Abstract
The prevalence of quinolone-resistant Neisseria gonorrhoeae (QRNG) in Greece remained low from 1997 to 2003 but increased dramatically from 11% to 56% between 2004 and 2007. N. gonorrhoeae multiantigen sequence typing (NG-MAST) and multilocus sequence typing (MLST) were used to investigate trends in quinolone resistance from 1997 to 2007 and explore the origins of the recent increase in QRNG. We characterized 295 QRNG isolates from the study period and 233 quinolone-susceptible (QS) gonococci from 2004 and 2005, when the rapid increase in QRNG occurred. From 1997 to 1999, an outbreak of QRNG was due to the dissemination of isolates of serovar Arst that belonged to two closely related genotypes. Few QRNG isolates, of diverse genotypes, were present between 2001 and 2003, whereas the sharp increase in QRNG from 2004 onwards was due to the appearance of serovar Bropyst isolates of several major NG-MAST sequence type (STs) that previously had not been identified in Greece. These isolates were shown by MLST to be variants of a single multiply antibiotic-resistant QRNG strain (ST1901) that appeared in Greece and rapidly diversified into 31 NG-MAST STs. There were no isolates of MLST ST1901 or any of the 31 NG-MAST STs among QS isolates from 2004 and 2005 or among 8 representatives of multiresistant but quinolone-susceptible serovar Bropyst isolates circulating in Greece during the 1990s, supporting the view that the recent increase in QRNG was due to importation of a QRNG strain(s) of MLST ST1901 into Greece. Recently, multiresistant QRNG isolates of ST1901 with reduced susceptibility to the newer cephalosporins have appeared in Greece.
INTRODUCTION
Resistance of Neisseria gonorrhoeae to fluoroquinolones has become widespread, and in many countries these antibiotics can no longer be used as the antibiotic of choice for uncomplicated gonorrhea (4, 5, 20). The Greek National Reference Center for N. gonorrhoeae (NRCNG) has conducted continuous surveillance of antimicrobial susceptibility trends since 1984 (24). NRCNG identified the first quinolone-resistant N. gonorrhoeae (QRNG) strain in Greece in 1990, from a patient infected in the Philippines (22). Gonococci with decreased susceptibility to fluoroquinolones emerged in 1996. These isolates had the Arst serovar, harbored the conjugative plasmid but lacked the cryptic gonococcal plasmid, and displayed a single restriction pattern on pulsed-field gel electrophoresis (PFGE) (12). However, isolates of this clone, which persisted up to 1999, exhibited resistance to nalidixic acid but only marginal nonsusceptibility to various fluoroquinolone compounds. QRNG isolates that were highly resistant to fluoroquinolones appeared again in 1997, initially as sporadic isolates but in increasing frequencies between the years 2000 and 2003 (18). Their prevalence has increased greatly since 2004, reaching a peak of 56% of all gonococcal isolates in 2007 (21, 23).
The increase in QRNG between 1997 and 2007 and particularly between 2004 and 2007 could be due to the development of resistance among quinolone-susceptible (QS) strains circulating in Greece or to the importation and establishment of resistant strains from other countries. Molecular typing can be used to explore the basis of the emergence of QRNG strains, and several methods have been used for typing gonococci (6, 9, 25). In recent years, sequence-based methods have been favored for molecular typing as they allow simple interlaboratory comparisons of isolates. Perhaps the most widely used current method of typing gonococci is multiantigen sequence typing (NG-MAST), which indexes nucleotide sequence variations in both porB and tbpB and assigns isolates to sequence types (STs) on the basis of the alleles present at these two loci (9).
Gonococci that are assigned to the same NG-MAST ST have identical sequences at both porB and tbpB. These genes encode cell surface antigens that are subject to selective pressures applied by the human immune response, and NG-MAST STs are therefore likely to change relatively rapidly as gonococcal strains spread. Thus, a single gonococcal strain will diversify rapidly into multiple NG-MAST STs by mutations (or recombinational replacements), first in one gene and then in the other (9). The rapid rate of change of NG-MAST STs is useful for short-term or fine-scale epidemiology (2), but for following the spread of strains over a period of years, rapid diversification can obscure the fact that several NG-MAST STs may be minor variants of the same gonococcal strain.
