Abstract
For the last decade China has occupied second place, after India, among the top five countries with high burdens of tuberculosis (TB). Heilongjiang Province is located in northeastern China. The prevalence of drug-resistant TB in Heilongjiang Province is higher than the average level in China. To determine the transmission characteristics of Mycobacterium tuberculosis strains isolated in this area and their genetic relationships, especially among the Beijing family strains, we investigated their genotypes. From May 2007 to October 2008, 200 M. tuberculosis isolates from patients presenting pulmonary TB were analyzed by molecular typing using PCR-based methods: spacer-oligonucleotide typing (spoligotyping), Beijing family-specific PCR (detection of the deletion of region of difference 105 [RD105]), and mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) analysis. Different combinations of MIRU-VNTR loci were evaluated to define the genotypes and clustering characteristics of the local strains. We found that Beijing family strains represented 89.5% of the isolates studied. However, the rates of multidrug-resistant (MDR) M. tuberculosis among Beijing and non-Beijing family strains were not statistically different. The 15-locus set is considered the optimal MIRU-VNTR locus combination for analyzing the M. tuberculosis strains epidemic in this area, while the 10-locus set is an ideal set for first-line molecular typing. We found that the clustering rate of all the M. tuberculosis isolates analyzed was 10.0% using the 15-locus set typing. We conclude that the Beijing family genotype is predominant and that highly epidemic TB and MDR TB are less likely associated with the active transmission of M. tuberculosis in the study area.
INTRODUCTION
Tuberculosis (TB) remains a major public health threat worldwide. China has occupied second place, after India, among the top five high-burden countries for the last decade (http://www.who.int/tb/en). Although both the incidence and prevalence of TB in China have shown a steady decline in recent years, it remains a leading notifiable infectious disease (http://www.moh.gov.cn). Currently, the spread of drug-resistant TB, especially multidrug-resistant (MDR) TB, in China, presents a major challenge. The most up-to-date data from the World Health Organization (WHO) indicates that the rate of MDR TB in China was 8.3% (the rates of primary and acquired MDR TB were 5.7% and 25.6%, respectively) in 2007 (50), significantly higher than the global average rate (3.6%). The higher levels of TB prevalence and drug resistance have become the main public health concern of the Chinese government.
Heilongjiang Province, located in northeastern China, is one of the regions where the prevalence of both TB and drug-resistant TB is higher than the average level in China. The most recently updated epidemiological data, from 2007 to 2008, show that the rates of primary and acquired MDR TB were 18.3% and 37.8%, respectively, in Heilongjiang Province (our unpublished data). The reasons for the high prevalence and drug resistance of TB in Heilongjiang Province are still unknown and should be investigated to facilitate control of the TB epidemic in this area and throughout China.
In several Asian countries with high TB rates, a unique genotype of Mycobacterium tuberculosis, known as the Beijing family genotype, has been found to be the dominant genotype (3, 17, 34). During the last decade, Beijing family strains have been spreading in various geographic locations worldwide and now account for more than a quarter of all TB cases worldwide (12). Possible associations of the epidemic caused by this genotype with its drug resistance (1, 41) and its high adaptability to the host intracellular environment (8) have been reported. In China, Beijing family strains have spread widely; however, the proportion of Beijing family strains in Heilongjiang Province remains unknown. It is thus unclear if the high prevalence and high drug resistance of epidemic TB are directly related to the spread of Beijing family strains. The answers to these questions would be very helpful in achieving effective control of TB in Heilongjiang Province as well as throughout China.
To determine the spreading characteristics and the genetic relationships among the M. tuberculosis isolates in a given geographic region, a suitable genotyping method is the most important means, i.e., one that is discriminatory, economical, and reproducible. Spacer-oligonucleotide typing (spoligotyping) is a PCR-based method which is simple, highly reproducible, and applicable to a digital format (22). Spoligotyping possesses the most features of an ideal typing method and is the gold standard for identifying strains as belonging to the Beijing family. However, due to the genetic homogeneity of this family, this method is uninformative in the areas where the Beijing family is prevalent (47) and is therefore limited to use as a first-line typing method.
A genotyping method based on mycobacterial interspersed repetitive unit-variable-number tandem repeats (MIRU-VNTR), which is technically flexible to facilitate interlaboratory comparison and database management, is well accepted and has recently been adapted to high-throughput techniques (7, 39). The digital format of MIRU-VNTR patterns makes this method suitable for the global study of the molecular epidemiology of M. tuberculosis. A number of studies have investigated the usefulness of different VNTR loci for typing M. tuberculosis and have shown that the discriminatory power of this method may be comparable to that of IS6110 typing, which is the most reliable method for the genetic differentiation of M. tuberculosis isolates (33, 38). Because Beijing family strains are prevalent in many developing areas of Asia, where resources are definitely limited, MIRU-VNTR typing would be the best choice here to study the molecular epidemiology of M. tuberculosis strains.
Because TB, especially drug-resistant TB, constitutes a heavy burden in Heilongjiang Province, it is important to understand the possible spreading mechanisms of M. tuberculosis and particularly drug-resistant TB. In the present study, we conducted the genotyping of M. tuberculosis isolates to determine the prevalence of Beijing family strains and the genotypes of M. tuberculosis isolates in Heilongjiang Province using MIRU-VNTR analysis. The appropriate VNTR locus combination for genotyping the M. tuberculosis isolates in our area was also evaluated and optimized.
MATERIALS AND METHODS
Mycobacterial specimens.
The study included 200 M. tuberculosis samples isolated between May 2007 and October 2008 from 200 patients from various regions of Heilongjiang Province who had been diagnosed with pulmonary TB in the Harbin Chest Hospital. All the patients were HIV-1 negative. The average age of the patients was 49.1 years, and 71.5% (143/200) were male. M. tuberculosis H37Rv was used as the reference strain.
DNA extraction and molecular identification of M. tuberculosis isolates and RD105 deletion detection.
