Abstract
Rapidly growing mycobacteria (RGM) are respiratory pathogens in patients with cystic fibrosis (CF), but detection generally requires specialized cultures for acid-fast bacilli (AFB; AFB cultures). We determined that RGM could be recovered from routine cultures of samples from patients with CF by extending incubation of the Burkholderia cepacia selective agar (BCSA) from 5 to 14 days. To explore the impact of this modification, we compared results from routine and AFB cultures of samples from CF patients for 2 years before (4,212 samples by routine culture, 1,810 samples by AFB culture, 670 patients) and 2 years after (4,720 samples by routine culture, 2,179 samples by AFB culture, 695 patients) the change. Clinical relevance was assessed with samples from a subgroup of 340 patients followed regularly throughout both periods. Extending incubation of BCSA enhanced RGM recovery from routine cultures (0.7% before, 2.8% after; P < 0.001); recovery from AFB cultures was unchanged (5.5% before, 5.7% after). Estimates of RGM detection sensitivity by culture or patient-based methods ranged from ∼65 to 75% for routine cultures (nonsignificantly lower than the ∼80 to 85% for AFB cultures) and were adversely affected by coculture with mold or nonpseudomonal, nonfermenting Gram-negative rods. In the after period, 16 CF patients met the criteria for RGM infection by routine culture, including 4 who did not meet the criteria for RGM infection by AFB culture. We conclude that a simple methodological change enhanced recovery of RGM from routine cultures. The modified culture method could be utilized to improve screening for RGM in CF patients or as a simpler method to follow patients with known RGM infection. However, this method should be used cautiously in patients with certain coinfections.
INTRODUCTION
Rapidly growing mycobacteria (RGM) represent a subgroup of environmental mycobacteria that includes the commonly recognized pathogen Mycobacterium abscessus as well as other species, such as M. fortuitum and M. chelonae (1). Accumulating evidence suggests that RGM are significant respiratory pathogens within patients with chronic lung diseases such as cystic fibrosis (CF) (2, 8). A large prospective study showed that RGM respiratory infection was relatively common in CF patients (prevalence, 2.3%) (12), and CF patients with chronic RGM infection have been shown to have greater declines in lung function over time (3).
Although these data suggest that RGM are significant pathogens in patients with CF, detection of RGM respiratory infection poses considerable challenges. While RGM are capable of growth on many standard culture media, they grow relatively slowly (they are rapidly growing only relative to the rate of growth of other mycobacteria) and are typically overgrown by other common CF pathogens, such as Pseudomonas aeruginosa, on nonselective agar. As a result, detection of RGM in respiratory secretions from patients with CF is generally performed using culture for acid-fast bacilli (AFB; AFB culture) by techniques that involve a decontamination step to eliminate Pseudomonas and other competing pathogens (15). AFB cultures require considerable time and expense and are generally performed less often and more selectively than routine culture of respiratory secretions. Indeed, Cystic Fibrosis Foundation guidelines recommend more selective surveillance for nontuberculous mycobacteria, as opposed to the minimum quarterly surveillance for many other respiratory pathogens (14).
Our anecdotal evidence suggested that RGM could be recovered from Burkholderia cepacia selective agar (BCSA), one of the culture media utilized in routine cultures of respiratory secretions from CF patients. Furthermore, recovery was found to be enhanced if the incubation period for BCSA was extended from the standard 5 days to 14 days. To assess the potential impact of extending the incubation of respiratory secretions from CF patients in BCSA cultures, we implemented this change and examined results from all routine and AFB culture results for CF patients for the period 2 years before and after this modification.
MATERIALS AND METHODS
Culture methods.