Tracking the spread of gonococcal strains over longer time periods requires the use of a typing method where the genetic variation being indexed accumulates more slowly than that in genes encoding cell surface antigens. This can be achieved by multilocus sequence typing (MLST), which characterizes isolates using the sequences of seven housekeeping genes (1, 8, 17), and can identify which of the NG-MAST STs among a set of QRNG isolates are minor variants of the same strain. Housekeeping genes of gonococci have relatively few polymorphic sites, and MLST is therefore not highly discriminatory for gonococci (17). In this study, we used NG-MAST and made selective use of MLST to investigate the emergence and rapid recent increase of QRNG in Greece and provide evidence that they were due to the importation and spread of a single strain.
MATERIALS AND METHODS
A total of 536 gonococci of known antibiotic susceptibility and serovar from the collection of the NRCNG were studied. This sample included 295 out of 306 QRNG isolates that were submitted to the NRCNG during the period from 1997 to 2007, as well as 233 out of 239 QS isolates submitted in the years 2004 and 2005, during which the abrupt increase in the prevalence of QRNG isolates was observed in Greece. The 17 missing isolates could not be recovered from storage. Eight isolates representing the three most prevalent PFGE patterns among multiresistant, but QS, serovar Bropyst gonococci were also included, one from each of the years from 1996 to 2003. Antibiotic susceptibility testing and susceptibility categorization of the isolates had been performed as described previously, using the Etest and generally accepted criteria (18, 21). QRNG included all isolates that exhibited resistance to nalidixic acid (ciprofloxacin MIC range, 0.047 to >32 mg/liter).
Molecular typing was performed using NG-MAST as described previously (10). Alleles at porB and tbpB and NG-MAST STs were assigned at the NG-MAST website (www.ng-mast.net). One isolate of each NG-MAST ST was also characterized by MLST using the Neisseria MLST scheme hosted at www.pubmlst.org. PCR amplification and sequencing of internal fragments of the seven housekeeping loci were performed as described previously (1, 8), using the following pairs of primers: abcZ-P1 (5′-AATCGTTTATGTACCGCAGG-3′) and abcZ-S2 (5′-GAGAACGAGCCGGGATAGGA-3′), adk-UP (5′-GGCATTCCGCAAATCTCTAC-3′) and adk-DN (5′-GTGGGTACGACCTTTTCGAT-3′), aroE-UP (5′-ACAGGAAGCGTTTCATCTGG-3′) and aroE-DN (5′-GGTGATGATGTTGCCGTACA-3′), fumC-UP (5′-CCATCCCGAATACGCTGA-3′) and fumC-DN (5′-TAGCGCGGATGAACCATATC-3′), gdh-UP (5′-GCAAAGAAAGCCTGCAAAAC-3′) and gdh-DN (5′-TAAAGGCGGACGGATTCAT-3′), pdhC-UP (5′-TTATGACCCGACCTTCCAATA-3′) and pdhC-DN (5′-GGCTTTGATGCCGTATTTCT-3′), and pgm-UP (5′-CCTAATCACCACCCTGATCC-3′) and pgm-DN (5′-TTCAAAACGCAACACCAAAA-3′). Alleles and MLST STs were assigned using the Neisseria MLST website (www.pubmlst.org/neisseria). Alignments of the porB and tbpB nucleotide sequences were performed using the MEGA software, version 4.0 (19), and the most likely pathways between sequences were assessed by statistical parsimony, as implemented in the TCS program (3).
RESULTS
Overview of QRNG isolates recovered between 1997 and 2007.
The 306 QRNG isolates recovered between 1997 and 2007 were assigned to 29 different serovars, of which 8 included at least five isolates (data not shown). Although none of these serovars were represented exclusively by QRNG strains, 213 of the QRNG isolates (70%) belonged to only two serovars, Arst and Bropyst. The 295 QRNG isolates that were available for further characterization were assigned to 84 NG-MAST STs, 12 of which included at least five isolates (Table 1). MLST was carried out on one isolate of each of the 84 NG-MAST STs, grouping them into 30 MLST STs. Assuming that all isolates of the same NG-MAST ST belonged to the same MLST ST, the 12 major NG-MAST STs were grouped into 7 MLST STs (Table 1). The numbers of QRNG isolates belonging to the 12 major NG-MAST STs and the 7 major MLST STs in each year for the period from 1997 to 2007 are shown in Fig. 1 and 2. The numbers of QRNG isolates of all NG-MAST STs in each year and their corresponding MLST ST assignments are given in Table S1 in the supplemental material.
