Abstract
Glucosylceramide synthase (GlcT-1) catalyzes the synthesis of glucosylceramide (GlcCer), the core structure of major glycosphingolipids (GSLs). Obesity is a metabolic disorder caused by an imbalance between energy uptake and expenditure, resulting in excess stored body fat. Recent studies have shown that GSL levels are increased in obese rodents and that pharmacologically reducing GSL levels by inhibiting GlcCer synthesis improves adipocyte function. However, the molecular mechanism underlying these processes is still not clearly understood. Using Drosophila as a model animal, we report that GlcT-1 expression in the fat body, which is equivalent to mammalian adipose tissue, regulates energy metabolism. Overexpression of GlcT-1 increases stored nutrition (triacylglycerol and carbohydrate) levels. Conversely, reduced expression of GlcT-1 in the fat body causes a reduction of fat storage. This regulation occurs, at least in part, through the activation of p38-ATF2 signaling. Furthermore, we found that GlcCer is the sole GSL of the fat body, indicating that regulation of GlcCer synthesis by GlcT-1 in the fat body is responsible for regulating energy homeostasis. Both GlcT-1 and p38-ATF2 signaling are evolutionarily conserved, leading us to propose an evolutionary perspective in which GlcT-1 appears to be one of the key factors that control fat metabolism.
Keywords: glycosphingolipid, ceramide, P38 mitogen-activated protein kinase, activating transcription factor 2, adipocyte
Glucosylceramide synthase (GlcT-1) catalyzes the formation of glucosylceramide (GlcCer), the core structure of major glycosphingolipids (GSLs), from ceramide and uridine diphosphate-glucose. We have previously cloned cDNAs of human GlcT-1 (hGlcT-1) (1, 2), mouse GlcT-1 (mGlcT-1) (1), Drosophila GlcT-1 (dGlcT-1) (3), and Caenorhabditis elegans GlcT-1 (CGT1∼3) (4, 5). To date, GlcT-1 from mammals, fish, fungi, plants, and bacteria have been either cloned or identified (3). The fact that GlcT-1 is evolutionarily conserved (e.g., dGlcT-1 shares 47% identity with hGlcT-1) and that GlcCer is distributed widely in eukaryotes suggests that GlcCer synthesis plays some basic roles in the cellular machinery.
GlcT-1 has been thought to have at least two distinct functions. First, GlcT-1 can act as a negative regulator of ceramide-mediated reactions by modifying ceramide with glucose. Increased basal levels of ceramide cause apoptosis (6). In Drosophila and mice, ceramide-mediated apoptosis could be regulated by upregulating or downregulating GlcT-1 activities (3, 7). Second, GlcT-1 can catalyze the synthesis of the precursor lipid GlcCer for most GSLs. GSLs are major membrane components in lipid microdomains or lipid rafts, which have been implicated in various important cellular processes, such as differentiation, adhesion, proliferation, and cell-cell interactions (8). Thus, GlcT-1 is expected to play significant roles in a variety of biological processes by regulating both intracellular ceramide levels and the overall synthesis of GSLs. However, the physiological functions of GlcT-1 in vivo are not yet fully understood.
Recent analysis of GSLs in obese rodents revealed that the concentration of GlcCer in plasma is higher in obese animals than in control wild-type animals, although ceramide concentrations are similar (9). Treatment of rodent obesity models with a GlcT-1 inhibitor reverses the insulin resistance syndrome (10, 11). Reducing GSLs on adipose tissue by means of a GlcT-1 inhibitor improves adipogenesis and adiponectin expression, implicating the role of GSLs in the pathogenesis of obesity (9). Although all of these observations point to the important role of GlcT-1 in fat metabolism in adipocytes, it is difficult to understand the effects of adipocyte GlcT-1, since GlcT-1 inhibitor was administered orally.
GlcT-1 is involved also in the production of most complex GSLs such as gangliosides. For example, ganglioside GM3 plays a role in the pathogenesis of type 2 diabetes. Obesity increases GM3 levels, resulting in the exclusion of insulin receptor from lipid rafts or caveolae but the retention of caveolin and flotillin (12). Membrane structural changes lead to the inhibition of insulin metabolic signaling (12). Thus, it is difficult to know whether the effect of GlcT-1 in fat metabolism is caused by GlcCer or by complex GSLs.
Drosophila shares most of the same basic metabolic functions found in vertebrates (13). For example, components of the insulin/insulin-like growth factor (IGF) pathway, which plays a central role in growth and metabolism, are conserved (14). Drosophila stores fat in a specialized tissue called the fat body. The fat body resembles the adipose tissue and liver of mammals, and metabolizes and stores nutrients primarily as triacylglycerol (TAG) and glycogen. Using Drosophila as an in vivo model system, we manipulated dGlcT-1 expression in the fat body and examined the resulting effects on fat metabolism. We found that the level of dGlcT-1 expression in the fat body regulates fat and sugar metabolism. Moreover, GlcCer was shown to be the dominant GSL in the fat body. GlcT-1 is highly conserved throughout evolution. Thus, our results suggest that GlcCer itself functions in fat metabolism and that manipulating GlcT-1 expression in mammalian adipose tissue may constitute a therapeutic way to counteract obesity.
