Abstract
To identify genetic loci influencing blood lipid levels in Caribbean Hispanics, we first conducted a genome-wide linkage scan in 1,211 subjects from 100 Dominican families on five lipid quantitative traits: total cholesterol (TC), low density lipoprotein cholesterol (LDL-C), high density lipoprotein cholesterol (HDL-C), triglycerides (TG), and LDL-C/HDL-C ratio. We then investigated the association between blood lipid levels and 21,361 single nucleotide polymorphisms (SNP) under the 1-logarithm of odds (LOD) unit down regions of linkage peaks in an independent community-based subcohort (N = 814, 42% Dominican) from the Northern Manhattan Study (NOMAS). We found significant linkage evidence for LDL-C/HDL-C on 7p12 (multipoint LOD = 3.91) and for TC on 16q23 (LOD = 3.35). In addition, we identified suggestive linkage evidence of LOD > 2.0 on 15q23 for TG, 16q23 for LDL-C, 19q12 for TC and LDL-C, and 20p12 for LDL-C. In the association analysis of the linkage peaks, we found that seven SNPs near FLJ45974 were associated with LDL-C/HDL-C with a nominal P < 3.5 × 10−5, in addition to associations (P < 0.0001) for other lipid traits with SNPs in or near CDH13, SUMF2, TLE3, FAH, ARNT2, TSHZ3, ZNF343, RPL7AL2, and TMC3. Further studies are warranted to perform in-depth investigations of functional genetic variants in these regions.
Keywords: genetics, heritability, single nucleotide polymorphism
Cardiovascular disease (CVD) and stroke are the leading causes of death and disability in the United States and around the world (1–4). Atherosclerosis, an underlying pathology of CVD, is characterized by lipid accumulation, inflammatory response, cell death, and fibrosis in arterial walls, and it accounts for 75% of all deaths from CVD (5). Compelling evidence accumulated over decades has demonstrated that abnormalities of blood lipid levels are a major contributor to atherosclerotic CVD (6–12). A substantial body of evidence suggests that blood lipid traits have a significant genetic component, with most studies showing the estimates of heritability around 40–70% for total cholesterol (TC), 30–70% for low-density lipoprotein cholesterol (LDL-C), 30–60% for high-density lipoprotein cholesterol (HDL-C), 20–55% for triglycerides (TG), and 30–50% for LDL-C/HDL-C (5, 13–17). Identification of genetic determinants of blood lipid levels may facilitate discovery of novel targets for drug development.
Recently, a large meta-analysis in more than 100,000 individuals of European descent identified 95 loci associated with lipid traits at a level of genome-wide significance and showed that these loci also had an impact on lipid phenotypes in three non-European populations (18). Although these associations appear robust, they together only explain 10–12% of the total variance or 25–30% of the genetic variance for each lipid trait. Given that there are known ethnic disparities in lipid metabolism, atherosclerosis, and cardiovascular disease (19), more studies from a variety of racial/ethnic groups will help to advance our understanding of the heritability and genetic variations of these complex traits (20, 21).
In the United States, compared with non-Hispanic whites, Hispanics are the fastest growing and largest minority group, with a higher prevalence of lipid abnormalities (19); however, genetic studies of blood lipids have been sparse in non-Mexican Hispanics. To fill the gap of knowledge in this understudied population, we performed an autosomal genome-wide linkage analysis of blood lipid quantitative traits in 100 multigeneration Caribbean Hispanic families from the Family Study of Stroke Risk and Carotid Atherosclerosis (22), followed by a peak-wide association analysis in an independent prospective community-based subcohort from the Northern Manhattan Study (NOMAS) (23).
MATERIALS AND METHODS
Subjects
This study was based on two independent datasets. The first consisted of 1,211 participants with no history of taking cholesterol-lowering medication from 100 Dominican families for genome-wide linkage analysis. The second included a stroke-free NOMAS subcohort of 814 individuals with no history of taking cholesterol-lowering medication for the association analysis of single nucleotide polymorphisms (SNP) under the 1-logarithm of odds (LOD) unit down regions of linkage peaks. Informed consent was provided by the participants, and the study was approved by the Institutional Review Boards of Columbia University, the University of Miami, the National Bioethics Committee, and the Independent Ethics Committee of Instituto Oncologico Regional del Cibao in the Dominican Republic.
The research design and detailed ascertainment scheme for the family study and NOMAS were described previously (22–24). In brief, for the family study, probands were selected from the Caribbean Hispanic participants in NOMAS based on the following criteria: 1) reporting a sibling with a history of myocardial infarction or stroke; or 2) having two of three quantitative risk phenotypes (maximal carotid plaque thickness, left ventricular mass, or homocysteine level > 75th percentile in the NOMAS cohort). To be eligible for NOMAS, participants 1) had never been diagnosed with a stroke, 2) were at least 40 years of age, and 3) resided for at least three months in a household with a telephone in northern Manhattan. A total of 3,298 community subjects were enrolled in 1993-2001, and 199 unrelated household members were recruited in 2003-2008. In the association analysis of linkage peaks, however, we used a convenient subcohort of NOMAS (n = 814, 42% Dominican subset) independent of the family study, who had blood lipid measurements, no history of cholesterol-lowering medication, and genotype data from a genome-wide association study done primarily to study subclinical brain phenotypes.
Blood lipid phenotypes and covariates
At enrollment, all participants underwent a thorough evaluation, including comprehensive medical history, physical examination, review of medical records, and blood sample collection after an overnight fast. Plasma levels of TC and TG were measured using standardized enzymatic procedures with a Hitachi 705 automated spectrophotometer (Boehringer Mannheim, Mannheim, Germany). HDL-C was quantified after precipitation of plasma apoB-containing lipoproteins with phosphotungstic acid. LDL-C levels were derived from the Friedewald equation (25). LDL-C/HDL-C was calculated based on measured LDL-C and HDL-C.
Covariates in linkage and association analyses included demographic and lifestyle factors assessed at enrollment. Body mass index (BMI) was calculated as measured body weight (in kilograms) divided by the square of height (in meters), and waist-to-hip ratio (WHR) was calculated as measured waist circumference divided by hip circumference. Cigarette pack-years were calculated as number of years smoked multiplied by number of cigarettes smoked per day, then divided by 20. Alcohol drinking was defined as currently drinking more than one drink per month. Leisure-time physical activity was assessed by a questionnaire, and physical inactivity was defined as no moderate or rigorous exercise in a typical 14-day period (26).