Routine cultures for pathogens from CF patients and AFB cultures and smears were performed using standardized methods (6), except that the incubation period for BCSA was extended from 5 to 14 days in September 2007. The plates were incubated at 35°C for the initial 5 days and then at 30°C for the remainder of the 14 days, with additional plate reading at a minimum of 7, 10, and 14 days. Suspicious colonies were stained using Kinyoun stain, and acid-fast organisms were identified using 16S rRNA sequencing. The species of isolates classified as belonging to the M. chelonae/M. abscessus complex were determined using hsp65 sequencing. Because some strains of M. abscessus and M. massiliense share the same 16S rRNA and hsp65 sequences, for the purposes of this study, both species were classified as M. abscessus (3).
Patient records.
Culture results for patients with CF were abstracted from a database maintained by the Clinical Microbiology-Immunology Laboratories at the University of North Carolina Hospitals for the period from September 2005 through August 2007 (before) and the period from September 2007 through August 2009 (after). Data were obtained from all patients diagnosed with CF, and a subset of regularly followed patients was identified as those for whom at least one sample was cultured in each of the 4 years of the study. Infection with RGM was defined on the basis of American Thoracic Society microbiological criteria (7) as two or more samples culture positive for RGM during either 2-year study period. This study was approved by the Institutional Review Board at the University of North Carolina at Chapel Hill.
Statistical analysis.
Data are presented as means and standard deviations (SDs) for continuous variables and proportions for categorical variables. Generalized estimating equations (GEEs) with a compound symmetric covariance structure were used to compare recovery rates across time for the AFB and routine tests in the before and after periods. A bootstrap of the kappa coefficient conducted by sampling patient identification codes was used to assess the independence of the AFB test and the routine test during the after period, as well as the strength of their agreement. To estimate the relative sensitivity of the AFB and routine tests, a null-model GEE with log link and compound symmetric covariance, along with the assumption that the false-positive rate was very low, was used to obtain a point estimate and 95% confidence interval for the ratio of the sensitivity of the AFB test to that of the modified routine test. Statistical significance for all analyses was established at 0.05. All analyses were conducted in GraphPad Prism (La Jolla, CA), R, or SAS, version 9.2 (SAS, Cary, NC), software.
RESULTS
Recovery of RGM from all CF patients.
We examined microbiological culture data from all CF patients followed at our institution for the 2 years before and after a change in methodology designed to enhance recovery of RGM from routine cultures. A total of 4,212 samples for routine cultures and 1,810 samples for AFB culture from 670 patients were available for analysis in the before period, and 4,720 samples for routine culture and 2,179 samples for AFB culture from 695 patients were available in the after period (Table 1). The change in culture methodology increased the total number and proportion of routine cultures from which RGM were recovered, from 31 (0.7%) in the before period to 133 (2.8%) in the after period (P < 0.001). In contrast, there was no significant change in the proportion of AFB cultures from which RGM were recovered (5.5% before, 5.7% after; P = 0.93) (Table 1). Interestingly, the numbers of samples for both routine and AFB cultures obtained per patient were higher in the after period than in the before period.
Table 1.
Culture results, all patients
| Parameter | Before period | After period | P value |
|---|---|---|---|
| No. of patients | 670 | 695 | |
| Routine culture | |||
| Total no. of samples | 4,212 | 4,719 | |
| Avg no. of samples cultured per patient annually | 2.4 ± 2.9 | 2.7 ± 3 | <0.01 |
| No. (%) RGM-positive samples cultured | 31 (0.7) | 133 (2.8) | <0.001 |
| Culture for AFB | |||
| Total no. of samples | 1,810 | 2,179 | |
| Avg no. of samples cultured for AFB per patient annually | 1.0 ± 1.9 | 1.2 ± 2 | <0.01 |
| No. (%) of RGM-positive samples cultured | 99 (5.5) | 125 (5.7) | 0.93 |
Recovery of RGM from routine and AFB cultures.