Table 1.
Major NG-MAST STs among QRNG isolated from 1997 to 2007
| NG-MAST ST | porB allele | tbpB allele | MLST ST | Serovar | No. (%) of all QRNG isolates |
|---|---|---|---|---|---|
| 154 | 87 | 33 | 1587 | Bopyst | 8 (2.7) |
| 225 | 4 | 4 | 1901 | Bropyst | 29 (9.8) |
| 289 | 234 | 4 | 1901 | Bropst | 1 (0.3) |
| 289 | 234 | 4 | 1901 | Bropyst | 19 (6.4) |
| 2616 | 1602 | 4 | 1901 | Bopyst | 1 (0.3) |
| 2616 | 1602 | 4 | 1901 | Bopyvst | 1 (0.3) |
| 2616 | 1602 | 4 | 1901 | Bropst | 1 (0.3) |
| 2616 | 1602 | 4 | 1901 | Bropyst | 61 (20.7) |
| 2618 | 1603 | 4 | 1901 | Bropyst | 10 (3.4) |
| 2618 | 1603 | 4 | 1901 | Brpst | 1 (0.3) |
| 3127 | 1901 | 110 | 1901 | Bpyu | 1 (0.3) |
| 3127 | 1901 | 110 | 1901 | Bpyut | 6 (2.0) |
| 3128 | 1900 | 110 | 1901 | Bpyut | 8 (2.7) |
| 2265 | 1417 | 4 | 8110 | Bpyvut | 5 (1.7) |
| 2265 | 1417 | 4 | 8110 | Byvt | 1 (0.3) |
| 2265 | 1417 | 4 | 8110 | Byvut | 5 (1.7) |
| 3348 | 1518 | 96 | 8129 | Bropst | 5 (1.7) |
| 3348 | 1518 | 96 | 8129 | Bropyst | 2 (0.7) |
| 2202 | 1382 | 4 | 8137 | Bropyst | 6 (2.0) |
| 2594 | 947 | 165 | 8392 | Arst | 25 (8.5) |
| 2595 | 947 | 33 | 8393 | Arst | 5 (1.7) |
| Subtotal | 201 (68.1) | ||||
| QRNG totala | 295 (100.0) |
Includes NG-MAST STs that are not shown in the table, as they each included less than five isolates.
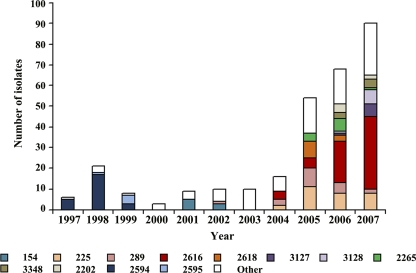
Fig. 1.
Number of isolates of gonococci of each NG-MAST ST submitted each year. The color coding for the major NG-MAST STs in the data set is shown at the bottom. Isolates of minor NG-MAST STs are grouped together and shown as white in the histogram.
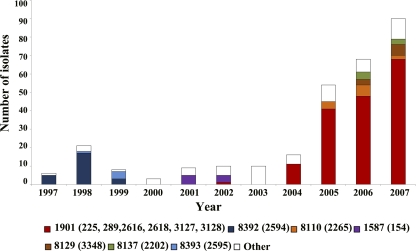
Fig. 2.
Number of isolates of gonococci of each MLST ST submitted each year. The color coding for the major MLST STs in the data set is shown at the bottom (NG-MAST STs are shown in parentheses). Isolates of minor MLST STs are grouped together and shown as white in the histogram.
QRNG isolates recovered between 1997 and 2003.