Genotyping
For the linkage study, genome-wide genotyping of 383 autosomal microsatellite markers was carried out by the Center for Inherited Disease Research at an average interval of 10 centimorgans (cM). Family structure was verified or adjusted through comparison of the putative relationship between pairs of individuals with those constructed based on the autosomal genotypes by performing a maximized log-likelihood ratio test using PREST (27). Relationships with P < 1.0 × 10−6 in a consistent manner across the family were considered an error. Mendelian errors were also checked on the final family structure using PEDCHECK (28).
For the NOMAS association study, genotyping of DNA samples was performed using the Affymetrix® Genome-Wide Human SNP Array 6.0 chips according to Affymetrix procedures at the Genotyping Core of the John P. Hussman Institute for Human Genomics at the University of Miami. Quality control was applied to both DNA samples and SNPs. Specifically, samples were removed from further analysis if they had call rates below 95%, relatedness or sex discrepancies, or were outliers beyond six SDs from the mean based on EIGENSTRAT analysis (29). SNPs were excluded if they were not in Hardy-Weinberg equilibrium (P < 1.0 × 10−6), had a genotyping call rate less than 95%, or had a minor allele frequency less than 5% as detected by PLINK (30).
Statistical analyses
In the linkage analysis, the outliers beyond mean ± 4 SD were first removed and log-transformation was then performed on all blood lipid phenotypes to reduce the skewness and kurtosis. Polygenic modeling with SOLAR was conducted to screen for a set of covariates: age, sex, age by sex, age2, age2 by sex, physical activity, smoking pack years, alcohol drinking, body mass index (BMI) and waist-to-hip ratio (WHR) based on a threshold of P < 0.1 for inclusion of any potentially significant covariates. For all traits, skewness and kurtosis were further checked for the residuals in the polygenic models and we did not observe departure from normality after adjustment for covariates. A maximum likelihood approach was then implemented in SOLAR to calculate heritability, proportion of alleles shared identical-by-descent (IBD), and multipoint LOD scores (31, 32). Empirical p-values for LOD scores were computed based on 15,000 replicates in which a fully-informative marker, unlinked to the specific trait, was simulated and used to compute possible LOD scores. Power analysis using SOLAR through simulation showed that with the final family sample (n = 1,211), we have at least 85% power detect a QTL with a heritability of 0.27 at a LOD score of 3 or of 0.23 at a LOD score of 2 for a lipid trait, given that the total genetic heritability for the lipid trait is 0.4.
In the association analysis of linkage peaks, population stratification was controlled for with a principal component analysis (PCA) using EIGENSTRAT. We used a subset of approximately 30K independent (r2 ≥ 0.25) SNPs across the entire genome to infer the principal components composing the axes of genetic variation in our NOMAS subcohort (supplemental Fig. I). Besides the covariates screened in the linkage study analysis, three top principal components of PCA (PCA1, PCA2, and PCA3) were screened using a stepwise selection procedure with SAS 9.2 (SAS Institute Inc., Cary, NC). Any covariates with P < 0.10 were kept in the model. Assuming an additive genetic model, multiple linear regression analysis was performed with PLINK to investigate the association between lipid levels and 21,361 SNPs located within the 1-LOD support intervals of the five linkage peaks on 7p12 (5,861), 15p23 (5,932), 16q23 (2,578), 19q12 (2,969), and 20p12 (4,021), after adjusting for the selected covariates. In addition, the analysis was carried out in the Dominican subset to check whether the observed associations in the whole NOMAS subcohort remained similar and significant after controlling for population structure using the three top PCAs as covariates. As many SNPs tested within the five regions are highly correlated due to linkage disequilibrium (LD), the effective number of tests in each region was determined based on the sum of singleton SNPs plus LD blocks identified using Gabriel's method (33). A Bonferroni procedure was then applied to adjust for multiple testing in each region as suggested by Nicodemus et al. (34).
RESULTS
Table 1 summarizes the demographic, lifestyle, and phenotypic characteristics of the study samples. The mean age of subjects was 44.1 ± 17.2 and 63.5 ± 8.4 years, respectively, for the Dominican family sample and community-based NOMAS subcohort. Both samples had more female subjects (60% in the family sample and 58% in the NOMAS subcohort) and a comparable average BMI. With an older age distribution, the NOMAS subcohort had a greater mean of TC, LDL-C, TG, and LDL-C/HDL-C, but a lower average HDL-C compared with the family sample.
TABLE 1.
Characteristics of Study Samples
| NOMAS Subcohort |
|||
| Characteristic | Dominican Family Sample (n = 1,211) | All (n = 814) | Dominican Subset (n = 343) |
| Age (years) | 44.1 ± 17.2 | 63.5 ± 8.4 | 61.6 ± 7.7 |
| Female | 61% | 58% | 62% |
| Hispanic | 100% | 65% | 100% |
| Physical inactivity | 55% | 43% | 52% |
| More than 1 alcohol drink/month | 57% | 50% | 43% |
| Cigarette pack-years | 3.3 ± 9.0 | 11.2 ± 21.7 | 7.7 ± 14.7 |
| Body mass index (kg/m ) | 28.5 ± 5.8 | 28.0 ± 4.8 | 28.2 ± 4.4 |
| Waist-to-hip ratio | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 |
| TC (mg/dl) | 183.5 ± 40.5 | 202.8 ± 38.0 | 201.9 ± 36.2 |
| LDL-C (mg/dl) | 109.5 ± 33.7 | 129.9 ± 34.3 | 131.2 ± 32.9 |
| HDL-C (mg/dl) | 50.3 ± 13.3 | 47.0 ± 14.8 | 43.8 ± 11.9 |
| TG (mg/dl) | 119.2 ± 86.7 | 131.5 ± 81.3 | 134.9 ± 69.6 |
| LDL-C/HDL-C | 2.3 ± 1.0 | 3.0 ± 1.2 | 3.2 ± 1.1 |
Values are means ± standard deviation or percentage.
Table 2 presents the effects of demographic and lifestyle factors on five blood lipid traits (all were log-transformed) and the adjusted heritability estimates for quantitative lipid traits in Dominican families. Both age and age2 were correlated with all lipid traits except HDL-C. Sex was associated with HDL-C, TG, and LDL-C/HDL-C, but not TC and LDL-C. BMI and WHR were the two covariates associated with all five lipid traits. In addition, cigarette pack-years and alcohol drinking were associated with HDL-C but not other lipid traits. No significant effect of leisure-time physical activity or sex-by-age interaction was found on these lipid traits. The variance explained by all covariates was just 11-20% for each of the five lipid traits. After adjustment for the associated covariates, SOLAR polygenic analyses support that lipid traits are under significant genetic influence, with the estimated heritabilities ranging from 0.41 to 0.57 for these lipid traits (P ≤ 1.2 × 10−15).
TABLE 2.