Mycobacterial cultures were obtained from clinical specimens after incubation in a BACTEC Mycobacterium Growth Indicator Tube (MGIT) 960 Automated System (Becton Dickinson). The cultures were inactivated by 70% ethanol for 2 h (9). DNA was extracted using lysozyme and the phenol-chloroform method (46). Molecular identification of the mycobacterial isolates was performed based on PCR amplification of the 16S rRNA gene and the Rv0577, Rv2073c, and Rv3120 genes (15). Rv2073c amplification was modified as follows: PrimeSTAR HS DNA polymerase (TakaRa, China) was used instead of Taq polymerase, and 2 μl of dimethyl sulfoxide was added to ensure good amplification. The amplification cycle was 5 min at 98°C followed by 25 cycles of 1 min at 98°C, 5 s at 65°C, and 40 s at 72°C, with a final 10 min at 72°C. The identification of genomic deletions in region of difference 105 (RD105) was performed by PCR as previously described (14, 26). Briefly, each PCR mixture was prepared in a volume of 20 μl containing 50 ng of DNA, 0.25 U of Taq polymerase (TakaRa, China), a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), and 0.2 μM (each) primer. The amplification cycle was 10 min at 94°C followed by 25 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, with a final step for 10 min at 72°C.
Spoligotyping.
Spoligotyping of the isolates was performed as described by Kamerbeek et al. (22). The direct repeat (DR) region was amplified with the primer pair, and the PCR products were hybridized to a set of 43 oligonucleotide probes corresponding to each spacer, which were covalently bound to a membrane (22). Spoligotypes in binary format were compared with the SpolDB4 database, and the spoligotype international type (SIT) numbers and the clades were also determined (4).
MIRU-VNTR typing.
To identify a suitable MIRU-VNTR locus set for genotyping M. tuberculosis isolates in this area, 19 loci were selected for analyzing the first set of 44 M. tuberculosis isolates (38). The PCR mixture and conditions were the same those for the RD105 deletion identification described above. Genomic DNA of the H37Rv strain and sterile distilled water were used as the positive and negative controls, respectively. PCR products were analyzed on a 1.5% agarose gel against a 100-bp DNA ladder (TakaRa, China), and the copy number at each locus was calculated using a Quantity 1 gel imaging system (Tanon, China). The MIRU-VNTR allelic diversity (h) at a given locus was calculated as follows: h = 1 − Σ xi2[n/(n − 1)], where xi is the frequency of the ith allele at the locus, and n is the number of isolates (35). The discrimination of the locus combination was calculated using the Hunter-Gaston discriminatory index (HDGI) (16):
where N is the total number of isolates in the typing method, s is the number of distinct patterns discriminated by MIRU-VNTR, and nj is the number of isolates belonging to the jth pattern.
Phylogenetic and cluster analysis.
We used the R software, version 2.11.1 (http://cran.r-project.org), for phylogenetic and cluster analysis. A dendrogram was produced from the MIRU-VNTR genotypes of the 200 M. tuberculosis isolates. First, the repeat numbers of MIRU-VNTR genotypes were standardized based on a z-score normalization. Then, a similarity coefficient matrix of the M. tuberculosis isolates was obtained by calculating the Euclidean distances between isolates from the standardized data. Finally, clustering was performed, and a phylogenetic tree was constructed using Ward's parameter with the matrix. The M. tuberculosis isolates analyzed in this study were classified into two groups, characterized by clustered and nonclustered M. tuberculosis isolates. A molecular cluster was defined as two or more M. tuberculosis isolates having identical genetic patterns as determined by MIRU-VNTR genotyping. The isolates with unmatched genetic profiles were considered nonclustered strains. Assuming that one patient from each cluster corresponded to the index case at the origin of infection, the clustering rate was calculated using the following formula: clustering rate = (nc − c)/n, where nc is the total number of clustered isolates, c is the number of isolate clusters, and n is the total number of isolates in the sample (37).
Statistical analysis.
Associations among multiple categorical variables were assessed using R, version 2.11.1, by a chi-square test or Fisher's exact test when the theoretical frequency was less than five. Two-by-two tables were assessed by a chi-square test (here, Yates' continuity correction was needed when the value was less than five), and results were expressed as odds ratios (OR) with 95% confidence intervals (95% CI). The agreement between spoligotyping and RD105 deletion typing was assessed using kappa statistics; the agreement was considered good for values of kappa above 0.75. P values of <0.05 were considered statistically significant.
RESULTS
Epidemic of Beijing family strains in Heilongjiang Province.
During the study period, 200 M. tuberculosis isolates identified using both the BACTEC 960 automated system and molecular methods were collected. First, we analyzed the correlation between spoligotyping and RD105 deletion for the identification of the Beijing genotype using 44 isolates collected from May 2007 to November 2007. Among the 44 M. tuberculosis isolates, spoligotypes of 41 isolates were classified into three designated SITs according to the SpolDB4 database (Table 1). Among these, 40 isolates were Beijing family strains. The most frequent genotype (39/41) was the typical Beijing spoligotype SIT1, which has only spacers 35 to 43; the only other Beijing genotype belonged to spoligotype SIT190. One isolate, with an SIT number of 1793, was not designated in the database. The remaining three isolates showed new spoligotypes, which were not registered in SpolDB4 database. Interestingly, one isolate (2460) showed a unique genotype with only two spacers, 35 and 36. We found that 40 isolates lacking RD105 exhibited Beijing family spoligotypes (Table 1). The M. tuberculosis isolate 2460 also lacked RD105. The results of the kappa statistics analysis showed that the agreement of spoligotyping and RD105 deletion detection in identifying the Beijing family genotype was high (κ = 0.8451). Subsequently, instead of using spoligotyping, RD105 deletions in the other 156 M. tuberculosis strains were examined, and we found that 179 of the 200 isolates (89.5%) had the Beijing family genotype, while 21 (10.5%) were non-Beijing family strains.
Table 1.
The profile of spoligotypes and RD105 deletion of the M. tuberculosis isolates (n = 44)
—, unassigned in the SpolDB4 database.
The black and white boxes indicate the presence and absence of the specific spacer, respectively.
The identification number of the isolate is given in parentheses.
Optimal combination of MIRU-VNTR loci for genotyping M. tuberculosis isolates in Heilongjiang Province.
First, to evaluate and determine the most suitable loci for genotyping the M. tuberculosis isolates epidemic in Heilongjiang Province, we analyzed 19 MIRU-VNTR loci, which had been previously identified as a suitable locus combination for genotyping M. tuberculosis isolates in the regions where the Beijing family is dominant (19, 27) (Table 2). The allelic diversity (h) of the first set of 44 M. tuberculosis isolates at each MIRU-VNTR locus varied significantly. Among the 19 loci, the allelic diversity for 2 loci (QUB11b and QUB26) exceeded 0.6, suggesting that they are highly discriminating (30). Seven loci (MIRU4, MIRU16, MIRU26, MIRU31, MIRU40, Mtub21, and Mtub4) showed moderate discrimination (0.3 ≤ h ≤ 0.6), but ETR C (h = 0.068) and ETR B (h = 0.066) were less polymorphic. Diversity was not observed for the MIRU23 locus (h = 0). Thus, the loci ETR C, ETR B, and MIRU23, having discriminatory powers of less than 0.1, were excluded from the subsequent MIRU-VNTR analysis.
