.
In this study, only five specimens (0.44%) remained discrepant after retesting, and all were discrepant in C. trachomatis detection. Sequencing analysis of the five C. trachomatis discrepant samples determined that three were Versant CT/GC assay false negative, one was AC2 assay false negative, and one was AC2 assay false positive.
Analytical performance studies demonstrate that the Versant CT/GC assay detects C. trachomatis in the range of 0.067 to 1 IFU per assay and N. gonorrhoeae in the range of 1 to 50 copies per assay. The C. trachomatis cryptic plasmid is present in all C. trachomatis serovars at 7 to 10 copies per elementary body of C. trachomatis (13). The N. gonorrhoeae assay targets the multiple-copy pivNG gene locus of N. gonorrhoeae, and studies (1) have shown that approximately 8 copies of the pivNG gene are found in the N. gonorrhoeae genome.
In a method comparison study, of 79 positive FCU specimens and 74 positive swab specimens, the lowest DNA copy number is 93 copies/ml (equivalent to 6 copies per assay) in C. trachomatis FCU specimens (data not shown), which were detected by the Versant CT/GC assay. Using Siemens proprietary magnetic silica beads, nonspecific nucleic acids (total RNAs and DNAs) were captured in an extremely efficient manner, and recoveries of 80 to 100% efficiency were achieved. This method does not rely on a centrifugation step to concentrate elementary bodies before elution and includes nucleic acid isolation prior to amplification, which leads to better assay sensitivity. The Versant CT/GC assay's analytical specificity was challenged against a battery of 136 potential cross-reacting organisms, and no Versant CT/GC assay false-positive reactions due to cross-reactivity were seen in this clinical study.
C. trachomatis/N. gonorrhoeae NAATs that employ nucleic acid isolation prior to amplification have consistently been shown to be the most sensitive. Since these assays are in fact very similar in sensitivity and specificity, there are other assay features that should be considered important factors in selecting the best C. trachomatis/N. gonorrhoeae NAAT. One such factor has to do with the economics related to throughput and time to results for all laboratories, whether they are high-, medium-, or low-volume testing laboratories. The Versant CT/GC assay's time to results of 5.5 to 6 h allows the processing of large numbers of specimens in a relatively short period of time. Literally hundreds of specimens can be processed weekly with a single, small-footprint Versant kPCR molecular system, leading to a 5-day-a-week run plan that can produce results from nearly 25,000 specimens annually. For laboratories receiving larger numbers of specimens, two runs per 9-hour shift can be accommodated, a scenario that would increase throughput to 188 specimens per day or nearly 50,000 specimens annually.
The Versant CT/GC assay can be recommended for use with urine and swab specimens as a screening test for either symptomatic or asymptomatic men and women. This new assay provides clinicians and laboratories with one of the necessary tools to significantly reduce the prevalence and incidence of C. trachomatis and N. gonorrhoeae infections in sexually active men and women, as well as prevent their costly and serious sequelae.
In conclusion, this study demonstrates the excellent performance of the Versant CT/GC assay for detecting C. trachomatis and N. gonorrhoeae in both FCU and swab specimens when it was compared to the Gen-Probe AC2 assay. The Versant CT/GC assay offers a rapid, accurate, and reliable alternative for detecting C. trachomatis and N. gonorrhoeae targets in a single endocervical or urethral swab and in male and female urine specimens. Its ability to simultaneously detect C. trachomatis and N. gonorrhoeae in a single swab and in FCU specimens with no cross-interference recommend the Versant CT/GC assay for use in large-scale screening programs.
ACKNOWLEDGMENTS
We thank Apurva Unyial, LaShawnda Royal, and Staeci Morita of the Los Angeles County Department of Public Health STD Program and Ruel Torres and Mary Beth Duke of the Los Angeles County Department of Public Health Public Health Laboratory.
Footnotes
Published ahead of print on 9 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Carrick C. S., Fyfe J. A. M., Davies J. K. 1998. Neisseria gonorrhoeae contains multiple copies of a gene that may encode a site-specific recombinase and is associated with DNA rearrangements. Gene 220:21–29 [DOI] [PubMed] [Google Scholar]
- 2. Cates W., Jr., Wasserheit J. N. 1991. Genital chlamydial infections: epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771–1781 [DOI] [PubMed] [Google Scholar]
- 3. CDC 2009. Sexually transmitted disease surveillance, 2008. U.S. Department of Health and Human Services, Atlanta, GA: http://www.cdc.gov/std/stats08/ [Google Scholar]
- 4. Crotchfelt K. A., Welsh L. E., DeBonville D., Rosenstraus M., Quinn T. C. 1997. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in genitourinary specimens from men and women by a coamplification PCR assay. J. Clin. Microbiol. 35:1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feinstein A. R., Cicchetti D. V. 1990. High agreement but low kappa. I. The problems of two paradoxes. J. Clin. Epidemiol. 43:543–549 [DOI] [PubMed] [Google Scholar]
- 6. Feinstein A. R., Cicchetti D. V. 1990. High agreement but low kappa. II. Resolving paradoxes. J. Clin. Epidemiol. 43:551–558 [DOI] [PubMed] [Google Scholar]
- 7. Ferrero D. V., Meyers H., Willis S., Schultz D. 1998. Performance of the Gen-Probe AMPLIFIED Chlamydia trachomatis assay in detecting Chlamydia trachomatis in endocervical and urine specimens from women and urethral and urine specimens from men attending sexually transmitted disease and family planning clinics. J. Clin. Microbiol. 36:3230–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fleming D. T., Wasserheit J. N. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 75:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaydos C., Ferrero D. V., Papp J. 2008. Laboratory aspects of screening men for Chlamydia trachomatis in the new millennium. Sex. Transm. Dis. 35:S45–S50 [DOI] [PubMed] [Google Scholar]
- 10. Goessens W. H. F., et al. 1997. Comparison of three commercially available amplification assays, AMP CT, LCx, and COBAS AMPLICOR, for detection of Chlamydia trachomatis in first-void urine. J. Clin. Microbiol. 35:2628–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hook E. W., III, Handsfield H. H. 1999. Gonococcal infections in the adult, p. 451–466 In Holmes K. K., et al. (ed.), Sexually transmitted diseases, 3rd ed. McGraw-Hill, New York, NY [Google Scholar]
- 12. Institute of Medicine, Division of Health Promotion and Disease Prevention 1997. In Eng T. R., Butler W. T. (ed.), Hidden epidemic: confronting sexually transmitted diseases. National Academy Press, Washington, DC [Google Scholar]
- 13. Palmer L., Falkow S. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52–62 [DOI] [PubMed] [Google Scholar]
- 14. Schachter J. 1989. Why we need a program for the control of Chlamydia trachomatis. N. Engl. J. Med. 320:802–804 [DOI] [PubMed] [Google Scholar]
- 15. Stamm W. E. 1999. Chlamydia trachomatis infections of the adult, p. 407–422 In Holmes K. K., et al. (ed.), Sexually transmitted diseases, 3rd ed. McGraw-Hill, New York, NY [Google Scholar]
- 16. Van Der Pol B., et al. 2001. Multicenter evaluation of the BDProbeTec system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 39:1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wallin K. L., et al. 2002. A population-based prospective study of Chlamydia trachomatis infection and cervical carcinoma. Int. J. Cancer 101:371–374 [DOI] [PubMed] [Google Scholar]
- 18. Westrom L., Eschenbach D. 1999. Pelvic inflammatory disease, p. 783–809 In Holmes K. K., et al. (ed.), Sexually transmitted diseases, 3rd ed. McGraw-Hill, New York, NY [Google Scholar]