EXPERIMENTAL PROCEDURES
Drosophila stocks
The Drosophila melanogaster strains used in this study were w1118 (wild-type), UAS-dGlcT-1, USA-dGlcT-1IR (VDRC Stock Center; Vienna, Austria); UAS-dATF2 (kind gift from Dr. S. Ishii); FB-GAL4 (kind gift from Dr. R. Kuhnlein); and Lsp2-GAL4 and hs-GAl4 (Bloomington Drosophila Stock Center; Bloomington, IN). FB-GAL4 and Lsp2-GAL4 were used to express transgenes in the fat body.
Fat body analysis by fluorescence microscopy
Larval fat bodies were dissected from third (L3) instar larvae and examined. Adult flies or dissected larvae were fixed in 4% paraformaldehyde for 25 min at room temperature and washed four times in PBS. To visualize lipid droplets, we stained the fixed fat bodies with BODIPY 493/503 (Molecular Probes; Carlsbad, CA) or Nile Red (Sigma; St. Louis, MO). BODIPY 493/503 was dissolved in ethanol at 1 mg/ml, and Nile Red was dissolved in acetone at 10 mg/ml. Then BODIPY 493/503 or Nile Red was diluted (BODIPY 493/503, 1:100,000; Nile Red, 1:2,500) in PBS and applied to tissue samples. After incubation for 1 h, tissues were washed three times with PBS. Mitotic clones of dGlcT-1 knockdown cells were generated by heat shock using the following genotypes: hs-flp UAS-GFP; tub-Gal4/UAS-dGlcT-1 IR; FRT82B/FRT82B Gal80 (15).
Nutrient measurements
Nutrient measurements were performed as described previously (16). Data are averages ± SEM from at least three independent experiments. P values presented are from Student's t-tests. To measure total body TAG, we extracted lipids from the sample (five flies or larvae) with 0.5 ml of chloroform-methanol (2:1, v/v). Aliquots of the extract (2 μl) were analyzed by TLC (Silica gel 60 plate; Merck, Darmstadt, Germany) and a solvent system consisting of chloroform-methanol-acetic acid (98:1:1, v/v/v). TAG was visualized with charring reagent (Ceric ammonium nitrate/H2SO4 solution), and TAG levels were estimated with an LAS 3000 (Fujifilm; Tokyo, Japan) set to digitized mode. TAG purified from Drosophila larvae was used as a TAG standard.
To measure the levels of carbohydrates (glucose, trehalose, and glycogen), 10 flies were homogenized in 100 μl of 0.1% Tween-20 on ice, heated at 70°C for 5 min to inactivate endogenous enzymes, and centrifuged. Five microliters of homogenate were used in each of the assays. Glucose (HK) assay kit (Sigma) and trehalase (Sigma) were used to determine glucose and trehalose levels. The Starch UV method (Roche) was used to determine glycogen levels.
Quantitative RT-PCR
Total RNAs were extracted from the fat bodies of L3 larvae and analyzed with SYBR Green (Power SYBR Green PCR Master Mix; Applied Biosystems) and an Applied Biosystems 7900HT fast real-time PCR System, as described previously (16). See supplementary experimental procedures online for primers used.
TLC analysis
Total GSLs were extracted from the fat bodies of L3 larvae and analyzed by TLC. Total lipid extracts corresponding to two larvae were applied on a TLC plate (Merck), and the plate was developed with a solvent system of chloroform-methanol-water (65:35:4, v/v/v). GSLs were visualized with orcinol/H2SO4 reagent.
MS
Total lipids were extracted from the fat bodies of 30 larvae, as described previously (13), and treated with 0.1 M KOH solution (chloroform-methanol [2:1, v/v]) to cleave potentially interfering glycerolipids. LC-MS was performed with a Nanofrontier LD system (Hitachi High Technologies; Tokyo, Japan). GSL was reconstituted in 40 μl of chloroform-methanol (9:1). One hundred nanoliters of sample solution was injected onto an Si column (Inertsil SIL-100A, 3 µm, 0.075 mm inner diameter and 150 mm length; GL Science, Tokyo, Japan) at 200 nL/min. Solvent A consisted of 5 mM ammonium formate (pH 7) in chloroform-methanol-2-propanol-water (80:12.5:7:0.5, v/v/v/v), and solvent B consisted of 5 mM ammonium formate (pH 7) in methanol-2-propanol-water (92.5:7:0.5, v/v/v). The sample was eluted through a 20 min gradient: 0–100% solvent B over 20 min, and maintained for 10 min. The instrument was run in the negative-ion mode and cycled through the full scan (m/z 200–2,000), followed by data-dependent MS/MS scanning.