Between 1997 and 1999, the majority of the QRNG isolates were serovar Arst, which corresponded to the serovar of an outbreak strain that was circulating at the beginning of the sampling period (12). The 30 serovar Arst QRNG isolates were assigned by NG-MAST mostly to ST2594 (first identified in 1997), with 5 being ST2595 (first identified in 1998) (Table 1; Fig. 1). The isolates of these two NG-MAST STs appear to be closely related, as they had the same porB allele and differed only at tbpB and they differed by MLST at only one of the seven loci (MLST STs 8392 and 8393). As previously reported, these isolates have only marginally decreased susceptibility to fluoroquinolones. Isolates of these NG-MAST STs were not found after 1999 (Fig. 1; see Table S1 in the supplemental material). Only 3 QRNG isolates, of two novel NG-MAST STs, were identified in 2000, and the 31 isolates recovered from 2001 to 2003 were of diverse genotypes, with the exception of ST154 (serovar Bopyst), which was found five times in 2001 and three times in 2002 but not subsequently (Fig. 1; see Table S1 in the supplemental material).
QRNG isolates recovered between 2004 and 2007.
The increase in prevalence of QRNG in Greece from 2004 to 2007 was associated with the emergence and spread of multiresistant QRNG isolates of serovar Bropyst. From December 2006, an additional cluster associated with quinolone resistance began to accumulate, consisting of multiresistant QRNG isolates that exhibited the Bpyut serovar and reduced susceptibility to the newer cephalosporins (21, 23). Molecular typing showed that the rapid spread of the Bropyst isolates was due to the appearance of a number of major NG-MAST STs of this serovar (Fig. 1), including STs 225, 289, and 2616 and, to a lesser extent, ST2618, whereas the multiresistant QRNG Bpyut isolates were STs 3127 and 3128.
Characterization by MLST of one isolate of each NG-MAST ST showed that all of the above NG-MAST STs were variants of a single QRNG strain of MLST ST1901 (Table 2; Fig. 2). Furthermore, 25 minor NG-MAST STs (each present between 2004 and 2007 as only one or two QRNG isolates) were also MLST ST1901 (Table 2). Altogether, the isolates of the 31 NG-MAST STs that were assigned to MLST ST1901 included 76, 71, and 76% of all QRNG isolates characterized from the years 2005, 2006, and 2007, respectively; 83% of these isolates were serovar Bropyst.
Table 2.
Distribution per year of QRNG isolates of NG-MAST STs belonging to MLST ST1901a
| NG-MAST ST | porB allele | tbpB allele | No. of isolates of each ST |
No. of isolates | ||||
|---|---|---|---|---|---|---|---|---|
| 2002 | 2004 | 2005 | 2006 | 2007 | ||||
| 225 | 4 | 4 | 0 | 2 | 11 | 8 | 8 | 29 |
| 289 | 234 | 4 | 1 | 3 | 9 | 5 | 2 | 20 |
| 383 | 281 | 127 | 0 | 0 | 0 | 0 | 1 | 1 |
| 891 | 603 | 4 | 0 | 0 | 1 | 0 | 0 | 1 |
| 964 | 30 | 4 | 0 | 0 | 1 | 1 | 0 | 2 |
| 972 | 91 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 1365 | 876 | 4 | 0 | 0 | 1 | 1 | 0 | 2 |
| 1634 | 4 | 35 | 0 | 0 | 0 | 0 | 1 | 1 |
| 2616 | 1602 | 4 | 0 | 4 | 5 | 20 | 35 | 64 |
| 2618 | 1603 | 4 | 0 | 0 | 8 | 3 | 0 | 11 |
| 2619 | 234 | 35 | 0 | 0 | 1 | 0 | 0 | 1 |
| 2624 | 1604 | 321 | 0 | 1 | 0 | 0 | 0 | 1 |
| 2625 | 281 | 4 | 0 | 0 | 2 | 0 | 0 | 2 |
| 2628 | 1610 | 4 | 0 | 0 | 1 | 0 | 0 | 1 |
| 2694 | 4 | 25 | 0 | 0 | 0 | 0 | 1 | 1 |
| 2706 | 232 | 633 | 0 | 0 | 1 | 0 | 0 | 1 |
| 3056 | 1601 | 4 | 0 | 1 | 0 | 0 | 0 | 1 |
| 3127 | 1901 | 110 | 0 | 0 | 0 | 1 | 6 | 7 |
| 3128 | 1900 | 110 | 0 | 0 | 0 | 1 | 7 | 8 |
| 3150 | 1907 | 4 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3163 | 4 | 708 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3166 | 1919 | 4 | 0 | 0 | 0 | 0 | 2 | 2 |
| 3347 | 1690 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3721 | 2253 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3722 | 2254 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3723 | 2255 | 4 | 0 | 0 | 0 | 2 | 0 | 2 |
| 3730 | 2246 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3731 | 2251 | 4 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3733 | 2252 | 4 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3735 | 2249 | 4 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3921 | 2252 | 854 | 0 | 0 | 0 | 0 | 1 | 1 |
| Total | 1 | 11 | 41 | 48 | 68 | 169 | ||
NG-MAST STs represented by at least five isolates are denoted in boldface. No isolates of NG-MAST STs assigned to MLST ST1901 were submitted in 2003.