To estimate the rates of recovery of RGM from routine and AFB cultures, we examined the subset of cultures from the after period in which both routine and AFB cultures were performed with the same respiratory sample (defined as a sample collected from the same patient on the same date, n = 2,109). We observed that 139 (6.6%) of these respiratory samples were RGM positive, defined as having RGM recovered from either routine culture or AFB culture (or both). Of these 139 RGM-positive respiratory samples, RGM were recovered less frequently from routine cultures (92/139, 66%) than from AFB cultures (114/139, 82%); 48% (67/139) of the samples were positive by both routine and AFB cultures. The estimate for the kappa test of agreement was 0.62 (95% confidence interval = 0.49 to 0.75, P < 0.001), suggesting moderate to substantial agreement between the two tests. To examine whether RGM organism burden influenced detection rates, we identified the subset of RGM-positive respiratory samples for which the stain for AFB was also positive (78 of the 139 RGM-positive samples, 56%), since staining positive for AFB may reflect a larger quantity of organisms. For these 78 respiratory samples, detection sensitivity in routine culture remained similar (53/78, 68%), whereas the rate of detection in AFB cultures was higher (75/78, 96%).
To determine whether other pathogens present in the respiratory sample impacted recovery of RGM from routine or AFB cultures, we examined the influence of Staphylococcus aureus, Pseudomonas aeruginosa, nonpseudomonal, nonfermenting Gram-negative rods (Stenotrophomonas, Achromobacter, or Burkholderia species), and mold on recovery of RGM from routine or AFB cultures. Using a generalized estimating equation model, we found that the presence of nonpseudomonal Gram-negative rods and/or mold decreased the probability of recovery of RGM from routine cultures relative to that from AFB cultures (odds ratio, 0.68; P < 0.05). Of note, both nonpseudomonal Gram-negative rods and mold were observed to be capable of growth on BCSA and were found growing on BCSA plates for which BCSA cultures were negative for RGM but AFB cultures were positive.
RGM recovery in regularly followed patients.
The clinical impact of changing the culture methodology was assessed with a subset of 340 CF patients followed regularly, defined as having at least one sample cultured during each of the 4 years of the study. From these patients, 3,154 samples for routine culture and 1,199 samples for AFB culture were obtained in the before period, and 3,381 samples for routine culture and 1,545 samples for AFB culture were obtained in the after period. Consistent with the previous findings, the change in culture methodology significantly increased the number of routine cultures from which RGM were recovered in the regularly followed patients from 25 (3.2%) to 111 (8.2%, P < 0.001). In the after period, 28 regularly followed patients had at least one sample from which RGM was recovered by routine culture, and 16 met microbiological criteria for RGM infection on the basis of the routine culture results alone. Both of these numbers were significantly higher than those in the before period (Table 2) and were similar to the results from AFB cultures: 22 individual patients had at least one sample RGM positive by AFB culture in the after period, and 16 patients met the microbiological criteria for RGM infection on the basis of AFB culture results alone. Of note, the results from AFB cultures were similar in the before and after periods (Table 2).
Table 2.
Culture results, regularly followed patients
| Parameter | Before period | After period | P value |
|---|---|---|---|
| No. of patients | 340 | 340 | |
| Routine cultures | 3,154 | 3,381 | |
| Total no. of samples | 4.6 ± 2.6 | 5.0 ± 2.8 | <0.01 |
| Avg no. of samples cultured per patient annually | |||
| No. (%) of RGM-positive samples | 25 (3.2) | 111 (8.2) | <0.001 |
| No. (%) of patients with any RGM | 11 (3.2) | 28 (8.2) | <0.01 |
| No. (%) of patients with RGM infectiona | 5 (1.5) | 16 (4.7) | 0.024 |
| Cultures for AFB | |||
| Total no. of samples | 1,199 | 1,545 | |
| Avg no. of samples cultured per patient annually | 1.8 ± 2.1 | 2.3 ± 2.5 | <0.001 |
| No. (%) of RGM-positive samples | 80 (6.7) | 102 (6.6) | 0.81 |
| No. (%) of patients with any RGM | 19 (5.6) | 22 (6.5) | 0.75 |
| No. (%) of patients with RGM infection | 14 (4.1) | 16 (4.7) | 0.85 |
RGM infection was defined as two or more RGM-positive cultures for an individual patient within a 2-year study period (before or after).