RESULTS
dGlcT-1 controls stored nutrient levels
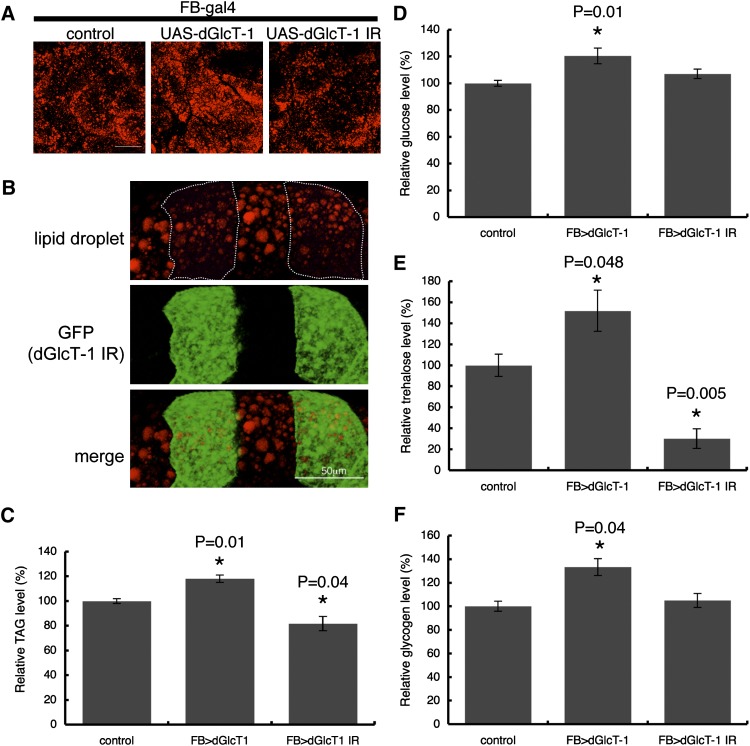
To characterize the function of dGlcT-1 in the fat body, we first manipulated dGlcT-1 expression in the fat body using a fat body-specific driver, FB-GAL4 (see supplementary Fig. I). This driver enabled us to generate dGlcT-1 transgenic flies that overexpress dGlcT-1 in the fat body and those that downregulate dGlcT-1 expression in the fat body (see supplementary Fig. II). Drosophila stores energy predominantly as triglycerides (TAGs) in the fat body. Thus, to evaluate intracellular TAG content, we stained the fat body with lipophilic dyes that detect lipid droplets. Lipid droplet formation was enhanced in fat bodies overexpressing dGlcT-1 (FB>dGlcT-1) compared with controls (Fig. 1A). By contrast, lipid droplet formation was reduced in fat bodies in which dGlcT-1 was knocked down (FB>dGlcT-1 IR) compared with controls (Fig. 1A).
Fig. 1.
Altered dGlcT-1 expression in the fat body results in impaired energy homeostasis. A: Staining of lipid droplets that store TAG in the fat bodies of adult flies. Scale bar, 50 μm. Genotypes were as follows: control (FB-Gal4/+; +/+), UAS-dGlcT-1 (FB-Gal4/+; UAS-dGlcT-1/+), UAS-dGlcT-1 IR (FB-Gal4/+; UAS-dGlcT-1 IR/+). B: Drosophila GlcT-1 (dGlcT-1) knockdown clones (GFP-positive) harbored fewer and smaller lipid droplets (red) than controls (GFP negative). The Flp/FRT system was used to make dGlcT-1 knockdown clones in the fat body. dGlcT-1 knockdown clones were marked with GFP (hs-flp, UAS-GFP; tub-Gal4/UAS-dGlcT-1 IR; FRT82B Gal80/FRT82B). Scale bar, 50 μm. C: Measurement of whole-body TAG levels of 6 day-old male flies; n = 5 for each genotype. D, E, F: Stored carbohydrate levels were measured: glucose (D), trehalose (E), and glycogen (F); n = 8 for each genotype. The error bars represent SD.
To determine whether the regulatory function of GlcT-1 in lipid metabolism is cell autonomous, we generated mitotic clones of dGlcT-1 knockdown cells and examined lipid droplet formation. We observed that the dGlcT-1 knockdown clones contained fewer and smaller lipid droplets than the neighboring control cells (Fig. 1B), confirming that dGlcT-1 affects lipid droplet formation in a cell-autonomous manner.
Fat body TAG levels were ∼18% higher in dGlcT-1-overexpressing flies and ∼19% lower in dGlcT-1 knockdown flies than in wild-type flies (Fig. 1C and supplementary Fig. III). We observed similar results when we manipulated dGlcT-1 expression using another fat body-specific driver, Lsp2-Gal4 (see supplementary Figs. I, IV).