The numbers of isolates assigned to the NG-MAST STs of MLST ST1901 in each year are shown in Table 2. By 2004, STs 225, 289, and 2616 were all present and were recovered in all subsequent years, with ST2616 increasing in prevalence each year to represent 39% of all QRNG isolates in 2007. No isolates of these STs were identified in 2003, although a single isolate of ST289 was present in 2002. ST2618 was identified only in 2005 and 2006. As mentioned, the QRNG isolates of serovar Bpyut with reduced susceptibility to cephalosporins that were assigned to NG-MAST STs 3127 and 3128 appeared only in 2006.
Among the 31 NG-MAST STs of MLST ST1901, there were 25 porB and 9 tbpB alleles (Table 2). The alignment of the nucleotide sequences revealed that there were 14 polymorphic sites among 20 of the 25 porB alleles, and similarly, there were 5 polymorphic sites in 5 out of 9 tbpB alleles (data not shown). The alignment of the concatenated sequences of porB and tbpB from the 31 NG-MAST STs revealed that there were 17 polymorphic sites among 21 NG-MAST STs (data not shown). The other 10 NG-MAST STs differed much more extensively in either porB or tbpB.
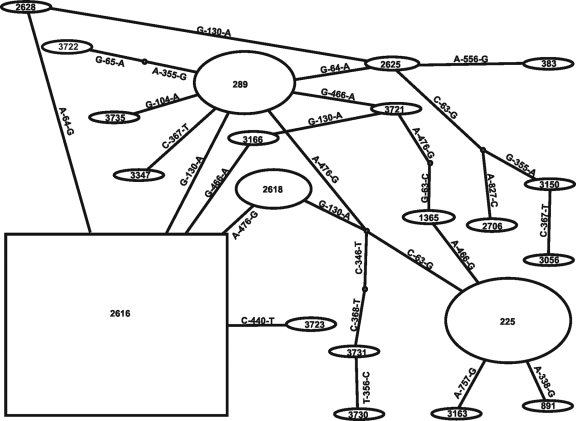
Statistical parsimony analysis was performed for the concatenated porB-tbpB nucleotide sequences of 21 NG-MAST STs. The NG-MAST STs with divergent sequences were excluded from the analysis as they probably have arisen by recombinational replacements rather than point mutations. The resulting network is shown in Fig. 3. The TCS program assigns different weights to each haplotype according to the haplotype frequency in the sample, assuming that the most frequent haplotype is ancestral on the basis of coalescent theory. NG-MAST ST2616 was the most frequent ST and was thus assigned to be the putative ancestor (displayed as a square in the TCS diagram). However, the assumption of ancestry on the basis of frequency is problematic with gonococci, as individual genotypes can increase in frequency rapidly if they enter dense sexual networks.
Fig. 3.
Most likely evolutionary pathways between NG-MAST STs. Predicted pathways were obtained using statistical parsimony as implemented in the TCS program (3). The prevalence of isolates of NG-MAST STs is shown by the size of the ovals and the square. The latter indicates the predicted ancestral ST. Nucleotide substitutions between connected STs are indicated.
An alternative approach is to consider the ST with the most variants that differ by a single-step link to be the putative ancestor. Using this criterion, NG-MAST ST289 was the putative ancestor, as it had the greatest number of single-step links to other NG-MAST STs (five, plus two links to hypothetical intermediate genotypes), including the most prevalent NG-MAST ST (ST2616), which appeared in 2004. The suggestion that NG-MAST ST289 may have been the ancestral genotype introduced into Greece is consistent with the fact that this was the genotype of the earliest QRNG isolate of MLST ST1901. ST2616 linked directly to ST2618, the fourth most prevalent NG-MAST ST, which appeared in 2005. There was no single-step path from ST225 (the second most prevalent NG-MAST ST, which appeared in 2004) to ST289 or to any of the other prevalent STs. However, ST225 could have emerged from ST289, as they differ only at two nucleotide sites, but we cannot exclude the possibility of two separate introductions of these NG-MAST STs of MLST ST1901 into the Greek gonococcal population.