A total of 20 patients in the after period were identified as having RGM infection by routine culture, AFB culture, or both. Twelve of these patients (Table 3, patients 1 to 12) were identified as having RGM infection from both the routine and AFB culture results. Four patients (Table 3, patients 13 to 16) were identified as having RGM infection from the routine culture results but not the AFB culture results. At least three samples from each of these four patients were cultured for AFB during the after period, and for two of the four patients, a single sample was RGM positive by AFB culture but they did not meet the definition of infection. An additional four patients (Table 3, patients 17 to 20) had RGM infection identified by AFB cultures but not by routine cultures. Between 3 and 15 samples from these four patients underwent routine cultures in the after period, and three of the four had one RGM-positive routine culture result. Interestingly, of the four patients in whom RGM infection was identified by AFB culture but not routine culture, three were colonized with nonpseudomonal Gram-negative rods. Among all 20 patients with RGM infection, only 3 patients had infection with RGM identified by one culture method but no RGM-positive cultures by the other. Each of these patients had a total of only two or three samples RGM positive by culture, suggesting a relatively modest burden of infection.
Table 3.
All patients meeting criteria for RGM infection by either AFB culture, routine culture, or both in the after period
| Culture meeting infection criteria | Patient no. | No. (%) of patients |
% nonpseud GNR+b routine cultures | |||
|---|---|---|---|---|---|---|
| Routine culture |
AFB culture |
|||||
| Cultures performed | RGM+a | Cultures performed | RGM+ | |||
| Both AFB and routine cultures | 1 | 22 | 4 (18) | 20 | 3 (15) | 91 |
| 2 | 8 | 2 (25) | 8 | 2 (25) | 25 | |
| 3 | 11 | 3 (27) | 7 | 6 (86) | 0 | |
| 4 | 9 | 3 (33) | 7 | 5 (71) | 67 | |
| 5 | 16 | 6 (38) | 6 | 4 (67) | 19 | |
| 6 | 22 | 11 (50) | 15 | 11 (73) | 50 | |
| 7 | 7 | 4 (57) | 6 | 5 (83) | 100 | |
| 8 | 16 | 10 (63) | 15 | 9 (60) | 0 | |
| 9 | 12 | 9 (75) | 11 | 10 (91) | 83 | |
| 10 | 14 | 11 (79) | 7 | 4 (57) | 0 | |
| 11 | 14 | 12 (86) | 12 | 12 (100) | 0 | |
| 12 | 9 | 9 (100) | 10 | 10 (100) | 0 | |
| Routine cultures only | 13 | 9 | 4 (44) | 4 | 1 (25) | 0 |
| 14 | 7 | 3 (43) | 3 | 0 (0) | 14 | |
| 15 | 13 | 6 (46) | 3 | 1 (33) | 92 | |
| 16 | 13 | 2 (15) | 3 | 0 (0) | 15 | |
| AFB cultures only | 17 | 15 | 1 (7) | 13 | 2 (15) | 87 |
| 18 | 8 | 1 (13) | 3 | 2 (67) | 88 | |
| 19 | 3 | 0 (0) | 3 | 2 (67) | 0 | |
| 20 | 13 | 1 (8) | 13 | 12 (92) | 100 | |
RGM+, RGM positive.
Nonpseud GNR+, nonpseudomonal Gram-negative rod (Stenotrophomonas, Achromobacter, or Burkholderia species) positive.
Estimated sensitivity of RGM recovery from regularly followed CF patients.
To expand upon our earlier estimates of RGM detection sensitivity, we examined all cultures of samples from regularly followed patients in the after period to assess the relationship between the percentage of cultures positive for RGM and the total number of RGM-positive cultures. We reasoned that patients with a large total number of samples positive by culture likely had persistent infection and that the fraction of cultures in which RGM were actually recovered for these patients would serve as an estimate of the detection sensitivity. Among the patients with the highest number of positive cultures (9 or more), rates of recovery of RGM were 75.3% for routine cultures and 83.0% for AFB cultures. These values are consistent with a plateau observed with nonlinear curve fitting of the entire data set (Fig. 1). A GEE model comparing the sensitivity of AFB culture to that of the modified routine culture for recovery of RGM yielded a ratio estimate of 1.50, consistent with a higher sensitivity of the AFB culture method, although the 95% confidence interval was wide (0.94, 2.42) and the possibility of equivalence could not be rejected.