Drosophila flies store nutrients not only in the form of lipids (TAG), but also in the form of carbohydrates (glucose, trehalose, and glycogen). Because carbohydrates are tightly regulated, we also measured stored carbohydrate levels in whole-fly homogenates. Flies overexpressing dGlcT-1 stored significantly more carbohydrates than did control flies (Fig. 1D–1F). However, dGlcT-1 knockdown flies displayed markedly decreased levels of trehalose compared with control flies, even though glucose and glycogen levels remained comparable. These observations suggest that dGlcT-1 expression in the fat body affects stored energy levels.
dGlcT-1 alters the expression of key energy metabolism genes
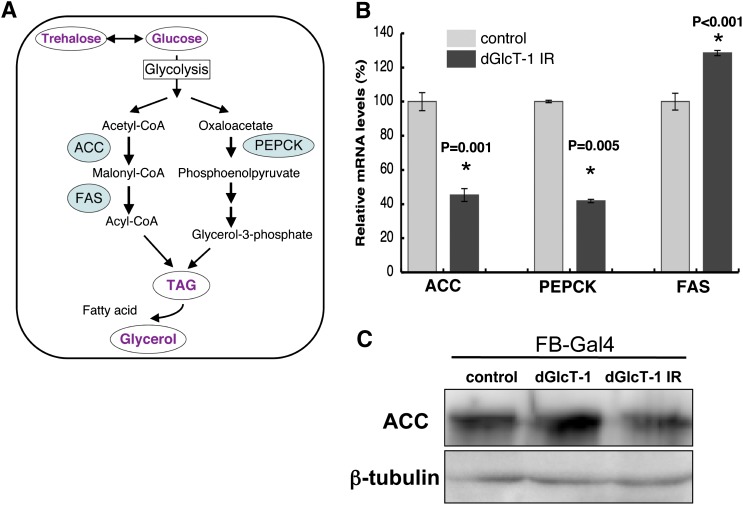
Because dGlcT-1 expression in the fat body affected stored energy levels, we assessed the expression of key genes involved in energy storage (Fig. 2A). Acetyl-CoA carboxylase (ACC) is one of the rate-limiting enzymes in FA synthesis, a source of TAG. We quantified the changes in ACC mRNA expression levels in the fat bodies of L3 larvae. We used larval fat bodies for the quantitative RT-PCR (qRT-PCR) assay because adult fat body cells are attached to the cuticle, and it is difficult to isolate the fat body without contamination from other tissues. We found that ACC mRNA expression was dramatically decreased in dGlcT-1 knockdown fat bodies (Fig. 2B). Western blot analysis using anti-ACC antibody revealed that ACC protein expression was also decreased in dGlcT-1 knockdown fat bodies (Fig. 2C).
Fig. 2.
ACC, FAS, and PEPCK mRNA expression is altered in dGlcT-1 knockdown fat bodies. A: Schematic diagram of the pathway and critical enzymes for TAG and glycerol biosynthesis. B: qRT-PCR was used to measure ACC, FAS, and PEPCK mRNAs involved in TAG and glycerol biosynthesis. FB-Gal4 was used to express UAS-dGlcT-1 IR specifically in third instar larval fat body; n = 3 for each genotype. C: Western blot analysis using anti-ACC indicated that changes in ACC mRNA expression are in good agreement with the levels of ACC enzyme protein. Proteins were collected from third instar larval fat body. The error bars represent SD.
We also examined other rate-limiting enzymes of FA synthesis and found that decreased phosphoenolpyruvate carboxykinase (PEPCK) mRNA expression increased fatty acid synthase (FAS) expression in dGlcT-1 knockdown fat bodies (Fig. 2B). Thus, decreased ACC and PEPCK expression may, in part, underlie the diminished energy storage phenotype of dGlcT-1 knockdown flies.