Characterization of quinolone-susceptible isolates.
QS gonococci isolated in Greece during the period from 2004 to 2005 were studied, as it was within this period that the QRNG strain defined by MLST ST1901 appeared and expanded. The 233 QS isolates were distributed into 36 MLST STs and 55 NG-MAST STs. Ten NG-MAST STs and eight MLST STs included at least 5 isolates, and 117 of the 233 (50%) QS isolates were either MLST ST1590 or ST8122 (Table 3; 7 of the isolates of MLST ST1590 are not shown, as they were minor NG-MAST STs). The great majority of the 46 MLST ST1590 isolates (85%) belonged to NG-MAST ST40, and all but 2 of these were serovar Bpyust. Similarly, the great majority of MLST ST8122 isolates (91%) corresponded to NG-MAST ST210, and all but one of these were serovar Bpyvut (Table 3). These two NG-MAST STs included 17% and 28% of all 233 QS isolates examined from 2004 to 2005, respectively.
Table 3.
Major NG-MAST STs among QS gonococci isolated during 2004 and 2005
| NG-MAST ST | porB allele | tbpB allele | MLST ST | Serovar | No. (%) of all QS isolates |
|---|---|---|---|---|---|
| 40 | 30 | 35 | 1590 | Bpyust | 37 (15.9) |
| 40 | 30 | 35 | 1590 | Brpyust | 1 (0.4) |
| 40 | 30 | 35 | 1590 | Byust | 1 (0.4) |
| 495 | 91 | 32 | 1902 | Boys | 8 (3.4) |
| 2609 | 1625 | 32 | 1902 | Bropyst | 4 (1.7) |
| 2609 | 1625 | 32 | 1902 | Broyst | 1 (0.4) |
| 1979 | 1264 | 4 | 7367 | Bopyst | 5 (2.1) |
| 1979 | 1264 | 4 | 7367 | Bropyst | 4 (1.7) |
| 2599 | 1164 | 60 | 8112 | Bpyvut | 8 (3.4) |
| 943 | 640 | 29 | 8113 | Bropt | 5 (2.1) |
| 2705 | 1614 | 321 | 8121 | Broput | 4 (1.7) |
| 2705 | 1614 | 321 | 8121 | Brput | 1 (0.4) |
| 210 | 59 | 4 | 8122 | Bpyvut | 64 (27.5) |
| 210 | 59 | 4 | 8122 | Byvut | 1 (0.4) |
| 2612 | 1628 | 4 | 8122 | Bpyvut | 6 (2.6) |
| 654 | 3 | 29 | 8127 | Brpyut | 8 (3.4) |
| 654 | 3 | 29 | 8127 | Bryut | 3 (1.3) |
| Subtotal | 161 (69.1) | ||||
| QS totala | 233 (100) |
Includes NG-MAST STs that are not shown in the table, as they each included less than five isolates.
Only one NG-MAST ST included both QS and QRNG isolates (ST1979); all of these were from 2005, and only 1 of the 10 isolates was QRNG. Five MLST STs included both QS and QRNG isolates, and in all but one case the resistant and susceptible isolates of the same MLST ST were different by NG-MAST. Importantly, no isolates of the predominant QRNG strain, MLST ST1901, or of any of the 31 NG-MAST STs within MLST ST1901 were found among the QS isolates from 2004 or 2005. Serovar Bropyst isolates with multiple resistance to antibiotics other than quinolones were commonly recovered in Greece between 1991 and 1998 (11) and could have been the QS progenitors of the recent QRNG isolates of this serovar. The majority of these isolates showed three different pulsed-field gel electrophoresis patterns, and eight isolates representative of these patterns, isolated between 1996 and 2003, were characterized by MLST and NG-MAST; none of these were MLST ST1901 (data not shown).