Covariates and Adjusted Heritability Estimates for Blood Lipid Traits
| Polygenic Model | TC | LDL-C | HDL-C | TG | LDL-C/HDL-C |
|---|---|---|---|---|---|
| Covariate screened, P value | |||||
| Sex | 0.74 | 0.15 | 9.99×10−15 | 7.96×10−6 | 2.91×10−8 |
| Age | 6.99×10−13 | 3.45×10−8 | 0.55 | 6.62×10−12 | 2.85×10−5 |
| Age2 | 6.34×10 | 2.79×10 | 0.9 | 1.17×10 | 0.0007 |
| Sex*age | 0.93 | 0.98 | 0.51 | 0.71 | 0.93 |
| Sex*age2 | 0.04 | 0.12 | 0.78 | 0.20 | 0.33 |
| Physical activity | 0.72 | 0.95 | 0.23 | 0.87 | 0.66 |
| Alcohol drinking | 0.13 | 0.27 | 0.01 | 0.82 | 0.8 |
| Cigarette pack-years | 0.34 | 0.15 | 0.06 | 0.46 | 0.11 |
| Body mass index | 0.01 | 0.0008 | 1.77×10−9 | 5.01×10−9 | 2.31×10−11 |
| Waist-to-hip ratio | 0.02 | 0.07 | 0.04 | 2.08×10−5 | 0.007 |
| Proportion of variance due to covariates | 0.15 | 0.11 | 0.15 | 0.20 | 0.15 |
| Trait residual skewness | −0.29 | −0.44 | −0.19 | 0.52 | −0.20 |
| Trait residual kurtosis | 0.65 | 0.79 | 0.36 | 0.63 | 0.39 |
| Adjusted heritability | |||||
| h2 ± SE | 0.41 ± 0.07 | 0.44 ± 0.06 | 0.54 ± 0.06 | 0.56 ± 0.06 | 0.57 ± 0.06 |
| P value | 1.20×10−15 | 1.42×10−17 | 9.27×10−26 | 1.21×10−27 | 2.33×10−27 |
Traits were log-transformed.
h2, heritability; SE, standard error.
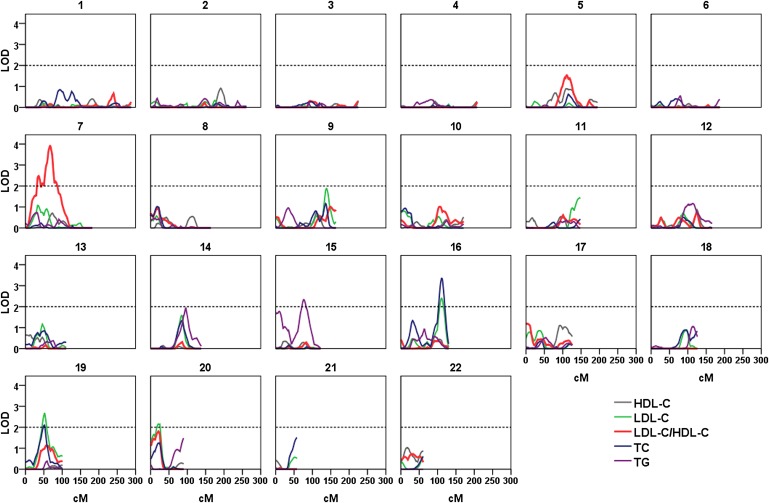
Figure 1 shows a plot of multipoint LOD scores for each lipid trait based on variance component linkage analysis, and Table 3 lists the chromosomal regions showing suggestive linkage with a multipoint LOD score > 2.0 (35). The strongest linkage was identified on chromosome 7p12 for LDL-C/HDL-C (LOD = 3.91 at 67 cM, empirical P = 0.00007, 1-LOD support interval: 37-60 Mb) and on 16q23 for TC (LOD = 3.35 at 111 cM, P < 00007, 1-LOD support interval: 79-84 Mb). Suggestive linkage was found for LDL-C (LOD = 2.40 at 111 cM on 16q23, LOD = 2.68 at 52 cM on 19q12, and LOD = 2.16 at 20 cM on 20p12), TC (LOD = 2.12 at 20 on 16q23), and TG (LOD = 2.33 at 76 on 15q23) (Table 3). In addition, weak linkage of a multipoint LOD score > 1.5 was found for LDL-C/HDL-C (LOD = 1.53 at 111 cM on chromosome 5, LOD = 1.81 at 21 cM on chromosome 20), LDL-C (LOD = 1.88 at 138 cM on chromosome 9, LOD = 1.58 at 84 cM on chromosome 14), and TG (LOD = 1.95 at 96 cM on chromosome 14, LOD = 1.77 at 10 cM on chromosome 15) (Fig. 1).
Fig. 1.
Results of autosomal genome scan for blood lipid quantitative traits in Caribbean Hispanic families. Multipoint LOD scores are shown in lines for SOLAR variance component analysis. All lipid traits were log-transformed.
TABLE 3.
Chromosomal Regions with Multipoint LOD score > 2.0 for Lipid Quantitative Traits
| Region | Traita | Nearest Marker | cM | h2q | LOD | Empirical Pb | 1-LOD Interval, Mb | Referencesc |
| 7p12 | LDL-C/HDL-C | D7S1818 | 67 | 0.36 | 3.91 | 0.00007 | 37-60 | 35–39 |
| 15q23-24 | TG | D15S131/D15S655 | 76 | 0.26 | 2.33 | 0.0004 | 65-91 | 17, 57–61 |
| 16q23 | TC | D16S3091 | 111 | 0.33 | 3.35 | <0.00007 | 79-84 | 36, 40–43 |
| LDL-C | D16S3091 | 111 | 0.26 | 2.40 | 0.0009 | 79-84 | ||
| 19q12 | TC | D19S433 | 52 | 0.23 | 2.12 | 0.001 | 12-39 | 5, 13, 16, 52–56 |
| LDL-C | D19S433 | 52 | 0.26 | 2.68 | 0.0005 | 16-39 | ||
| 20p12 | LDL-C | D20S851 | 21 | 0.28 | 2.16 | 0.001 | 00-11 | 54, 62 |
h2q, locus-specific heritability; Mb, megabase.
Traits were log-transformed.
Empirical P was based on 15,000 replicates for the specific trait.
Linkage was reported for lipid traits in previous studies.