Table 2.
Allelic diversity of different MIRU-VNTR locia
| MIRU-VNTR locus | No. of alleles (range) |
h |
||
|---|---|---|---|---|
| Isolate set 1 | Isolate set 2 | Isolate set 1 | Isolate set 2 | |
| QUB26 | 7 (4–10) | 8 (3–10) | 0.706 | 0.581 |
| QUB11b | 6 (3–8) | 9 (0–8) | 0.644 | 0.730 |
| MIRU31 | 6 (2–7) | 7 (2–8) | 0.599 | 0.500 |
| MIRU16 | 4 (2–6) | 4 (2–6) | 0.520 | 0.230 |
| MIRU4 | 5 (1–5) | 6 (0–5) | 0.517 | 0.260 |
| MIRU26 | 5 (4–9) | 8 (3–10) | 0.467 | 0.649 |
| Mtub4 | 4 (2–5) | 6 (0–5) | 0.413 | 0.463 |
| MIRU40 | 5 (1–5) | 6 (1–6) | 0.373 | 0.358 |
| Mtub21 | 6 (1–8) | 9 (1–9) | 0.307 | 0.493 |
| Mtub39 | 4 (1–6) | 6 (1–6) | 0.229 | 0.243 |
| MIRU10 | 4 (1–4) | 4 (1–4) | 0.229 | 0.300 |
| Mtub30 | 4 (2–5) | 4 (2–5) | 0.191 | 0.267 |
| Mtub29 | 3 (3–5) | 5 (2–6) | 0.189 | 0.138 |
| MIRU39 | 3 (1–3) | 4 (1–4) | 0.149 | 0.388 |
| ETR A | 2 (3–4) | 7 (0–6) | 0.146 | 0.329 |
| QUB4156 | 2 (2–4) | 5 (0–4) | 0.107 | 0.182 |
| ETR C | 3 (2–5) | 0.068 | ||
| ETR B | 2 (1–2) | 0.066 | ||
| MIRU23 | 1 (5) | 0 | ||
Set 1 consists of 44 isolates obtained from May 2007 to November 2007. Set 2 consists of 200 isolates obtained from May 2007 to October 2008.
Next, we analyzed the 200 M. tuberculosis isolates collected from May 2007 to October 2008 using the remaining 16 MIRU-VNTR loci. All 16 loci displayed an allelic diversity similar to the original 19 loci (Table 2). The highest diversity among the 200 isolates was observed at QUB11b (h = 0.730), and the lowest diversity was observed at Mtub29 (h = 0.138). The HGDI of the 16-locus set was as high as 0.9977. However, because the 16-locus procedure still did not meet the requirements of cost and labor expenditure for high-throughput genotyping, we then tried to optimize the locus combination while minimizing the number of loci. Based on the allelic diversity of each MIRU-VNTR locus, the cumulative HGDI of the locus combination by successive addition of a locus was compared (Table 3). The cumulative HGDI and clustering rate of the 10-locus set were equal to that of the 11-locus set (HGDI, 0.9950; clustering rate, 15.5%); they were also the same for the 15- and the 16-locus sets (HGDI, 0.9977; clustering rate, 10.0%). The set of the first seven loci with the highest allelic diversity gave an HGDI of 0.9913 and a clustering rate of 25.0%.
Table 3.
The cumulative HGDI with successive addition of each MIRU-VNTR locus
| Locus combinationa | VNTR locus (h)b | No. of patterns | No. of clusters | No. of clustered isolates | No. of isolates in each cluster | Clustering rate (%) | HGDI (cumulative) |
|---|---|---|---|---|---|---|---|
| 1 | QUB11b (0.730) | ||||||
| 2 | MIRU26 (0.649) | 35 | 19 | 184 | 2–41 | 82.5 | 0.9042 |
| 3 | QUB26 (0.581) | 84 | 33 | 149 | 2–23 | 58.0 | 0.9686 |
| 4 | MIRU31 (0.500) | 112 | 33 | 121 | 2–18 | 44.0 | 0.9808 |
| 5 | Mtub21 (0.493) | 126 | 33 | 107 | 2–16 | 37.0 | 0.9867 |
| 6 | Mtub4 (0.463) | 135 | 30 | 95 | 2–15 | 32.5 | 0.9888 |
| 7 | MIRU39 (0.388) | 150 | 23 | 73 | 2–14 | 25.0 | 0.9913 |
| 8 | MIRU40 (0.358) | 159 | 18 | 59 | 2–12 | 20.5 | 0.9935 |
| 9 | ETR A (0.329) | 164 | 16 | 52 | 2–12 | 18.0 | 0.9943 |
| 10 | MIRU10 (0.300) | 169 | 14 | 45 | 2–12 | 15.5 | 0.9950 |
| 11 | Mtub30 (0.267) | 169 | 14 | 45 | 2–12 | 15.5 | 0.9950 |
| 12 | MIRU4 (0.260) | 176 | 12 | 36 | 2–10 | 12.0 | 0.9967 |
| 13 | Mtub39 (0.243) | 178 | 10 | 32 | 2–10 | 11.0 | 0.9968 |
| 14 | MIRU16 (0.230) | 179 | 11 | 32 | 2–8 | 10.5 | 0.9976 |
| 15 | QUB4156 (0.182) | 180 | 10 | 30 | 2–8 | 10.0 | 0.9977 |
| 16 | Mtub29 (0.138) | 180 | 10 | 30 | 2–8 | 10.0 | 0.9977 |
The successive addition of each VNTR locus.
The h value represents the diversity determined from the 200 isolates.
VNTR profiles and genotypes of the M. tuberculosis isolates in Heilongjiang Province.
The MIRU-VNTR genotyping results showed that the 200 isolates were classified into 180 genotypes. A total of 170 isolates had unique patterns, while the remaining 30 isolates were in 10 clusters. A dendrogram was constructed based on the genotypes of 200 isolates using 16 loci (Fig. 1). The isolates were divided into four groups based on phylogenetic clustering and genotypic characteristics. Groups I to IV contained 21, 13, 21, and 145 isolates, respectively. Among the 179 Beijing family isolates, 144 (99.3%) were in group IV; the remaining 35 isolates were in groups I, II, and III (P < 0.0001), and all the clustered isolates were in group IV (P = 0.0018), suggesting that the distributions of the Beijing family isolates and clustered isolates were distinctive among the four groups (Table 4).