Fig. 1.
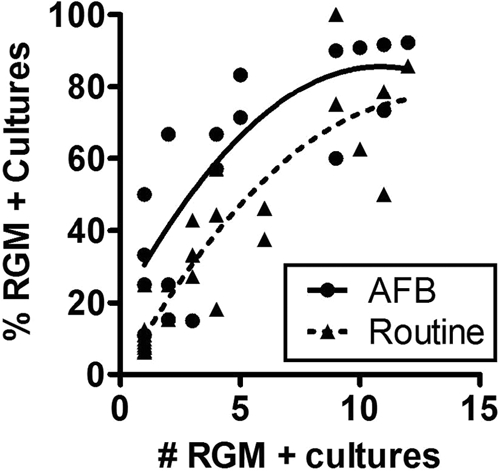
Comparison of positivity rates for routine and AFB cultures. The relationship between the number of RGM-positive cultures and the percentage of RGM-positive cultures was examined, with the assumption that the percentage of RGM-positive cultures of samples from patients with high numbers of positive cultures (suggesting persistent infection) would provide an estimate of sensitivity. Curve fitting suggested that the percentages of positive cultures plateaued at ∼85% for AFB cultures and ∼75% for routine cultures when the total number of RGM-positive cultures was ≥9.
DISCUSSION
A relatively simple change to the culture methodology significantly enhanced recovery of RGM from routine cultures of samples from CF patients, resulting in an ∼4-fold increase in the number of RGM-positive routine cultures. Detection sensitivities estimated by both culture-based (percent detection from all samples from which RGM were recovered) and patient-based (percent detection from samples from patients with clear evidence of persistent infection) methods yielded comparable results, suggesting a sensitivity for routine cultures of ∼65 to 75%. While it is lower than the estimated 80 to 85% sensitivity for AFB cultures in this study (which is consistent with previously reported values [5]), the reduced sensitivity was offset by the fact that samples for routine culture were obtained more than twice as often as samples for AFB culture for this cohort of CF patients.
In practice, both methods performed similarly for identification of RGM infection within a clinical population of regularly followed CF patients. In fact, the modified routine culture method resulted in identification of several patients with RGM infection that were not identified through AFB culture alone, although this method also failed to identify some patients. The group of patients that were not identified to have infection with RGM by one or the other method generally appeared to have a relatively modest burden of infection, and the failure of one method relative to the other may have reflected random chance. The discrepancies between culture methods highlight the fact that neither is 100% sensitive for detection of RGM in respiratory secretions.
Overall, these data suggest that application of the change in methodology for all routine cultures would improve identification of CF patients with RGM infection. The growing evidence that chronic RGM infection is associated with worse clinical outcomes (4, 10, 11) suggests that this change could convey clinical benefit, and improved screening would also be important, given the increasing use of chronic azithromycin therapy, for which infection with environmental mycobacteria is a contraindication (13). However, it is important to note that the total number of newly identified patients was relatively small and the burden of infection was relatively low, and definitively proving the clinical benefit of improved identification by this or any method would be exceedingly difficult without prolonged study of a large population of patients with CF.
We recognize that potential benefits of improved screening for RGM could likely be achieved through more frequent use of AFB cultures. However, such a change would entail additional cost that might be difficult to justify without strong evidence of clinical benefit. In contrast, the modest additional effort to detect RGM on routine culture, including an additional reading of plates at 7, 10, and 14 days of incubation, is significantly less than that required for a separate AFB culture. Indeed, reducing the cost of monitoring for RGM infection could be another rationale for using the modified routine culture methodology in lieu of AFB cultures for selected patients to follow RGM infection.