In the association analysis of 21361 SNPs within the 1-LOD unit down regions of the five linkage peaks, LD block analysis yielded an effective number of tests of 1,872 for 7p12; 2,620 for 15q23; 1,390 for 16q23; 1,129 for 19q12; and 1,740 for 20p12. Table 4 reports the lipid-associated SNPs with a nominal P < 0.0001 in the whole NOMAS subcohort and NOMAS Dominican subset. The most significant associations were found with LDL-C/HDL-C on chromosome 15 for rs4932506 (P = 5.05 × 10−6, multiple testing adjusted P = 0.013, near hypothetical gene LOC441732) and on chromosome 7 for rs7806473 [P = 9.24 × 10−6, multiple testing adjusted P = 0.017, an intergenic SNP between FLJ45974 protein (FLJ45974) and solute carrier family 25 member 5 pseudogene 3 (SLC25A5P3) genes]. Interestingly, seven other intergenic SNPs between FLJ45974 and SLC25A5P3 were also associated with LDL-C/HDL-C with a P < 8.5 × 10−5 in the NOMAS subcohort and P < 0.01 in NOMAS Dominican subset. Additionally, 10 other SNPs associated with any lipid trait in the entire NOMAS subcohort or only Dominican subset are located in or near eight known genes, including sulfatase modifying factor 2 (SUMF2), transducin-like enhancer of split 3 (TLE3), fumarylacetoacetate hydrolase (FAH), aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2), transmembrane channel-like 3 (TMC3), cadherin 13 (CDH13), teashirt zinc finger homeobox 3 (TSHZ3), and zinc finger protein 343 (ZNF343); and five are located in or near three hypothetical genes, LOC10013187, LOC100128139, and ribosomal protein L7a pseudogene 12 (RPL7AL2).
TABLE 4.
Lipid-associated SNPs with P < 0.0001 in the NOMAS Subcohort or Dominican Subset
| Chr | SNP | BP | Minor/Major Allele (MAF) | Lead Trait | Beta (SE)b | P c | Other Lipid Traitsa,d | Gene Nearby | |
| All NOMAS Subcohort | NOMAS Dominican Subset | ||||||||
| 7 | rs4947638 | 51794849 | T/C (0.26) | T/C (0.23) | LDL-C/HDL-C | −0.10 (0.02) | 3.27×10−5 | HDL-C; LDL-C/HDL-C, HDL-C | LOC100131871 |
| rs7785376 | 51923383 | G/A (0.17) | G/A (0.14) | HDL-C | 0.08 (0.02) | 2.25×10−5−5 | LDL-C/HDL-C, TG | LOC100131871 | |
| rs13235686 | 54095049 | G/A (0.12) | G/A (0.13) | LDL-C/HDL-C | 0.13 (0.03) | 3.34×10−5 | LDL-C; LDL-C/HDL-C LDL-C | FLJ45974/ SLC25A5P3 | |
| rs17560050 | 54095274 | C/A (0.14) | C/A (0.14) | LDL-C/HDL-C | 0.12 (0.03) | 2.60×10−5−5 | LDL-C; LDL-C/HDL-C, LDL-C | FLJ45974/ SLC25A5P3 | |
| rs17560056 | 54095333 | G/C (0.14) | G/C (0.14) | LDL-C/HDL-C | 0.12 (0.03) | 2.60×10−5−5 | LDL-C; LDL-C/HDL-C, LDL-C | FLJ45974/ SLC25A5P3 | |
| rs10499728 | 54098093 | A/G (0.14) | A/G (0.14) | LDL-C/HDL-C | 0.12 (0.03) | 8.45×10−5 | LDL-C; LDL-C/HDL-C | FLJ45974/ SLC25A5P3 | |
| rs13233885 | 54099495 | C/T (0.14) | C/T (0.14) | LDL-C/HDL-C | 0.12 (0.03) | 2.60×10−5−5 | LDL-C; LDL-C/HDL-C, LDL-C | FLJ45974/ SLC25A5P3 | |
| rs7806473 | 54100065 | C/T (0.11) | C/T (0.12) | LDL-C/HDL-C | 0.14 (0.03) | 9.24×10−6−5 | LDL-C, HDL-C; LDL-C, LDL-C/HDL-C | FLJ45974/ SLC25A5P3 | |
| rs953160 | 54104682 | G/A (0.13) | G/A (0.14) | LDL-C/HDL-C | 0.13 (0.03) | 2.52×10−5−5 | LDL-C; LDL-C/HDL-C, LDL-C | FLJ45974/ SLC25A5P3 | |
| rs17560210 | 54110101 | C/T (0.14) | C/T (0.15) | LDL-C/HDL-C | 0.12 (0.03) | 3.42×10−5 | LDL-C; LDL-C/HDL-C | FLJ45974/ SLC25A5P3 | |
| rs13238899 | 56108112 | C/A (0.09) | C/A (0.10) | LDL-C | −0.10 (0.03) | 7.02×10−5 | LDL-C/HDL-C, TC; LDL-C | SUMF2 | |
| 15 | rs17758593 | 68100336 | T/C (0.11) | T/C (0.11) | LDL-C | −0.12 (0.03) | 8.13×10−5 | TC, LDL-C/HDL-C | TLE3 |
| rs3858961 | 78394769 | A/G (0.15) | A/G (0.17) | HDL-C | −0.08 (0.02) | 6.29×10−5 | TC, LDL-C/HDL-C | FAH | |
| rs4778823 | 78649800 | G/T (0.14) | G/T (0.15) | LDL-C | −0.09 (0.02) | 6.35×10−5 | TC, LDL-C/HDL-C | ARNT2 | |
| rs1482937 | 79598554 | G/C (0.33) | G/C (0.30) | LDL-C | −0.06 (0.02) | 4.99×10−5 | TMC3 | ||
| rs7179520 | 89972904 | G/C (0.18) | G/C (0.16) | LDL-C/HDL-C | −0.11 (0.03) | 1.65×10−5−5 | HDL-C | LOC441732 | |
| rs4932506 | 89976260 | A/G (0.14) | A/G (0.11) | LDL-C/HDL-C | −0.14 (0.03) | 5.05×10−6−5 | HDL-C | LOC441732 | |
| 16 | rs4357934 | 82148718 | G/C (0.32) | G/C (0.32) | TC | 0.05 (0.01) | 9.13×10−5 | LDL-C, LDL-C/HDL-C | CDH13 |
| rs11645264 | 82159785 | T/C (0.34) | T/C (0.35) | TC | −0.05 (0.01) | 9.25×10−5 | LDL-C, LDL-C/HDL-C; TC, LDL-C | CDH13 | |
| 19 | rs1216119 | 22501685 | T/G (0.29) | T/G (0.27) | LDL-C/HDL-C | −0.12 (0.03) | 9.03×10−5 | TC, LDL-C, HDL-C, TG | LOC100128139 |
| rs7245570 | 36505242 | A/G (0.05) | A/G (0.06) | LDL-C | −0.13 (0.03) | 5.23×10−5 | TC, LDL-C/HDL-C; LDL-C, TC, LDL-C/HDL-C | TSHZ3 | |
| rs10417470 | 36896151 | C/T (0.48) | T/C (0.47) | LDL-C/HDL-C | 0.08 (0.02) | 7.27×10−5 | HDL-C | TSHZ3 | |
| 20 | rs1547063 | 2427198 | G/A (0.35) | G/A (0.35) | TC | 0.06 (0.01) | 6.15×10−5 | LDL-C, LDL-C/HDL-C; | ZNF343 |
| rs6037842 | 4366157 | G/A (0.10) | G/A (0.12) | LDL-C/HDL-C | 0.19 (0.04) | 1.61×10−5−5 | LDL-C, HDL-C, TG; HDL-C, LDL-C/HDL-C | RPL7AL2 | |
Chr, chromosome; BP, base pair.