dGlcT-1 interacts with the p38-ATF2 signaling pathway
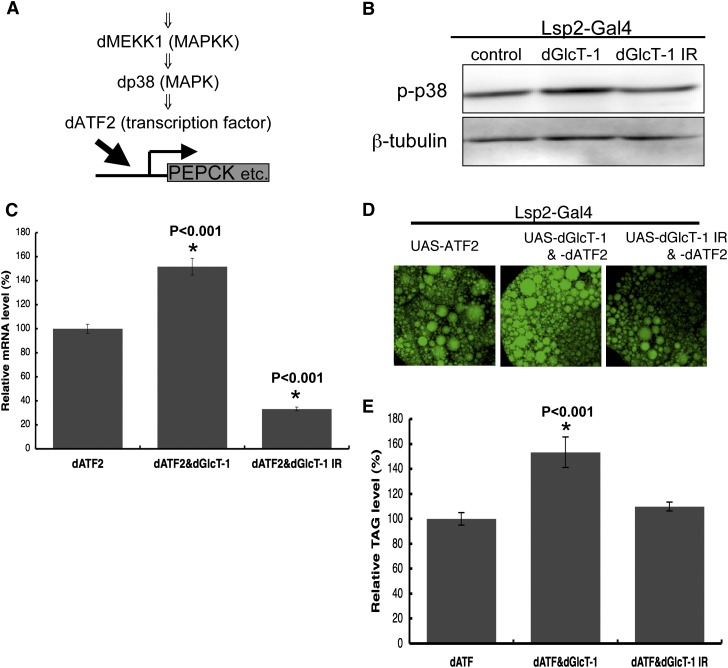
The Drosophila p38-ATF2 signaling pathway mediates fat metabolism through the activation of PEPCK transcription (Fig. 3B) (17). Increased FAS and decreased PEPCK mRNA expression phenotypes observed in dGlcT-1 knockdown flies were similar to that in dATF2 knockdown flies. Thus, GlcT-1 might regulate energy metabolism by regulating the p38-ATF2 signaling pathway. To examine this possibility, we first assessed the level of dp38 phosphorylation in the fat body. Phosphorylation of dp38 in the fat body increased when dGlcT-1 was overexpressed but decreased when GlcT-1 expression was reduced (Fig. 3A and supplementary Fig. VA). To confirm that dGlcT-1 does indeed regulate the p38-ATF2 signaling pathway, we also assessed the genetic interaction between dGlcT-1 and dATF2 in the wing, a site where the p38-ATF2 signaling pathway affects wing pattern formation. We found that dGlcT-1 did indeed regulate this signaling pathway such that dGlcT-1 overexpression or underexpression enhanced or suppressed, respectively, aberrant wing phenotypes (see supplementary Fig. VB). These results support the notion that dGlcT-1 activates the p38-ATF2 signaling pathway in Drosophila.
Fig. 3.
Genetic interaction between dGlcT-1 and the p38-ATF2 signaling pathway. A: Schematic of the p38-ATF2 signaling pathway. B: Lsp2-Gal4 was used to overexpress or suppress dGlcT-1 specifically in the fat body of larvae, and the level of dp38 phosphorylation was analyzed by Western blotting with phospho-p38 antibody. C–E: Effect of dGlcT-1 overexpression or knockdown on PEPCK mRNA (C) and on stored lipid levels (D, E) in dATF2-overexpressing fat bodies of larvae. As described above, Lsp2-Gal4 was used to express dATF2 specifically in the fat body. In C, PEPCK mRNA levels were measured by qRT-PCR; n = 3 for each genotype. In E, whole-body TAG levels of third (L3) instar larvae were measured; n = 5 for each genotype. The error bars represent SD.
Given that dGlcT-1 activates the p38-ATF2 signaling pathway in the fat body, it logically follows that dGlcT-1 expression may also affect ATF2 function in fat metabolism in the fat body. To test this possibility, we manipulated dGlcT-1 expression in dATF2-overexpressing fat bodies and assessed PEPCK mRNA levels by collecting mRNAs from fat bodies and performing qRT-PCR. As expected, we observed increased dPEPCK mRNA levels when dGlcT-1 and dATF2 were coexpressed but decreased dPEPCK mRNA levels when dGlcT-1 expression was knocked down (Fig. 3C).
Overexpression of dATF2 increases TAG levels, whereas dATF2 knockdown decreases TAG levels (17). Thus, we also examined whether dGlcT-1 expression affects dATF2-mediated effects on stored TAG levels by staining fat bodies with lipophilic dyes. Lipophilic dye signals were stronger when dGlcT-1 was overexpressed with dATF2 in the fat body than when only dATF2 was overexpressed in the fat body (Fig. 3D). Furthermore, whole-body TAG levels were ∼45% higher in flies that overexpressed dGlcT-1 in the fat body than in flies that overexpressed only dATF2 in the fat body (Fig. 3E). On the other hand, lipophilic dye signals were lower when dGlcT-1 expression was reduced (Fig. 3D), even though whole-body TAG levels were not affected (Fig. 3E). These results demonstrate that dGlcT-1 expression in the fat body regulates p38-ATF2 signaling activity.