Fig. 1.
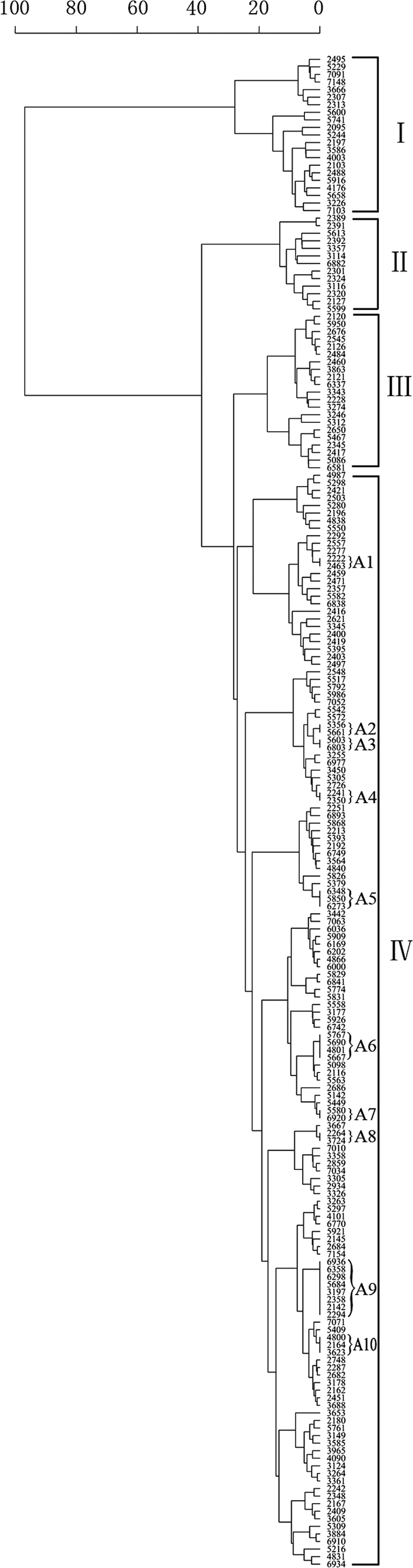
Dendrogram of 200 M. tuberculosis isolates from Heilongjiang Province. The phylogenetic tree was produced from the MIRU-VNTR genotypes which were derived from 16 of the 19 loci by excluding ETR B, ETR C, and MIRU23. A1 to A10, cluster names.
Table 4.
Differences of M. tuberculosis characteristics among the four subgroups
| Isolate characteristic | Total no. of isolates | No. (%) of isolates by subgroupa |
P valueb | |||
|---|---|---|---|---|---|---|
| I (n = 21) | II (n = 13) | III (n = 21) | IV (n = 145) | |||
| Resistance | ||||||
| Streptomycin | 85 | 9 (42.9) | 6 (46.2) | 8 (38.1) | 62 (42.8) | 0.9704* |
| Isoniazid | 92 | 9 (42.9) | 8 (61.5) | 7 (33.3) | 68 (46.9) | 0.4317* |
| Rifampin | 55 | 4 (19.0) | 7 (53.8) | 3 (14.3) | 41 (28.3) | 0.0815 |
| Ethambutol | 48 | 4 (19.0) | 6 (46.2) | 3 (14.3) | 35 (24.1) | 0.2070 |
| MDR | 51 | 4 (19.0) | 6 (46.2) | 2 (9.5) | 39 (26.9) | 0.1032 |
| Four-drug susceptibility | 77 | 8 (38.1) | 3 (23.1) | 9 (42.9) | 57 (39.3) | 0.6786* |
| Four-drug resistance | 23 | 2 (9.5) | 3 (23.1) | 1 (4.8) | 17 (11.7) | 0.4402 |
| Obtained from a patient with acquired TB | 126 | 17 (81.0) | 9 (69.2) | 11 (52.4) | 89 (61.4) | 0.2294 |
| Obtained from a patient with hemoptysis | 169 | 15 (71.4) | 9 (69.2) | 20 (95.2) | 125 (86.2) | 0.0562 |
| Beijing strain | 179 | 3 (14.3) | 12 (92.3) | 20 (95.2) | 144 (99.3) | <0.0001 |
| Clustered | 30 | 0 (0) | 0 (0) | 0 (0) | 30 (20.7) | 0.0018 |
n, number of isolates in the subgroup.
Values marked with an asterisk were determined by a chi-square test; other values were determined by a Fisher's exact test.
Characteristics of the clustered isolates.
Thirty Beijing family isolates (30/179, or 16.8% of the Beijing family strains) were determined to be in 10 clusters (A1 to A10), with the clustering rate of 10.0% based on 15-locus MIRU-VNTR patterns. In contrast, none of 21 non-Beijing family isolates were clustered (OR, 0; 95% CI, 0 to 1.024; P = 0.087) (Table 5). Most of the clusters were small: six (A1 to A4, A7, and A8) contained only two members; two (A5 and A10) contained three members; cluster A6 contained four members. The largest cluster, A9, contained eight members. In addition, the clustering rates of the two periods, May 2007 to May 2008 (106 isolates) and June 2008 to October 2008 (94 isolates), were 6.4% and 12.8%, respectively (OR, 0.3240; 95% CI, 0.1161 to 0.8265; P = 0.0088).
Table 5.
Differences of M. tuberculosis characteristics between Beijing and non-Beijing family
| Isolate characteristic | Total no. of isolates | No. (%) of isolatesa |
OR | 95% CI | P valueb | |
|---|---|---|---|---|---|---|
| Beijing (n = 179) | Non-Beijing (n = 21) | |||||
| Resistance | ||||||
| Streptomycin | 85 | 78 (43.6) | 7 (33.3) | 0.6488 | 0.2110–1.8176 | 0.3691 |
| Isoniazid | 92 | 84 (46.9) | 8 (38.1) | 0.6972 | 0.2381–1.9189 | 0.4423 |
| Rifampin | 55 | 52 (29.1) | 3 (14.3) | 0.4086 | 0.0739–1.4876 | 0.1517 |
| Ethambutol | 48 | 45 (25.1) | 3 (14.3) | 0.4978 | 0.0898–1.8235 | 0.4056* |
| MDR | 51 | 48 (26.8) | 3 (14.3) | 0.4564 | 0.0824–1.6670 | 0.2127 |
| Four-drug susceptibility | 77 | 66 (36.8) | 11 (52.4) | 1.8771 | 0.6828–5.2244 | 0.1670 |
| Four-drug resistance | 23 | 21 (11.7) | 2 (9.5) | 0.7928 | 0.0837–3.6902 | 0.9510* |
| Obtained from a patient with acquired TB | 126 | 109 (60.9) | 17 (90.0) | 2.7174 | 0.8390–11.5638 | 0.0717 |
| Obtained from a patient with hemoptysis | 169 | 153 (85.5) | 16 (76.2) | 1.8323 | 0.4830–5.8459 | 0.4275* |
| Clustered | 30 | 30 (16.8) | 0 (0) | 0 | 0–1.0241 | 0.0869* |
n, number of isolates in the group.