All lipid traits were log-transformed. Traits in italics apply only to the Dominican subset; otherwise, traits apply to the entire NOMAS subcohort.
Regression coefficient and its standard error (SE) were based on additive genetic effect model after adjustment for significant covariates.
P values are bolded if they had a Bonferroni-corrected P < 0.05 based on the effective number of tests within the linkage peak.
Traits were included if the associated P < 0.01 in the entire MONAS subcohort or if P < 0.02 in the NOMAS Dominican subset.
In addition, three loci (genes) reported by Teslovich et al. are located within the 1-LOD intervals of the five chromosomal regions showing suggestive linkage in Table 3: Niemann-Pick disease, type C1 gene (NPC1L1), c-Maf-inducing protein gene (CMIP), and CAP-GLY domain containing linker protein 2 gene (CLIP2). Within the three loci, we had genotype data on 2 NPC1L1 SNPs, 115 CMIP SNPs, and 2 CLIP2 SNPs, but we did not have genotype data on the top SNPs (rs2072183, rs217386, rs2925979, and rs10401969) from these three genes. For the 2 NPC1L1 SNPs (rs217429 and rs217426), we did not find an association with P < 0.05 for any of the lipid traits in either the entire NOMAS subcohort or the NOMAS Dominican subset. For the 2 CLIP2 SNPs, we observed an association between rs11670882 and TG in the Dominican subset (P = 0.03) and between rs2304097 and LDL-C/HDL-C in the whole NOMAS subcohort (P = 0.02). Among the 115 CMIP SNPs (from 80,041,701 to 80,298,964 bp), 6 SNPs (rs12934986, rs2966120, rs2911278, rs1471152, rs4074144, and rs4889327) were associated with a lipid trait with P < 0.01 in the Dominican subset or the entire NOMAS subcohort. The most significant association was found between rs12934986 and HDL-C in the Dominican subset (P = 0.002) and the entire NOMAS subcohort (P = 0.003).
DISCUSSION
Here we report a comprehensive genetic study of lipid quantitative traits in Caribbean Hispanics by combining a genome-wide linkage scan with peak-wide association. The present study is further complemented by the use of two independent samples: well-characterized Caribbean Hispanic multigeneration families and a community-based prospective cohort mainly composed of Hispanics. This unique resource allows us to search for the genetic loci that influence blood lipid concentrations in a well-characterized minority population in a systematic manner. In our polygenic analysis adjusted for covariates, the heritability estimates ranged from 41-57% for the five quantitative lipid traits in Caribbean families. These estimates are very comparable to those obtained in most reported family studies from different populations (5, 13–17) and, therefore, provide further evidence that blood lipids are largely under genetic control.
In this report, the largest linkage peak was mapped to chromosome 7p for LDL-C/HDL-C. In this region, there is evidence for both significant linkage with an LOD score of 3.9 and significant association with multiple genetic variants between FLJ45974 and SLC25A5P3 in an independent sample, after adjustment for multiple testing. Previously, several studies reported linkage for lipid-related traits in or near this region. For example, a study of 485 pairs of dizygotic Australian twins identified significant linkage in the same region (LOD = 2.9 at 65 cM for log-transformed TG, LOD = 2.6 at 60 cM for log-transformed TG/HDL-C ratio) (36); a meta-analysis of nine populations with families ascertained for type 2 diabetes revealed suggestive linkage for TG (LOD = 1.9) and TG/HDL-C ratio (LOD = 2.1) at 64-93 cM (37); a genome-wide linkage analysis from the Framingham Heart Study found linkage at 52 cM, with a LOD of 2.0 for TG/HDL-C ratio (38); and the San Antonio Family Diabetes Study (SAFADS) of Mexican Americans detected suggestive linkage near this region at 94 cM for log-transformed TG (LOD = 2.1) (39). In our association analysis, the most significantly associated SNPs are located between FLJ45974 and SLC25A5P3, but the function of these two genes is largely unknown. SLC25A5P3 has been proposed to participate in transport processes, encodes proteins similar to ADP/ATP translocase 2, and is localized in mitochondrion and membrane, whereas the in vivo function of FLJ45974 is yet unknown. SUMF2, a nearby gene with significant SNP association for LDL-C, is one member of the sulfatase-modifying factor family that can catalyze the oxidation of cysteine in the active site of sulfatases into C-alphaformylglycine, which is necessary for catalytic activities of the sulfatases. It has been shown that SUMF2 may interact with interleukin (IL)-13 and inhibit IL-13 secretion (40), but its link with lipid metabolism or atherosclerosis is unknown. These genes and others nearby, therefore, are subject to further investigation.
Our second significant linkage peak was observed at 111 cM on 16q23 (LOD = 3.35 for TC, and LOD = 2.40 for LDL-C). This region has been linked to lipid-related traits by scans from different studies, including a Mexican American family study (LOD = 3.7 at 92 cM for HDL-C) (41), a Finnish and Dutch family study (LOD = 3.2 at 99 cM for HDL-C) (42), an African American family study (LOD = 2.6 at 100 cM for TG) (43), a French Canadian family study (LOD = 2.6 at 98 cM for HDL-C) (44), and a meta-analysis (LOD = 1.5 at 98-130 cM for LDL-C/HDL-C) (37). In the present study, however, we did not find suggestive linkage for HDL-C. Instead, we observed the connection between 16q23 loci and TC (mainly LDL-C) in both linkage and association analysis in two independent Dominican samples. In the 1-LOD support interval (5 Mb) of this linkage peak, the most significant association was found between TC and SNP rs4357934 in the T-cadherin (CDH13) gene (P = 9.13 × 10−5) in a small independent NOMAS Dominican dataset (N = 343). Given the biological function of T-cadherin, this finding is of particular interest. T-cadherin is highly expressed in the cardiovascular and nervous system (45), and it interacts in vascular endothelial and smooth muscle cells with two different ligands, LDL-C and adiponectins, both of which play an important role in cardiovascular physiology (46, 47). Recently, CHD13 variants were found to be associated with CVD risk, adiponectin concentration, and blood pressure in several large genome-wide association studies (48–52). These features make CDH13 a promising candidate for further investigation to identify functional variants influencing lipid variations.