GlcCer is a prominent GSL in the fat body
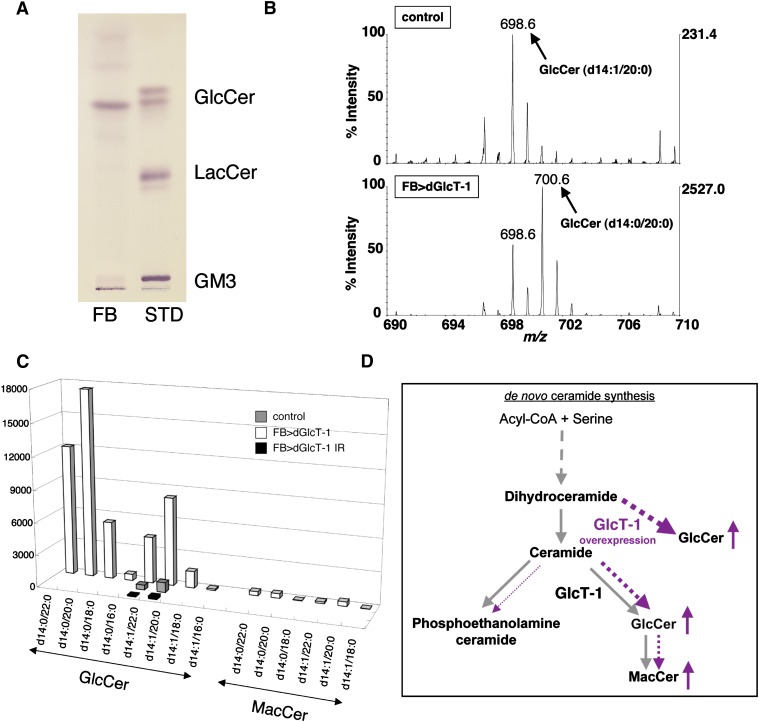
Although we demonstrated that dGlcT-1 expression levels affect stored nutrient levels, we needed to clarify whether this was due to the amount of GlcCer or due to more-glycosylated GSLs such as mactosylceramide (MacCer) (see supplementary Fig. VIA). Thus, we examined the GSL composition in the fat body by TLC and LC/MS of larvae fat bodies. We detected only GlcCer (Fig. 4A). In the wild-type fat body, however, GSLs such as MacCer or more-glycosylated GSLs were barely detectable. Consistent with this finding is that qRT-PCR detected very low levels of endogenous egghead (egh) mRNA expression in these fat bodies (see supplementary Fig. VIB). Egh protein catalyzes the transfer of mannose to GlcCer from GDP-mannose to produce MacCer. Furthermore, overexpression or underexpression of egh in the fat body did not affect stored TAG levels (see supplementary Fig. VIC). Therefore, these results strongly indicated that GlcCer itself is responsible for metabolic regulation in the fat body (see supplementary Fig. VII).
Fig. 4.
GlcCer is the sole GSL in the fat body. A: TLC analysis of GSLs from the fat body demonstrated that GlcCer is the sole GSL in the fat body. Total lipid extracts corresponding to two larvae were assayed (FB). Two micrograms of GlcCer, LacCer, and ganglioside GM3 were applied as standard GSLs (STD). B: LC/MS spectra from control (upper panel) and dGlcT-1-overexpressing fat bodies (FB>dGlcT-1; lower panel). Scans from m/z 690 to 710 are shown. Peaks of m/z 698.6 and 700.6 correspond to GlcCer having d14:1/20:0 and d14:0/20:0 conformations, respectively. C: Total GlcCer and MacCer levels in lipid extracts from fat bodies of control, FB>dGlcT-1, and FB>dGlcT-1 IR larvae are shown. D: Model showing the effect of overexpressing dGlcT-1 in the fat body. When dGlcT-1 is overexpressed in the fat body, dGlcT-1 adds glucose not only to regular ceramide but also to dihydroceramide (dotted line).
The fat body synthesizes GlcCer having d14:1/20:0 and d14:1/22:0 (Fig. 4C, D). This is consistent with previous studies showing that sphingosine (d14:1)-containing ceramide represents the most-abundant ceramide species in Drosophila (18, 19). In the present study, however, dGlcT-1 overexpression caused additional ceramide species [dihydroceramide (GlcCer having d14:0/20:0 and d14:0/22:0)] to be used (Fig. 4C, D). By contrast, phosphoethanolamine ceramide, another ceramide metabolite generated in the Golgi complex (20), contained no dihydroceramide, even in GlcT-1-overexpressing fat bodies (data not shown).
DISCUSSION
In this paper, we describe how GlcT-1 functions in energy storage. First, our data showed that GlcT-1 in the fat body regulates energy storage. Overexpression of dGlcT-1 in the fat body increased the levels of stored lipid (TAG) and carbohydrates (glucose, trehalose, and glycogen). On the other hand, dGlcT-1 knockdown flies displayed markedly decreased levels of TAG and trehalose.
Drosophila stores lipid (TAG) as a source of energy, mainly in the fat body, and reserves this energy until it is required for behavioral activities such as flight. In addition to TAG, trehalose released from the fat body is a major source of energy in the hemolymph and thorax muscles and is consumed during flight. Thus, our results suggest that GlcT-1 in the fat body may be one of the key factors involved in energy homeostasis.
A recent study has shown that p38-ATF2 signaling leads to the accumulation of stored energy in Drosophila (17). In the present study, we showed that dGlcT-1 activates p38-ATF2 signaling. Supporting our results is the finding that GlcT-1 activates p38 in mammalian cultured cells (21). Furthermore, dp38 mutant flies are more sensitive to starvation stress than are wild-type flies, which is indicative of dp38 involvement in energy metabolism (22). However, how GlcT-1 regulates dp38 is as yet unclear. We lack empirical evidence for a candidate molecule that interacts with newly synthesized GlcCer. Thus, further analysis is required to elucidate the upstream signaling mechanism that regulates the p38-ATF2 pathway.