Values marked with an asterisk were determined by a continuity-adjusted chi-square test; other values were determined by a chi-square test.
To determine if there was any correlation between the clustering characteristics and the geographical origins of the isolates, we investigated the home addresses of the patients in the clusters from the available medical records. We found that the isolates belonging to clusters A2 and A7 were scattered throughout the Heilongjiang Province while clusters A3 to A6, A9, and A10 were registered in Harbin City.
Drug susceptibility patterns of the M. tuberculosis isolates in Heilongjiang Province.
To determine the association between drug resistance patterns and genotypic characteristics, drug susceptibility to the four first-line antituberculosis drugs, i.e., streptomycin, isoniazid, rifampin, and ethambutol, was examined using an automated BACTEC MGIT 960 SIRE system (Becton Dickinson). A total of 77 isolates (38.5%) were susceptible to all four drugs; 123 (61.5%) were resistant to at least one drug, and 51 (41.5%) were MDR M. tuberculosis (Tables 4 and 5). The drug susceptibility patterns of the isolates among the four genotype groups were not significantly different (Table 4). Of the 51 MDR M. tuberculosis isolates, 48 isolates were found to be the Beijing family strains, and 3 were non-Beijing family strains. Of the Beijing family strains, 26.8% (48/179) were MDR, and 14.3% (3/21) of the non-Beijing family strains were MDR. The rates of MDR M. tuberculosis among Beijing and non-Beijing family strains were not statistically different (OR, 0.4564; 95% CI, 0.0824 to 1.6670; P = 0.2127). Resistance to at least one drug was observed more frequently among Beijing family strains (63.1%, or 113/179) than among non-Beijing family strains (47.6%, or 10/21), but the difference was not statistically significant (OR, 1.8771; 95% CI, 0.6828 to 5.2244; P = 0.1670) (Table 5).
DISCUSSION
The Beijing family strains currently prevail throughout China. RD105 deletion has recently been reported to serve as a genetic marker for Beijing family strains (44), and several studies have used this method to identify them (6, 26, 45). It is financially economical, labor saving, and especially suitable for high-throughput analysis. In this study, we found good agreement between RD105 deletion detection and spoligotyping. One strain (2460) showed a novel spoligotype containing only spacers 35 and 36 and an RD105 deletion. According to the definition of the Beijing family spoligotype, these strains contain at least three spacers among direct repeats 35 to 43; however, strain 2460 can be included in the Beijing family because it lacks RD105.
We found that 89.5% of the M. tuberculosis isolates in Heilongjiang Province were Beijing family strains. This genotype accounts for 80 to 90% of the M. tuberculosis strains currently epidemic in the Beijing area (19); it is also prevalent in Ningxia (67%), Shanghai (89%), Zhejiang (70%), Tianjin (91.7%), and Guangxi (55.3%) but less prevalent in Guangdong (25%) (5, 24, 25, 36, 48). Hence, Heilongjiang Province is one of the regions where the proportion of the Beijing genotype is the highest. This genotype is thought to be associated with drug resistance (1, 10, 23, 41). However, less association has been reported in other geographic settings (2, 3, 20, 43). In the present study, the statistical analysis showed that there was no difference between the Beijing and non-Beijing genotype strains in drug resistance patterns, indicating that the Beijing genotype is less likely to be associated with the high prevalence of drug resistance and M. tuberculosis TB in our area.
Molecular typing by MIRU-VNTR has been used in epidemiology studies, and its stability is adequate for tracking recent transmission and distinguishing relapses and reinfections (39). Currently, the system based on 12 loci (29) is most widely used among the different sets of MIRU-VNTR loci. However, it is not effective for the analysis of clustered isolates (7). Other sets of MIRU-VNTR loci, such as the 14-locus set and the 15-locus set, have improved the discrimination of unrelated isolates (23, 38). An optimized set of 24 loci has also been defined; however, not all 24 loci are required for genotyping M. tuberculosis strains in any given situation (38) as the number of loci required depends on the lineage known to be prevalent in the investigated area.
In the present study, we found that the 16 of the 19 loci had high discriminatory diversity. This 16-locus set showed strong discriminatory power in analyzing the M. tuberculosis strains in our area (HGDI of 0.9977). Because the ability of the different locus combinations to differentiate the M. tuberculosis strains varied, we evaluated various sets of MIRU-VNTR loci to identify a minimal subset that provided discrimination comparable to that of the 16 loci. We found that the locus Mtub29 could be excluded from the set because the HGDI and clustering rate of the remaining 15 loci were the same as those of the 16 loci. The HGDI and the clustering rate of a 10-locus set were comparable to those of the 16-locus set. Therefore, we suggest that this 10-locus set be used as a first-line set for genotyping M. tuberculosis isolates in Heilongjiang Province, especially for routine epidemiological investigation and large-scale genotyping. Comparing the HGDI and the clustering rate of this locus set with those of various locus sets reported in other areas of China, we found that the discriminatory power of the 15-locus set used in the present study was the highest and that the clustering rate was the lowest (Table 6).
Table 6.