In addition to the significant linkage on 7p12 and 16q23, suggestive linkage was found on 15q23 for TG, 19q12 for TC and LDL-C, and 20p12 for LDL-C. Among these three regions, 19q has been linked to lipid quantitative traits with the most consistent linkage evidence in the previous reports, with LOD scores ranging from 1.9 to 5.7 in the region at 50-70 cM among various populations (5, 13, 16, 53–57) (Table 3). Similarly, 15q has previously been implicated in several genome scans of lipid-related traits, with LOD scores from 1.8 to 4.2 in the region at 61-72 cM among various populations (17, 58–62) (Table 3). For 20p12, two studies have shown evidence for linkage in this region: a LOD score of 2.8 for TG at 28 cM in an African sibling sample of the Hypertension Genetic Epidemiology Network (HyperGEN) (63), and 1.6 for LDL-C in French Canadian families (55). In the association analysis of these three linkage peaks, the most significant associations were found between LDL-C/HDL-C and three SNPs (rs7179520 and rs4932506 near hypothetical gene LOC44173 on 15q, and rs6037842 near RPL7AL2 on 20p), with a multiple testing adjusted P < 0.05. In addition, seven SNPs in or near six genes (TLE3, FAH, ARNT2, TMC3, TSHZ3, and ZNF343) showed an association with a lipid trait with a nominal P < 0.0001. TLE3 may interact with SIRT1 to suppress the transcriptional activity of nuclear factor-kB and lead to anti-inflammatory activity (64). FAH deficiency is associated with Type 1 hereditary tyrosinemia. While the function of other associated genes in these regions is largely unknown, they may also be candidates for further investigation, given the consistent linkage evidence from independent studies as well as only a small proportion of variance of lipid traits explained by the identified genes.
We also found an association with nominal P < 0.01 between lipid traits and six SNPs in CMIP, one of the 95 most significant lipid-associated loci reported by Teslovich et al. (18) in the Dominican subset or the whole NOMAS subcohort. The strongest of the six significant associations was found in the Dominican subset between HDL-C and rs12934986, which is 18 kb from the lead SNP rs2925979 observed by Teslovich et al. and replicated in Europeans, Africans, and Asians. As Dominicans have both African and European ancestry, CMIP may be a candidate for further investigation.
There are several strengths and weaknesses in this study. The strengths include 1) well-characterized and extended Caribbean Hispanic families with relatively large family size, which minimizes genetic heterogeneity, 2) adjustment for lifestyle factors that were seldom controlled for in previous studies, and 3) follow-up fine mapping in an independent community-based (primarily Caribbean Hispanic) cohort with genomic control. The weaknesses include 1) lack of more accurate lipid phenotypes, such as LDL size and apolipoprotein measures, 2) small sample size of the Dominican dataset in the association analysis, and 3) our findings in this specific population may not be directly generalized to other populations.
In conclusion, we report linkage and association evidence for the presence of genetic variations on 7p, 15q, 16q, 19q, and 20p that influence the blood lipids in the Caribbean Hispanic population. The CDH13 gene may be of particular interest, given its biological function and association with CVD and other metabolic syndrome-related components. These findings will guide further in-depth investigations of the functional variants of the genes in these regions.
Supplementary Material
Acknowledgments
The authors thank study participants for their collaboration and all staff of the Northern Manhattan Study and Family Study for their energetic efforts in this study.
Footnotes
Abbreviations:
- BMI
- body mass index
- CVD
- cardiovascular disease
- HDL-C
- high density lipoprotein cholesterol
- LDL-C
- low density lipoprotein cholesterol
- LDL-C/HDL-C
- LDL-C-to-HDL-C ratio
- IL
- interleukin
- LD
- linkage disequilibrium
- LOD
- logarithm of odds
- MAF
- minor allele frequency
- NOMAS
- Northern Manhattan Study
- PCA
- principal component analysis
- SNP
- single nucleotide polymorphism
- TC
- total cholesterol
- TG
- triglyceride
- WHR
- waist-to-hip ratio
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Husten L. 1998. Global epidemic of cardiovascular disease predicted. Lancet. 352: 1530. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D., Adams R. J., Brown T. M., Carnethon M., Dai S., De Simone G., Ferguson T. B., Ford E., Furie K., Gillespie C., et al. 2010. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 121: e46–e215. [DOI] [PubMed] [Google Scholar]
- 3.Murray C. J., Lopez A. D. 1997. Global mortality, disability, and the contribution of risk factors: Global Burden of Disease Study. Lancet. 349: 1436–1442. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). 2007. Prevalence of stroke--United States, 2005. MMWR Morb. Mortal. Wkly. Rep. 56: 469–474. [PubMed] [Google Scholar]
- 5.Bielinski S. J., Tang W., Pankow J. S., Miller M. B., Mosley T. H., Boerwinkle E., Olshen R. A., Curb J. D., Jaquish C. E., Rao D. C., et al. 2006. Genome-wide linkage scans for loci affecting total cholesterol, HDL-C, and triglycerides: the Family Blood Pressure Program. Hum. Genet. 120: 371–380. [DOI] [PubMed] [Google Scholar]
- 6.Clarke R., Emberson J. R., Parish S., Palmer A., Shipley M., Linksted P., Sherliker P., Clark S., Armitage J., Fletcher A., et al. 2007. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch. Intern. Med. 167: 1373–1378. [DOI] [PubMed] [Google Scholar]
- 7.Gotto A. M., Jr, Brinton E. A. 2004. Assessing low levels of high-density lipoprotein cholesterol as a risk factor in coronary heart disease: a working group report and update. J. Am. Coll. Cardiol. 43: 717–724. [DOI] [PubMed] [Google Scholar]
- 8.Kuulasmaa K., Tunstall-Pedoe H., Dobson A., Fortmann S., Sans S., Tolonen H., Evans A., Ferrario M., Tuomilehto J. 2000. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 355: 675–687. [DOI] [PubMed] [Google Scholar]