DISCUSSION
NG-MAST was developed as a typing tool to help identify those individuals with gonorrhea within a local area who are in the same sexual network (2, 9, 10). However, the method has also been used, as it is here, to compare gonococcal isolates recovered from different years across whole countries (7, 13, 14, 16, 26). The method has some drawbacks for this purpose, as variant STs that differ at either porB or tbpB and then at both loci will rapidly be generated. They can then no longer be recognized as being minor variants of the same strain using NG-MAST, but they can be by combining NG-MAST with the selective use of MLST. The value of this approach was clearly demonstrated in the present study, where MLST established that the QRNG isolates assigned to all of the major NG-MAST STs that appeared since 2004 were minor variants of a single QRNG strain (MLST ST1901) that within 3 years had diversified into 31 different NG-MAST STs.
The sudden increase in QRNG isolates since 2004 could have been due to the importation and establishment of a quinolone-resistant isolate of MLST ST1901, with subsequent rapid spread within Greece, or to the development of resistance to quinolones among QS isolates already circulating within Greece. Statistical parsimony provided some tentative support for ST289 being the most likely ancestral QRNG NG-MAST ST in Greece, which is consistent with the fact that it was recognized 2 years before any of the other STs assigned to MLST ST1901. Isolates of both STs 225 and 289 from countries outside Greece (e.g., England, France, and Scotland) are already in the NG-MAST database (www.ng-mast.net) (2, 9, 10, 13, 16), whereas the three other STs that appeared in 2004 were novel and presumably arose within Greece.
There have been few studies of gonococci using MLST, as it lacks sufficient discrimination to address many of the kinds of epidemiological questions that arise with gonorrhea. Nevertheless, two isolates of ST1901, from South Korea and the United Kingdom, have been deposited in the Neisseria MLST database, although their dates of isolation are unclear. Interestingly, QRNG isolates of MLST ST1901, which also exhibit reduced susceptibility to cefixime, have been recovered in Japan from as early as 2000 (15; M. Ohnishi, personal communication). It is therefore possible that QRNG isolates of MLST ST1901 had spread internationally by the early 2000s and were first introduced into Greece from another country, subsequently spreading widely within Greece and diversifying into a large number of novel NG-MAST STs. Importation is not unlikely, given the large number of tourists that visit Greece each year, and this scenario is supported by the failure to identify QS isolates of MLST ST1901 in Greece in 2004 to 2005 or among multiresistant QS Bropyst isolates from 1996 to 2003 that could have been progenitors of the QRNG isolates of this ST. Importation of QRNG isolates of MLST ST1901 into Greece in about 2002 therefore appears to be a more likely possibility than the emergence of quinolone resistance within gonococci already circulating in the country. Whatever the source of the QRNG isolates of MLST ST1901, they have subsequently spread very rapidly and are the major contributor to the current high prevalence of quinolone resistance in Greece. The appearance in late 2006 of the QRNG multiresistant NG-MAST ST3127 and ST3128 variants of MLST ST1901 that also have decreased susceptibility to the newer cephalosporins currently used for treatment of gonorrhea is of concern and needs close monitoring.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Wellcome Trust (WT089472). B.G.S. is a Wellcome Trust Principal Research Fellow.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 19 January 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Bennett J. S., et al. 2007. Species status of Neisseria gonorrhoeae: evolutionary and epidemiological inferences from multilocus sequence typing. BMC Biol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Choudhury B., et al. 2006. Identification of individuals with gonorrhoea within sexual networks: a population-based study. Lancet 368:139–146 [DOI] [PubMed] [Google Scholar]
- 3. Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657–1660 [DOI] [PubMed] [Google Scholar]
- 4. Farhi D., Dupin N. 2006. The rise of fluoroquinolone resistant Neisseria gonorrhoeae. Implications for treatment guidelines. Swiss Med. Wkly. 138:223–224 [DOI] [PubMed] [Google Scholar]