Discriminatory index of different locus sets used in various regions of China and the clustering rates
| Area | Locus set | Clustering rate (%) | HGDI | Reference |
|---|---|---|---|---|
| Hong Kong | 17 loci | 17.4 | 0.9900 | 23 |
| Shanghai | 16 loci | 16.1 | 0.9982 | 52 |
| 7 loci | 25.0 | 0.9957 | ||
| Beijing | 24 loci | 15.3 | 0.9920 | 19 |
| 15 loci | 18.1 | 0.9900 | ||
| 12 loci | 59.7 | 0.7880 | ||
| Fujian | 12 loci | 17.1 | 0.9808 | 18 |
| Gansu | 15 loci | 42.1 | 42 | |
| Zhejiang | 15 loci | 30.0 | 0.9905 | 49 |
| Eight regions | 12 loci | 33.5 | 0.9780 | 13 |
| Five regions | 19 loci | 15.7 | 0.9949 | 27 |
| Heilongjiang | 15 loci | 10.0 | 0.9977 | This study |
| 10 loci | 15.5 | 0.9950 | ||
| 7 loci | 25.0 | 0.9913 |
However, MIRU-VNTR loci showed variation in the ability to differentiate Beijing genotype strains from different geographical areas. Trying to explore the loci showing high discriminatory power among Beijing genotype strains in various areas of the world (Table 7), we found that at least 11 and 14 loci showed high enough diversity among the locally circulating Beijing genotype strains in China and Japan, respectively. Therefore, we recommend them as the predominant candidates (Table 7, underlined median h value for China and Japan). Since the Beijing genotype is dominant in China and Japan, we also suggest taking the 14 loci that show high diversity among the strains epidemic in the two countries as the predominant candidates for Asia (Table 7, underlined Asian median values). Meanwhile, the loci showing very low diversity (Table 7, boldface), 11 from China and 9 from Japan, may not need to be included for future studies. However, there are still some loci that showed high variation in differentiating Beijing genotype strains. For example, the loci MIRU10 and MIRU16 showed moderate diversity in Hong Kong and Gansu but low diversity in the other areas of China. The locus VNTR4120 was highly discriminatory in Japan (h of 0.902) but less discriminatory in China (h of 0.092).
Table 7.
Allelic diversity of different MIRU-VNTR loci for differentiating M. tuberculosis Beijing family strains in different areas
| Locus | Allelic diversity (h) by regiona |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Russiab |
Japanc |
Chinad |
Median for Asia | ||||||||||||
| St. Petersburg (n = 48) | West Siberia (n = 51) | Median | Kobe (n = 181) | Japan (n = 240) | Chiba (n = 185) | Median | Beijing (n = 72) | Shanghai (n = 189)e | Hong Kong group 1 (n = 51) | Hong Kong group 2 (n = 243) | Gansu (n = 202) | Heilongjiang (n = 179) | Median | ||
| VNTR4120 | 0.370 | 0.370 | 0.902 | 0.902 | 0.882 | 0.902 | 0.092* | 0.092 | 0.892 | ||||||
| QUB3232 | 0.729 | 0.729 | 0.880 | 0.909 | 0.813 | 0.880 | 0.804 | 0.804 | 0.847 | ||||||
| VNTR3820 | 0.542 | 0.542 | 0.800 | 0.871 | 0.817 | 0.817 | 0.821 | 0.821 | 0.819 | ||||||
| QUB11b | 0.205 | 0.210 | 0.208 | 0.772 | 0.815 | 0.763 | 0.772 | 0.651 | 0.689 | 0.618 | 0.669 | 0.704 | 0.669 | 0.697 | |
| QUB18 | 0.740 | 0.740 | 0.629 | 0.629 | 0.607 | 0.740 | 0.488 | 0.607 | 0.618 | ||||||
| Mtub24 | 0.591 | 0.614 | 0.603 | 0.223 | 0.223 | 0.591 | |||||||||
| QUB26 | 0.636 | 0.780 | 0.708 | 0.741 | 0.764 | 0.215 | 0.741 | 0.518 | 0.630 | 0.299 | 0.314 | 0.607 | 0.518 | 0.563 | |
| Mtub21 | 0.330 | 0.110 | 0.220 | 0.393 | 0.598 | 0.537 | 0.537 | 0.556 | 0.544 | 0.690 | 0.396 | 0.550 | 0.544 | ||
| QUB11a | 0.685 | 0.752 | 0.535 | 0.685 | 0.538 | 0.384 | 0.514 | 0.514 | 0.537 | ||||||
| QUB3336 | 0.487 | 0.642 | 0.482 | 0.487 | 0.214 | 0.214 | 0.485 | ||||||||
| QUB4156 | 0.082 | 0.082 | 0.611 | 0.623 | 0.603 | 0.611 | 0.395 | 0.469 | 0.167 | 0.182 | 0.289 | 0.469 | |||
| Mtub4 | 0.000 | 0.000 | 0.459 | 0.468 | 0.581 | 0.468 | 0.306 | 0.266 | 0.391 | 0.306 | 0.425 | ||||
| MIRU26 | 0.520 | 0.520 | 0.383 | 0.314 | 0.283 | 0.314 | 0.353 | 0.614 | 0.200 | 0.560 | 0.596 | 0.560 | 0.368 | ||
| QUB1895 | 0.364 | 0.337 | 0.468 | 0.364 | 0.365 | 0.229 | 0.206 | 0.229 | 0.351 | ||||||
| VNTR2372 | 0.595 | 0.345 | 0.470 | 0.177* | 0.177 | 0.345 | |||||||||
| QUB15 | 0.537 | 0.629 | 0.583 | 0.032* | 0.132 | 0.082 | 0.335 | ||||||||
| MIRU31 | 0.160 | 0.000 | 0.080 | 0.322 | 0.270 | 0.379 | 0.322 | 0.169 | 0.328 | 0.156 | 0.370 | 0.395 | 0.328 | 0.325 | |
| ETR F | 0.237 | 0.499 | 0.368 | 0.290 | 0.290 | 0.290 | |||||||||
| MIRU10 | 0.082 | 0.082 | 0.419 | 0.431 | 0.291 | 0.419 | 0.144 | 0.239 | 0.377 | 0.160 | 0.154 | 0.160 | 0.265 | ||
| MIRU40 | 0.122 | 0.390 | 0.256 | 0.327 | 0.229 | 0.473 | 0.327 | 0.194 | 0.147* | 0.196 | 0.350 | 0.292 | 0.196 | 0.261 | |
| MIRU16 | 0.082 | 0.082 | 0.310 | 0.258 | 0.421 | 0.310 | 0.068 | 0.131 | 0.058 | 0.580 | 0.200 | 0.131 | 0.229 | ||
| ETR A | 0.158 | 0.000 | 0.079 | 0.147 | 0.223 | 0.165 | 0.165 | 0.232 | 0.031* | 0.201 | 0.188 | 0.280 | 0.238 | 0.217 | 0.201 |