- 9.Law M. R., Wald N. J., Rudnicka A. R. 2003. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ. 326: 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewington S., Whitlock G., Clarke R., Sherliker P., Emberson J., Halsey J., Qizilbash N., Peto R., Collins R. 2007. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 370: 1829–1839. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd-Jones D. M., Leip E. P., Larson M. G., D'Agostino R. B., Beiser A., Wilson P. W., Wolf P. A., Levy D. 2006. Prediction of lifetime risk for cardiovascular disease by risk factor burden at 50 years of age. Circulation. 113: 791–798. [DOI] [PubMed] [Google Scholar]
- 12.Pencina M. J., D'Agostino R. B., Sr, Larson M. G., Massaro J. M., Vasan R. S. 2009. Predicting the 30-year risk of cardiovascular disease: the Framingham Heart Study. Circulation. 119: 3078–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adeyemo A. A., Johnson T., Acheampong J., Oli J., Okafor G., Amoah A., Owusu S., Agyenim-Boateng K., Eghan B. A., Jr, Abbiyesuku F., et al. 2005. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis. 181: 389–397. [DOI] [PubMed] [Google Scholar]
- 14.Elbein S. C., Hasstedt S. J. 2002. Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes. 51: 528–535. [DOI] [PubMed] [Google Scholar]
- 15.Hasstedt S. J., Hanis C. L., Elbein S. C. 2010. Univariate and bivariate linkage analysis identifies pleiotropic loci underlying lipid levels and type 2 diabetes risk. Ann. Hum. Genet. 74: 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malhotra A., Wolford J. K. 2005. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 54: 3007–3014. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y., Wyszynski D. F., Waterworth D. M., Wilton S. D., Barter P. J., Kesaniemi Y. A., Mahley R. W., McPherson R., Waeber G., Bersot T. P., et al. 2005. Multiple QTLs influencing triglyceride and HDL and total cholesterol levels identified in families with atherogenic dyslipidemia. J. Lipid Res. 46: 2202–2213. [DOI] [PubMed] [Google Scholar]
- 18.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuller L. H. 2004. Ethnic differences in atherosclerosis, cardiovascular disease and lipid metabolism. Curr. Opin. Lipidol. 15: 109–113. [DOI] [PubMed] [Google Scholar]
- 20.Musunuru K., Lettre G., Young T., Farlow D. N., Pirruccello J. P., Ejebe K. G., Keating B. J., Yang Q., Chen M. H., Lapchyk N., et al. 2010. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ. Cardiovasc. Genet. 3: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler C. A., Nelson G. W., Smith M. W. 2010. Admixture mapping comes of age. Annu. Rev. Genomics Hum. Genet. 11: 65–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacco R. L., Sabala E. A., Rundek T., Juo S. H., Huang J. S., DiTullio M., Homma S., Almonte K., Lithgow C. G., Boden-Albala B. 2007. Design of a family study among high-risk Caribbean Hispanics: the Northern Manhattan Family Study. Ethn. Dis. 17: 351–357. [PMC free article] [PubMed] [Google Scholar]
- 23.Sacco R. L., Khatri M., Rundek T., Xu Q., Gardener H., Boden-Albala B., Di Tullio M. R., Homma S., Elkind M. S., Paik M. C. 2009. Improving global vascular risk prediction with behavioral and anthropometric factors. The multiethnic NOMAS (Northern Manhattan Cohort Study). J. Am. Coll. Cardiol. 54: 2303–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacco R. L., Blanton S. H., Slifer S., Beecham A., Glover K., Gardener H., Wang L., Sabala E., Juo S. H., Rundek T. 2009. Heritability and linkage analysis for carotid intima-media thickness: the family study of stroke risk and carotid atherosclerosis. Stroke. 40: 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedewald W. T., Levy R. I., Fredrickson D. S. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18: 499–502. [PubMed] [Google Scholar]
- 26.Sacco R. L., Gan R., Boden-Albala B., Lin I. F., Kargman D. E., Hauser W. A., Shea S., Paik M. C. 1998. Leisure-time physical activity and ischemic stroke risk: the Northern Manhattan Stroke Study. Stroke. 29: 380–387. [DOI] [PubMed] [Google Scholar]
- 27.Sun L., Wilder K., McPeek M. S. 2002. Enhanced pedigree error detection. Hum. Hered. 54: 99–110. [DOI] [PubMed] [Google Scholar]
- 28.O'Connell J. R., Weeks D. E. 1998. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almasy L., Blangero J. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amos C. I., Krushkal J., Thiel T. J., Young A., Zhu D. K., Boerwinkle E., de Andrade M. 1997. Comparison of model-free linkage mapping strategies for the study of a complex trait. Genet. Epidemiol. 14: 743–748. [DOI] [PubMed] [Google Scholar]
- 33.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. 2002. The structure of haplotype blocks in the human genome. Science. 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 34.Nicodemus K. K., Liu W., Chase G. A., Tsai Y. Y., Fallin M. D. 2005. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 6 (Suppl. 1): S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander E., Kruglyak L. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- 36.Middelberg R. P., Martin N. G., Montgomery G. W., Whitfield J. B. 2006. Genome-wide linkage scan for loci influencing plasma triglycerides. Clin. Chim. Acta. 374: 87–92. [DOI] [PubMed] [Google Scholar]
- 37.Malhotra A., Elbein S. C., Ng M. C., Duggirala R., Arya R., Imperatore G., Adeyemo A., Pollin T. I., Hsueh W. C., Chan J. C., et al. 2007. Meta-analysis of genome-wide linkage studies of quantitative lipid traits in families ascertained for type 2 diabetes. Diabetes. 56: 890–896. [DOI] [PubMed] [Google Scholar]
- 38.Horne B. D., Malhotra A., Camp N. J. 2003. Comparison of linkage analysis methods for genome-wide scanning of extended pedigrees, with application to the TG/HDL-C ratio in the Framingham Heart Study. BMC Genet. 4 (Suppl. 1): S93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duggirala R., Blangero J., Almasy L., Dyer T. D., Williams K. L., Leach R. J., O'Connell P., Stern M. P. 2000. A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am. J. Hum. Genet. 66: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang H., Li Z., Xue L., Jiang X., Liu F. 2009. SUMF2 interacts with interleukin-13 and inhibits interleukin-13 secretion in bronchial smooth muscle cells. J. Cell. Biochem. 108: 1076–1083. [DOI] [PubMed] [Google Scholar]
- 41.Mahaney M. C., Almasy L., Rainwater D. L., VandeBerg J. L., Cole S. A., Hixson J. E., Blangero J., MacCluer J. W. 2003. A quantitative trait locus on chromosome 16q influences variation in plasma HDL-C levels in Mexican Americans. Arterioscler. Thromb. Vasc. Biol. 23: 339–345. [DOI] [PubMed] [Google Scholar]