Drosophila GlcT-1 generates GlcCer, a precursor glycolipid of most GSLs, which are important components of lipid rafts. Lipid rafts have been suggested to be involved in a variety of dynamic membrane processes such as signal transduction, membrane trafficking, and stress sensing (23). Thus, GlcT-1 may affect not only p38-ATF2 signaling but also other signaling pathways that regulate energy metabolism. This may account for the differences in the phenotypes between GlcT-1- and ATF2-transgenic flies. Indeed, regulation of body trehalose levels varies across different transgenic flies (14). Knockdown of dATF2 did not change trehalose levels, but knockdown of dGlcT-1 did. Furthermore, we observed increased FAS mRNA levels in dGlcT-1 knockdown flies, even though TAG levels decreased. FAS gene transcription is under tight nutritional and hormonal control. Insulin, sterol regulatory element binding protein, and upstream stimulatory factor have been reported to be regulatory elements of FAS transcription (24). Further studies are necessary to elucidate the specific contribution of fat body dGlcT-1 in energy homeostasis.
We showed that GlcCer is a prominent GSL in the fat body. Indeed, MacCer or more-glycosylated GSLs are barely detectable in the fat body. Altering the expression of egh, which catalyzes the reaction that produces MacCer from GlcCer, did not affect stored TAG levels. Thus, it seems likely that GlcCer expression in the fat body is sufficient to regulate stored energy levels.
We observed that dGlcT-1 overexpression caused the formation of an additional ceramide species, dihydroceramide (GlcCer having d14:0/20:0 and d14:0/22:0). Why was such an unusual GlcCer formed? A plausible explanation for this phenomenon is that excess dGlcT-1 protein localizes to the endoplasmic reticulum (ER) when dGlcT-1 is overexpressed. Under normal conditions, dihydroceramide is an intermediate lipid that is quickly converted to ceramide by desaturase, an enzyme found in ER membranes. Possibly, dGlcT-1 overexpressed in the ER utilizes dihydroceramide as substrate. We have shown previously that dGlcT-1 is expressed not only in Golgi membranes but also in the ER in photoreceptor cells (3).
Finally, it has been reported that the effects of schlank, a Drosophila ceramide synthase, on TAG metabolism are independent of its synthetic function (25). Therefore, it is possible that GlcT-1 regulates TAG metabolism not through GlcCer synthetic activity but through another unidentified function. However, because treatment with GlcT-1-specific inhibitors that block GlcCer synthesis improves the obesity phenotype (9), GlcT-1 may indeed affect TAG metabolism mainly through GlcCer synthetic activity.
Recent drug and in vitro studies have suggested that suppression of GlcT-1 function may present a novel strategy for treating diseases such as obesity (9–11). In Drosophila and mammals, GlcT-1 function and p38-ATF2 signaling are conserved. Thus, we believe that our in vivo data will be useful for future studies examining the detailed molecular mechanisms underlying the GlcT-1-mediated regulation of energy metabolism.
Supplementary Material
Acknowledgments
The authors thank Y. Shinoda, E. Oshima, and T. Shimizu for their technical assistance.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- ER
- endoplasmic reticulum
- GlcCer
- glucosylceramide
- GlcT-1
- glucosylceramide synthase
- GSL
- glycosphingolipid
- MacCer
- mactosylceramide
- PEPCK
- phosphoenolpyruvate carboxykinase
- TAG
- triacylglycerol
This work was partially supported by Core Research for Evolutional Science and Technology (CREST) of the Japan Science Technology Agency (JST) from 2005 to 2010.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures.