| Mtub39 | 0.000 | 0.000 | 0.000 | 0.186 | 0.215 | 0.271 | 0.215 | 0.171 | 0.061* | 0.120 | 0.174 | 0.146 | 0.174 | ||
| MIRU39 | 0.000 | 0.000 | 0.221 | 0.156 | 0.160 | 0.160 | 0.119 | 0.141 | 0.320 | 0.040 | 0.100 | 0.290 | 0.141 | 0.158 | |
| Mtub30 | 0.042 | 0.042 | 0.403 | 0.379 | 0.210 | 0.379 | 0.068 | 0.091* | 0.090 | 0.133 | 0.090 | 0.133 | |||
| Mtub29 | 0.087 | 0.180 | 0.134 | 0.043 | 0.095 | 0.103 | 0.095 | 0.119 | 0.061* | 0.123 | 0.119 | 0.103 | |||
| MIRU23 | 0.000 | 0.000 | 0.000 | 0.176 | 0.158 | 0.124 | 0.158 | 0.014 | 0.061* | 0.030 | 0.030 | 0.093 | |||
| QUB5(MIRU27) | 0.000 | 0.000 | 0.115 | 0.081 | 0.074 | 0.081 | 0.014 | 0.031* | 0.100 | 0.031 | 0.078 | ||||
| MIRU4 | 0.000 | 0.000 | 0.086 | 0.049 | 0.000 | 0.049 | 0.120 | 0.061 | 0.019 | 0.072 | 0.212 | 0.061 | 0.067 | ||
| MIRU20 | 0.120 | 0.120 | 0.022 | 0.065 | 0.063 | 0.063 | 0.014 | 0.061* | 0.038 | 0.061 | |||||
| ETR C | 0.042 | 0.000 | 0.021 | 0.022 | 0.057 | 0.063 | 0.057 | 0.094 | 0.165 | 0.057 | 0.000 | 0.076 | 0.057 | ||
| Mtub34 | 0.000 | 0.000 | 0.066 | 0.033 | 0.000 | 0.033 | 0.014 | 0.089* | 0.052 | 0.033 | |||||
| QUB23 | 0.025 | 0.025 | 0.016 | 0.016 | 0.021 | ||||||||||
| ETR B | 0.000 | 0.000 | 0.033 | 0.017 | 0.032 | 0.032 | 0.014 | 0.000* | 0.000 | 0.064 | 0.020 | 0.014 | 0.019 | ||
| VNTR0569 | 0.011 | 0.011 | 0.000* | 0.000 | 0.006 | ||||||||||
| QUB1451 | 0.033 | 0.033 | 0.000* | 0.008 | 0.004 | 0.008 | |||||||||
| MIRU2 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000* | 0.000 | 0.000 | |||||
| MIRU24 | 0.000 | 0.000 | 0.000 | 0.000 | 0.042 | 0.000 | 0.000 | 0.000* | 0.000 | 0.000 | |||||
| ETR E | 0.000 | 0.000 | 0.000 | ||||||||||||
n, number of isolates; underlining, corresponding locus was recommended; boldface, corresponding locus was not recommended.
See the following references: for Kobe, 17; for Japan, 32; for Chiba, 51. The Japan strains were from a drug resistance survey in Japan in 2002.
See the following references: for Beijing, 19; for Shanghai, 52; for Hong Kong group 1, 23; for Hong Kong group 2, 21; and for Gansu, 42. Hong Kong strains were collected in 2001 (group 1) or 2001 to 2003 (group 2).
For values marked with an asterisk, the number of samples was 65.
In Japan, most of the loci reported showed comparatively high discriminatory power; therefore, considering labor and cost, some loci with moderate h values (>0.3) may not need to be included. Russia is much different from Japan and China in allelic diversity of the MIRU-VNTR loci, and the h values of most loci are much lower than those in China and Japan. This difference may imply that the loci which are suitable for genotyping the isolates epidemic in Asia may not be suitable for genotyping isolates in Russia.
Active transmission of drug-resistant M. tuberculosis strains in a community is an emerging problem. It is generally assumed that the proportion of clustered strains in a population reflects the level of active transmission (11, 28). The present study using 15 loci showed that the clustering rate in Heilongjiang Province is 10.0%, which is lower than the rates reported in other areas (Table 6). Though some loci that show moderate or high discriminatory power in other areas were not included in the present study, omitting them will not increase the clustering rate in this area because the loci may decrease the clustering trend of the strains by decreasing the diversity. The comparatively low rate and the small size of the clusters suggest that the high resistance of M. tuberculosis in Heilongjiang Province is not related to recent transmission but, rather, may be related to reactivation or inappropriate therapy. However, the clustering rate is still increasing and was much higher in late 2008 (12.8%) than in 2007 (6.4%), suggesting that more effective control strategies are needed.
This is the first report describing the molecular epidemiology of M. tuberculosis isolated from patients with pulmonary TB in Heilongjiang Province, China. The low clustering rate in our area indicates that only mild active transmission occurred in the time period studied. We defined the most suitable MIRU-VNTR locus set for analyzing the M. tuberculosis isolates in Heilongjiang Province, where Beijing family strains are prevalent. In our hands, the 15-locus set provided a high degree of discrimination; the 10-locus set was shown to be ideal for use in first-line molecular typing in future research although we still need to examine the discriminatory power of the rest of the recommended loci.
ACKNOWLEDGMENTS
This study was supported by a Doctoral Grant Program Foundation award to J.W. from Harbin Medical University (HCXB2010020), by a Grants-in-Aid Program award to Y.S. by the Founding Research Center for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (MEXT), and by Grants-in-Aid for Scientific Research awards to Y.S. and C.N. from the Japanese Society for the Promotion of Science.