While it is beneficial, the modified routine culture methodology does have limitations. This methodology would not eliminate the need for screening AFB cultures, which would still be necessary to detect other environmental mycobacterial species, such as M. avium complex, that cannot be recovered from BCSA or other media utilized for routine culture of samples from CF patients. Furthermore, our data suggested that the routine culture method for detection of RGM must be utilized cautiously with patients infected with mold or nonpseudomonal Gram-negative rods, such as Burkholderia or Stenotrophomonas species. These pathogens are capable of growth on BCSA agar and may outcompete RGM when they are present in the respiratory sample. The negative relationship between recovery of RGM from routine culture and recovery of nonpseudomonal, nonfermenting Gram-negative rods or mold is a concern, since these pathogens have been recovered more frequently from patients with environmental myocbacterial infection in previous studies (4, 9).
In summary, we report a simple change in culture methodology that allows recovery of RGM from routine cultures of respiratory specimens from CF patients. Adoption of this method, either universally or for selected patients, could improve detection and management of RGM infections in patients with CF.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Colombo R. E., Olivier K. N. 2008. Diagnosis and treatment of infections caused by rapidly growing mycobacteria. Semin. Respir. Crit. Care Med. 29:577–588 [DOI] [PubMed] [Google Scholar]
- 2. Daley C. L., Griffith D. E. 2002. Pulmonary disease caused by rapidly growing mycobacteria. Clin. Chest Med. 23:623–632, vii [DOI] [PubMed] [Google Scholar]
- 3. Esther C. R., Jr., Esserman D. A., Gilligan P., Kerr A., Noone P. G. 2010. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J. Cyst Fibros. 9:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esther C. R., Jr., Henry M. M., Molina P. L., Leigh M. W. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Pediatr. Pulmonol. 40:39–44 [DOI] [PubMed] [Google Scholar]
- 5. Ferroni A., et al. 2006. Value of the chlorhexidine decontamination method for recovery of nontuberculous mycobacteria from sputum samples of patients with cystic fibrosis. J. Clin. Microbiol. 44:2237–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilligan P. H., Kiska D. L., Applebaum M. A. 2006. Cumitech 43, Cystic fibrosis microbiology. ASM Press, Washington, DC [Google Scholar]
- 7. Griffith D. E., et al. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 8. Griffith D. E., Girard W. M., Wallace R. J., Jr 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am. Rev. Respir. Dis. 147:1271–1278 [DOI] [PubMed] [Google Scholar]
- 9. Levy I., et al. 2008. Multicenter cross-sectional study of nontuberculous mycobacterial infections among cystic fibrosis patients, Israel. Emerg. Infect. Dis. 14:378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olivier K. N. 2004. The natural history of nontuberculous mycobacteria in patients with cystic fibrosis. Paediatr. Respir. Rev. 5(Suppl. A):S213–S216 [DOI] [PubMed] [Google Scholar]
- 11. Olivier K. N., et al. 2003. Nontuberculous mycobacteria. II. Nested-cohort study of impact on cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 167:835–840 [DOI] [PubMed] [Google Scholar]
- 12. Olivier K. N., et al. 2003. Nontuberculous mycobacteria. I. Multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:828–834 [DOI] [PubMed] [Google Scholar]
- 13. Saiman L., et al. 2003. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290:1749–1756 [DOI] [PubMed] [Google Scholar]
- 14. Saiman L., Siegel J. 2003. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Infect. Control Hosp. Epidemiol. 24:S6–S52 [DOI] [PubMed] [Google Scholar]
- 15. Whittier S., Olivier K., Gilligan P., Knowles M., Della-Latta P. 1997. Proficiency testing of clinical microbiology laboratories using modified decontamination procedures for detection of nontuberculous mycobacteria in sputum samples from cystic fibrosis patients. The Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. J. Clin. Microbiol. 35:2706–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]