- 5. Goold P. C., Bignell C. J. 2006. No way back for quinolones in the treatment of gonorrhoea. Sex. Transm. Infect. 82:225–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kolader M. E., et al. 2006. Molecular epidemiology of Neisseria gonorrhoeae in Amsterdam, The Netherlands, shows distinct heterosexual and homosexual networks. J. Clin. Microbiol. 44:2689–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao M., et al. 2009. Comparison of Neisseria gonorrhoeae multiantigen sequence typing and porB sequence analysis for identification of clusters of N. gonorrhoeae isolates. J. Clin. Microbiol. 47:489–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maiden M. C., et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin I. M., Ison C. A., Aanensen D. M., Fenton K. A., Spratt B. G. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 189:1497–1505 [DOI] [PubMed] [Google Scholar]
- 10. Martin I. M., Ison C. A., Aanensen D. M., Fenton K. A., Spratt B. G. 2005. Changing epidemiologic profile of quinolone-resistant Neisseria gonorrhoeae in London. J. Infect. Dis. 192:1191–1195 [DOI] [PubMed] [Google Scholar]
- 11. Mavroidi A., et al. 2001. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrob. Agents Chemother. 45:2651–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mavroidi A., et al. 2000. Characterization of Neisseria gonorrhoeae strains with decreased susceptibility to fluoroquinolones isolated in Greece from 1996 to 1999. J. Clin. Microbiol. 38:3489–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monfort L., et al. 2009. First Neisseria gonorrhoeae genotyping analysis in France: identification of a strain cluster with reduced susceptibility to ceftriaxone. J. Clin. Microbiol. 47:3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moodley P., Martin I. M., Pillay K., Ison C. A., Sturm A. W. 2006. Molecular epidemiology of recently emergent ciprofloxacin-resistant Neisseria gonorrhoeae in South Africa. Sex. Transm. Dis. 33:357–360 [DOI] [PubMed] [Google Scholar]
- 15. Ohnishi M., et al. 2010. Spread of a chromosomal cefixime-resistant penA gene among different Neisseria gonorrhoeae lineages. Antimicrob. Agents Chemother. 54:1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palmer H. M., Young H., Martin I. M., Ison C. A., Spratt B. G. 2005. The epidemiology of ciprofloxacin resistant isolates of Neisseria gonorrhoeae in Scotland 2002: a comparison of phenotypic and genotypic analysis. Sex. Transm. Infect. 81:403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pérez-Losada M., Viscidi R. P., Demma J. C., Zenilman J., Crandall K. A. 2005. Population genetics of Neisseria gonorrhoeae in a high-prevalence community using a hypervariable outer membrane porB and 13 slowly evolving housekeeping genes. Mol. Biol. Evol. 22:1887–1902 [DOI] [PubMed] [Google Scholar]
- 18. Stathi M., et al. 2006. Antimicrobial susceptibility of Neisseria gonorrhoeae in Greece: data for the years 1994-2004. J. Antimicrob. Chemother. 57:775–779 [DOI] [PubMed] [Google Scholar]
- 19. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis MEGA software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 20. Tapsall J. W. 2006. What management is there for gonorrhea in the postquinolone era? Sex. Transm. Dis. 33:8–10 [DOI] [PubMed] [Google Scholar]
- 21. Tzelepi E., et al. 2010. Changing figures of antimicrobial susceptibility and serovar distribution in Neisseria gonorrhoeae isolated in Greece. Sex. Transm. Dis. 37:115–120 [DOI] [PubMed] [Google Scholar]
- 22. Tzelepi E., et al. 1997. Antimicrobial susceptibility and types of Neisseria gonorrhoeae in Greece. Data for the period 1990 to 1993. Sex. Transm. Dis. 24:378–385 [DOI] [PubMed] [Google Scholar]
- 23. Tzelepi E., et al. 2008. Cluster of multidrug-resistant Neisseria gonorrhoeae with reduced susceptibility to the newer cephalosporins in northern Greece. J. Antimicrob. Chemother. 62:637–639 [DOI] [PubMed] [Google Scholar]
- 24. Tzelepi E., Fragouli E., Athanassopoulou V., Tzanakaki G., Tseliou P. 1991. Neisseria gonorrhoeae in Athens, Greece. Epidemiologic classification and antimicrobial susceptibility patterns of strains isolated between 1986 and 1989. Sex. Transm. Dis. 18:238–244 [PubMed] [Google Scholar]
- 25. Unemo M., Olcén P., Berglund T., Albert J., Fredlund H. 2002. Molecular epidemiology of Neisseria gonorrhoeae: sequence analysis of the porB gene confirms presence of two circulating strains. J. Clin. Microbiol. 40:3741–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Unemo M., et al. 2007. Molecular characterization of Neisseria gonorrhoeae identifies transmission and resistance of one ciprofloxacin-resistant strain. APMIS 115:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.