REFERENCES
- 1.Ichikawa S., Ozawa K., Hirabayashi Y. 1998. Molecular cloning and expression of mouse ceramide glucosyltransferase. Biochem. Mol. Biol. Int. 44: 1193–1202. [DOI] [PubMed] [Google Scholar]
- 2.Ichikawa S., Sakiyama H., Suzuki G., Hidari K. I., Hirabayashi Y. 1996. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. USA. 93: 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohyama-Koganeya A., Sasamura T., Oshima E., Suzuki E., Nishihara S., Ueda R., Hirabayashi Y. 2004. Drosophila glucosylceramide synthase: a negative regulator of cell death mediated by proapoptotic factors. J. Biol. Chem. 279: 35995–36002. [DOI] [PubMed] [Google Scholar]
- 4.Nomura K. H., Murata D., Hayashi Y., Dejima K., Mizuguchi S., Kage-Nakadai E., Gengyo-Ando K., Mitani S., Hirabayashi Y., Ito M., et al. 2011. Ceramide glucosyltransferase of the nematode Caenorhabditis elegans is involved in oocyte formation and in early embryonic cell division. Glycobiology. 116: 834–848. [DOI] [PubMed] [Google Scholar]
- 5.Ichikawa S., Hirabayashi Y. 1998. Glucosylceramide synthase and glycosphingolipid synthesis. Trends Cell Biol. 8: 198–202. [DOI] [PubMed] [Google Scholar]
- 6.Kolesnick R., Hannun Y. A. 1999. Ceramide and apoptosis. Trends Biochem. Sci. 24: 224–225. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita T., Wada R., Proia R. L. 2002. Early developmental expression of the gene encoding glucosylceramide synthase, the enzyme controlling the first committed step of glycosphingolipid synthesis. Biochim. Biophys. Acta. 1573: 236–240. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe S., Endo S., Oshima E., Hoshi T., Higashi H., Yamada K., Tohyama K., Yamashita T., Hirabayashi Y. 2010. Glycosphingolipid synthesis in cerebellar Purkinje neurons: roles in myelin formation and axonal homeostasis. Glia. 58: 1197–1207. [DOI] [PubMed] [Google Scholar]
- 9.Van Eijk M., Aten J., Bijl N., Ottenhoff R., van Roomen C. P., Dubbelhuis P. F., Seeman I., Ghauharali-van der Vlugt K., Overkleeft H. S., Arbeeny C., et al. 2009. Reducing glycosphingolipid content in adipose tissue of obese mice restores insulin sensitivity, adipogenesis and reduces inflammation. PLoS ONE. 4: e4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aerts J. M., Ottenhoff R., Powlson A. S., Grefhorst A., van Eijk M., Dubbelhuis P. F., Aten J., Kuipers F., Serlie M. J., Wennekes T., et al. 2007. Pharmacological inhibition of glucosylceramide synthase enhances insulin sensitivity. Diabetes. 56: 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., Przybylska M., Wu I. H., Zhang J., Maniatis P., Pacheco J., Piepenhagen P., Copeland D., Arbeeny C., Shayman J. A., et al. 2009. Inhibiting glycosphingolipid synthesis ameliorates hepatic steatosis in obese mice. Hepatology. 50: 85–93. [DOI] [PubMed] [Google Scholar]
- 12.Kabayama K., Sato T., Kitamura F., Uemura S., Kang B. W., Igarashi Y., Inokuchi J. 2005. TNFalpha-induced insulin resistance in adipocytes as a membrane microdomain disorder: involvement of ganglioside GM3. Glycobiology. 15: 21–29. [DOI] [PubMed] [Google Scholar]
- 13.Baker K. D., Thummel C. S. 2007. Diabetic larvae and obese flies: emerging studies of metabolism in Drosophila. Cell Metab. 6: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porte D., Jr, Baskin D. G., Schwartz M. W. 2005. Insulin signaling in the central nervous system: a critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes. 54: 1264–1276. [DOI] [PubMed] [Google Scholar]
- 15.Xu T., Rubin G. M. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- 16.Kohyama-Koganeya A., Kim Y. J., Miura M., Hirabayashi Y. 2008. A Drosophila orphan G protein-coupled receptor BOSS functions as a glucose-responding receptor: loss of boss causes abnormal energy metabolism. Proc. Natl. Acad. Sci. USA. 105: 15328–15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura T., Shimizu H., Nagao T., Ueda R., Ishii S. 2007. ATF-2 regulates fat metabolism in Drosophila. Mol. Biol. Cell. 18: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fyrst H., Herr D. R., Harris G. L. 2004. Characterization of free endogenous C14 and C16 sphingoid bases from Drosophila melanogaster. J. Lipid Res. 45: 54–62. [DOI] [PubMed] [Google Scholar]
- 19.Acharya U., Acharya J. K. 2005. Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell. Mol. Life Sci. 62: 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Jr, Acharya U., Acharya J. K. 2007. Ceramide transfer protein function is essential for normal oxidative stress response and lifespan. Proc. Natl. Acad. Sci. USA. 104: 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitatani K., Sheldon K., Anelli V., Jenkins R. W., Sun Y., Grabowski G. A., Obeid L. M., Hannun Y. A. 2009. Acid beta-glucosidase 1 counteracts p38delta-dependent induction of interleukin-6: possible role for ceramide as an anti-inflammatory lipid. J. Biol. Chem. 284: 12979–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craig C. R., Fink J. L., Yagi Y., Ip Y. T., Cagan R. L. 2004. A Drosophila p38 orthologue is required for environmental stress responses. EMBO Rep. 5: 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Roy C., Wrana J. L. 2005. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6: 112–126. [DOI] [PubMed] [Google Scholar]
- 24.Griffin M. J., Wong R. H., Pandya N., Sul H. S. 2007. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J. Biol. Chem. 282: 5453–5467. [DOI] [PubMed] [Google Scholar]
- 25.Bauer R., Voelzmann A., Breiden B., Schepers U., Farwanah H., Hahn I., Eckardt F., Sandhoff K., Hoch M. 2009. Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 28: 3706–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.