We thank Yu Zhang for assistance in collecting M. tuberculosis isolates.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Almeida D., et al. 2005. High incidence of the Beijing genotype among multidrug-resistant isolates of Mycobacterium tuberculosis in a tertiary care center in Mumbai, India. Clin. Infect. Dis. 40:881–886 [DOI] [PubMed] [Google Scholar]
- 2. Alonso M., et al. 2010. Characterization of Mycobacterium tuberculosis Beijing isolates from the Mediterranean area. BMC Microbiol. 10:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anh D. D., et al. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brudey K., et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chai L. Q., et al. 2007. Study on the genotype of Mycobacterium tuberculosis isolates from hospitals in Tianjin. Zhonghua Liu Xing Bing Xue Za Zhi 28:785–788 [PubMed] [Google Scholar]
- 6. Chen J., et al. 2007. Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 87:446–449 [DOI] [PubMed] [Google Scholar]
- 7. Cowan L. S., et al. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ebrahimi-Rad M., et al. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elbir H., Abdel-Muhsin A. M., Babiker A. 2008. A one-step DNA PCR-based method for the detection of Mycobacterium tuberculosis complex grown on Lowenstein-Jensen media. Am. J. Trop. Med. Hyg. 78:316–317 [PubMed] [Google Scholar]
- 10. Ghebremichael S., et al. 2010. Drug resistant Mycobacterium tuberculosis of the Beijing genotype does not spread in Sweden. PLoS One 5:e10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glynn J. R., et al. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. European Concerted Action on Molecular Epidemiology and Control of Tuberculosis. Int. J. Tuberc. Lung Dis. 3:1055–1060 [PubMed] [Google Scholar]
- 12. Glynn J. R., Whiteley J., Bifani P. J., Kremer K., van Soolingen D. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo Y. L., Liu Y., Wang S. M., Li C. Y. 2005. The identification of Mycobacterium tuberculosis isolates by DNA typing technique. Chin J. Epidemiol. 26:361–365 (In Chinese.) [PubMed] [Google Scholar]
- 14. Hanekom M., et al. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45:1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huard R. C., Lazzarini L. C., Butler W. R., van Soolingen D., Ho J. L. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunter P. R., Gaston M. A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwamoto T., et al. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 270:67–74 [DOI] [PubMed] [Google Scholar]
- 18. Jiang Y., et al. 2007. Preliminary genotyping in 105 strains of Mycobacterium tuberculosis isolated from Fujian province by variable number tandem repeat analysis. Chinese J. Zoonoses 23:1–4 (In Chinese.) [Google Scholar]
- 19. Jiao W. W., et al. 2008. Evaluation of new variable-number tandem-repeat systems for typing Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J. Clin. Microbiol. 46:1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jou R., Chiang C. Y., Huang W. L. 2005. Distribution of the Beijing family genotypes of Mycobacterium tuberculosis in Taiwan. J. Clin. Microbiol. 43:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kam K. M., et al. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256:258–265 [DOI] [PubMed] [Google Scholar]
- 22. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kremer K., et al. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43:314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W. M., et al. 2003. DNA fingerprinting of Mycobacterium tuberculosis strains from Beijing, Guangdong and Ningxia. Zhonghua Liu Xing Bing Xue Za Zhi 24:381–384 (In Chinese.) [PubMed] [Google Scholar]
- 25. Liu F. Y., et al. 2007. Genotyping study of 208 Mycobacterium tuberculosis clinical isolates from Guangxi with Spoligotyping. Chinese J. Zoonoses 23:1226–1230 (In Chinese.) [Google Scholar]
- 26. Liu J. H., et al. 2008. A new method for the identification of the “Beijing family” strain of Mycobacterium tuberculosis. Chin. J. Microbiol. Immunol. 28:172–175 (In Chinese.) [Google Scholar]
- 27. Lv B., et al. 2009. Analysis on genotyping of 159 strains of Mycobacterium tuberculosis isolated clinically in some areas of China with MLVA-19 loci. Dis. Surveill. 24:359–362 (In Chinese.) [Google Scholar]
- 28. Maguire H., et al. 2002. Molecular epidemiology of tuberculosis in London 1995–7 showing low rate of active transmission. Thorax 57:617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mazars E., et al. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mokrousov I., et al. 2004. Analysis of the allelic diversity of the mycobacterial interspersed repetitive units in Mycobacterium tuberculosis strains of the Beijing family: practical implications and evolutionary considerations. J. Clin. Microbiol. 42:2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mokrousov I., et al. 2008. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J. Clin. Microbiol. 46:3576–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murase Y., Mitarai S., Sugawara I., Kato S., Maeda S. 2008. Promising loci of variable numbers of tandem repeats for typing Beijing family Mycobacterium tuberculosis. J. Med. Microbiol. 57:873–880 [DOI] [PubMed] [Google Scholar]
- 33. Oelemann M. C., et al. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prodinger W. M., Bunyaratvej P., Prachaktam R., Pavlic M. 2001. Mycobacterium tuberculosis isolates of Beijing genotype in Thailand. Emerg. Infect. Dis. 7:483–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selander R. K., et al. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen G. M., et al. 2005. Evaluation of the mycobacterial interspersed repetitive units typing as a practical approach in molecular epidemiology of Mycobacterium tuberculosis. Zhonghua Jie He He Hu Xi Za Zhi 28:292–296 (In Chinese.) [PubMed] [Google Scholar]
- 37. Small P. M., et al. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703–1709 [DOI] [PubMed] [Google Scholar]
- 38. Supply P., et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Supply P., et al. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surikova O. V., et al. 2005. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur. J. Epidemiol. 20:963–974 [DOI] [PubMed] [Google Scholar]
- 41. Tanveer M., et al. 2008. Genotyping and drug resistance patterns of M. tuberculosis strains in Pakistan. BMC Infect. Dis. 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tong C. X., et al. 2010. Characterization of Mycobacterium tuberculosis isolates from patients in Gansu by multiple locus variable numbers of tandem repeats. Chin. J. Zoonoses 26:627–630 (In Chinese.) [Google Scholar]
- 43. Toungoussova O. S., Mariandyshev A., Bjune G., Sandven P., Caugant D. A. 2003. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates in the Archangel prison in Russia: predominance of the W-Beijing clone family. Clin. Infect. Dis. 37:665–672 [DOI] [PubMed] [Google Scholar]
- 44. Tsolaki A. G., et al. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 43:3185–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsolaki A. G., et al. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. U. S. A. 101:4865–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Soolingen D., Hermans P. W., de Haas P. E., Soll D. R., van Embden J. D. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Soolingen D., et al. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X. M., et al. 2008. Analysis on the Genotyping of 70 Mycobacterium Tuberculosis clinical strains isolated from Zhejiang province with spoligotyping. China Prev. Med. 9:946–949 (In Chinese.) [Google Scholar]
- 49. Wang X. M., et al. 2008. Genotyping of Mycobacterium Tuberculosis Clinical Isolates in Zhejiang province with spacer oligonucleotide typing and multiple loci variable number tandem repeat analysis. Chinese J. Zoonoses 24:1090–1094 (In Chinese.) [Google Scholar]
- 50. World Health Organization 2010. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response. WHO/HTM/TB/2010.3. World Health Organization, Geneva, Switzerland [Google Scholar]
- 51. Yokoyama E., Kishida K., Uchimura M., Ichinohe S. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7:499–508 [DOI] [PubMed] [Google Scholar]
- 52. Zhang L., et al. 2008. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol. Lett. 282:22–31 [DOI] [PubMed] [Google Scholar]