- 42.Pajukanta P., Allayee H., Krass K. L., Kuraishy A., Soro A., Lilja H. E., Mar R., Taskinen M. R., Nuotio I., Laakso M., et al. 2003. Combined analysis of genome scans of Dutch and Finnish families reveals a susceptibility locus for high-density lipoprotein cholesterol on chromosome 16q. Am. J. Hum. Genet. 72: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W. D., Dong C., Li D., Garrigan C., Price R. A. 2005. A genome scan for serum triglyceride in obese nuclear families. J. Lipid Res. 46: 432–438. [DOI] [PubMed] [Google Scholar]
- 44.Dastani Z., Pajukanta P., Marcil M., Rudzicz N., Ruel I., Bailey S. D., Lee J. C., Lemire M., Faith J., Platko J., et al. 2010. Fine mapping and association studies of a high-density lipoprotein cholesterol linkage region on chromosome 16 in French-Canadian subjects. Eur. J. Hum. Genet. 18: 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov D., Philippova M., Antropova J., Gubaeva F., Iljinskaya O., Tararak E., Bochkov V., Erne P., Resink T., Tkachuk V. 2001. Expression of cell adhesion molecule T-cadherin in the human vasculature. Histochem. Cell Biol. 115: 231–242. [DOI] [PubMed] [Google Scholar]
- 46.Hug C., Wang J., Ahmad N. S., Bogan J. S., Tsao T. S., Lodish H. F. 2004. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. USA. 101: 10308–10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubina K., Talovskaya E., Cherenkov V., Ivanov D., Stambolsky D., Storozhevykh T., Pinelis V., Shevelev A., Parfyonova Y., Resink T., et al. 2005. LDL induces intracellular signalling and cell migration via atypical LDL-binding protein T-cadherin. Mol. Cell. Biochem. 273: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wellcome Trust Case Control Consortium (WTCCC). 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bressler J., Folsom A. R., Couper D. J., Volcik K. A., Boerwinkle E. 2010. Genetic variants identified in a European genome-wide association study that were found to predict incident coronary heart disease in the Atherosclerosis Risk in Communities Study. Am. J. Epidemiol. 171: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy D., Larson M. G., Benjamin E. J., Newton-Cheh C., Wang T. J., Hwang S. J., Vasan R. S., Mitchell G. F. 2007. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med. Genet. 8 (Suppl. 1): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling H., Waterworth D. M., Stirnadel H. A., Pollin T. I., Barter P. J., Kesaniemi Y. A., Mahley R. W., McPherson R., Waeber G., Bersot T. P., et al. 2009. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring). 17: 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J., Province M. A., Coon H., Hunt S. C., Eckfeldt J. H., Arnett D. K., Heiss G., Lewis C. E., Ellison R. C., Rao D. C., et al. 2007. An investigation of the effects of lipid-lowering medications: genome-wide linkage analysis of lipids in the HyperGEN study. BMC Genet. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aberg K., Dai F., Sun G., Keighley E., Indugula S. R., Bausserman L., Viali S., Tuitele J., Deka R., Weeks D. E., et al. 2008. A genome-wide linkage scan identifies multiple chromosomal regions influencing serum lipid levels in the population on the Samoan islands. J. Lipid Res. 49: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beekman M., Heijmans B. T., Martin N. G., Whitfield J. B., Pedersen N. L., DeFaire U., Snieder H., Lakenberg N., Suchiman H. E., de Knijff P., et al. 2003. Evidence for a QTL on chromosome 19 influencing LDL cholesterol levels in the general population. Eur. J. Hum. Genet. 11: 845–850. [DOI] [PubMed] [Google Scholar]
- 55.Bosse Y., Chagnon Y. C., Despres J. P., Rice T., Rao D. C., Bouchard C., Perusse L., Vohl M. C. 2004. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J. Lipid Res. 45: 419–426. [DOI] [PubMed] [Google Scholar]
- 56.Ober C., Abney M., McPeek M. S. 2001. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 69: 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rainwater D. L., Almasy L., Blangero J., Cole S. A., VandeBerg J. L., MacCluer J. W., Hixson J. E. 1999. A genome search identifies major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler. Thromb. Vasc. Biol. 19: 777–783. [DOI] [PubMed] [Google Scholar]
- 58.Almasy L., Hixson J. E., Rainwater D. L., Cole S., Williams J. T., Mahaney M. C., VandeBerg J. L., Stern M. P., MacCluer J. W., Blangero J. 1999. Human pedigree-based quantitative-trait-locus mapping: localization of two genes influencing HDL-cholesterol metabolism. Am. J. Hum. Genet. 64: 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arnett D. K., Miller M. B., Coon H., Ellison R. C., North K. E., Province M., Leppert M., Eckfeldt J. H. 2004. Genome-wide linkage analysis replicates susceptibility locus for fasting plasma triglycerides: NHLBI Family Heart Study. Hum. Genet. 115: 468–474. [DOI] [PubMed] [Google Scholar]
- 60.Austin M. A., Edwards K. L., Monks S. A., Koprowicz K. M., Brunzell J. D., Motulsky A. G., Mahaney M. C., Hixson J. E. 2003. Genome-wide scan for quantitative trait loci influencing LDL size and plasma triglyceride in familial hypertriglyceridemia. J. Lipid Res. 44: 2161–2168. [DOI] [PubMed] [Google Scholar]
- 61.Li X., Monda K. L., Goring H. H., Haack K., Cole S. A., Diego V. P., Almasy L., Laston S., Howard B. V., Shara N. M., et al. 2009. Genome-wide linkage scan for plasma high density lipoprotein cholesterol, apolipoprotein A-1 and triglyceride variation among American Indian populations: the Strong Heart Family Study. J. Med. Genet. 46: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang R., Li L., Seidelmann S. B., Shen G. Q., Sharma S., Rao S., Abdullah K. G., Mackinlay K. G., Elston R. C., Chen Q., et al. 2010. A genome-wide linkage scan identifies multiple quantitative trait loci for HDL-cholesterol levels in families with premature CAD and MI. J. Lipid Res. 51: 1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coon H., Leppert M. F., Eckfeldt J. H., Oberman A., Myers R. H., Peacock J. M., Province M. A., Hopkins P. N., Heiss G. 2001. Genome-wide linkage analysis of lipids in the Hypertension Genetic Epidemiology Network (HyperGEN) Blood Pressure Study. Arterioscler. Thromb. Vasc. Biol. 21: 1969–1976. [DOI] [PubMed] [Google Scholar]
- 64.Lavu S., Boss O., Elliott P. J., Lambert P. D. 2008. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat. Rev. Drug Discov. 7: 841–853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.