Abstract
Abstract Niemann-Pick disease, type C (NP-C), often associated with Niemann-Pick disease, type C1 (NPC1) mutations, is a cholesterol-storage disorder characterized by cellular lipid accumulation, neurodegeneration, and reduced steroid production. To study NPC1 function in vivo, we cloned zebrafish npc1 and analyzed its gene expression and activity by reducing Npc1 protein with morpholino (MO)-oligonucleotides. Filipin staining in npc1-morphant cells was punctate, suggesting abnormal accumulation of cholesterol. Developmentally, reducing Npc1 did not disrupt early cell fate or survival; however, early morphogenetic movements were delayed, and the actin cytoskeleton network was abnormal. MO-induced defects were rescued with ectopic expression of mouse NPC1, demonstrating functional gene conservation, and by treatments with steroids pregnenolone or dexamethasone, suggesting that reduced steroidogenesis contributed to abnormal cell movements. Cell death was found in anterior tissues of npc1 morphants at later stages, consistent with findings in mammals. Collectively, these studies show that npc1 is required early for proper cell movement and cholesterol localization and later for cell survival.
Keywords: Niemann-Pick, cholesterol, steroid, epiboly, gastrulation
Niemann-Pick disease, type C (NP-C) is a rare, autosomal-recessive disorder marked by progressive neurodegeneration and lysosomal storage deficits with an estimated incidence of between 1:120,000 and 1:150,000 live births (1, 2). NP-C symptoms typically appear in childhood, although infant onsets are possible. By the time the disorder's symptoms manifest, significant cellular damage has already occurred. At the cellular level, NP-C is characterized by an aberrant accumulation of unesterified cholesterol and glycolipids in the late endosomes and lysosomes of the endocytotic pathway (3–6). The disease is caused by mutations in one of two genes, Niemann-Pick disease, type C1 (NPC1), which accounts for 95% of cases, or Niemann-Pick disease, type C2 (NPC2), which accounts for 4% of cases. The remaining 1% of cases involves patients with confirmed biochemical defects without an accompanying identified mutation (7–10).
The function of NPC1 involves regulating the movement of LDL-derived cholesterol and plasma-derived glycolipids from the endocytotic pathway to the endoplasmic reticulum/trans-Golgi network (ER/TGN) and the mitochondrial membrane (11–15). Despite reductions in sterol exit from the endocytotic pathway, NPC1-deficient cells display normal sterol uptake, proper sterol delivery to the endocytotic organelles, and unperturbed levels of cholesterol ester hydrolysis (3, 16, 17). Excessive sterol content in the liver and spleen likely accounts for reports of hepato-splenomegaly, whereas sterol accumulation in the brain may contribute to the neurological deterioration in NP-C patients (16, 18–20).
Recent studies have established that NP-C pathology is enhanced by a physiological deficiency state (21–23), as cholesterol sequestration in late endosomes and lysosomes consequently limits substrates for steroid biosynthesis (23). Cholesterol-derived steroid hormones include mineralocorticoids, glucocorticoids, sex hormones, and neurosteroids that collectively control important physiological functions, such as electrolyte balance, glucose homeostasis, and sexual development (24, 25). Normal steroid hormone metabolism is disrupted in flies and mice where Npc1 and Npc2 have been genetically inactivated (21–23, 26). Steroid hormone replacement therapy may be an option for alleviating NP-C patient symptoms as this method of treatment has proven to be successful in studies involving Npc1-deficient mice. Injection of allopregnanolone perenterally to Npc1-deficient mouse embryos results in delayed onset of symptoms, prolonged life span, and improved neurological function (21, 27), possibly by quenching elevated levels of reactive oxygen species in the brain (28). Moreover, chronic treatment with the sex steroid estradiol in Npc1-deficient mice improves defective pituitary development and is capable of reversing ovarian defects and infertility in Npc1-deficient females (29, 30). These studies suggest that further insight into the pathophysiology of NP-C may be gained by having a better understanding of cholesterol metabolism and steroid hormone action during development and adulthood.
The Npc1 gene has been found to be strongly conserved in many experimental model organisms (31), playing a critical role in some aspect of sterol homeostasis in each species examined to date. Structurally, Npc1 is a 13 transmembrane-spanning protein (32), containing a sterol-sensing domain (SSD) composed of five transmembrane helices. The SSD is found in a number of different proteins, some of which are also involved in binding cholesterol and in sterol metabolism [e.g., sterol regulatory element-binding protein cleavage-activating protein (SCAP)], and others of which are involved in moving and binding to the cholesterol-modified protein Sonic hedgehog (e.g., Dispatched and Patched) (33). Despite this, the mechanistic role of this family of proteins in vertebrate development has not been widely examined. Our interest in the contribution of Npc1 to vertebrate development stems from our overall interest in the role of SSD-containing proteins during development (34). To this point, most studies concerning Npc1 function have focused on either the cell biology of sterol movements within a cell or the neuropathological outcomes that result from disruption of this process. Less is known about the contribution of this protein to vertebrate development.
Because it is a vertebrate animal and genetic morphants can be produced in large numbers, the zebrafish embryo is an ideal organism to examine the role of Npc1 in development. Additionally, zebrafish closely resemble mammals in their development and in cholesterol metabolism (reviewed in Ref. 35), making them highly amenable for studying proteins involved in lipid and sterol metabolism. In zebrafish development, early studies have revealed a requirement for cholesterol in promoting cell migration during epiboly, one of the earliest morphogenetic movements of gastrulation that ultimately generate the embryo's complex body plan (36, 37). The morphogenetic process of epiboly involves coordinated movements of each of the embryonic cell layers that are present during late blastula (1): the deep cell layer, which gives rise to the embryo proper (2); the enveloping layer (EVL), an extra-embryonic superficial epithelial layer covering the deep cells; and (3) the yolk syncytial layer (YSL), an extra-embryonic cytoplasmic cell layer within the yolk cell (38, 39). Epiboly commences when the yolk cell bulges toward the animal pole and the deep cell blastomeres radially intercalate. This process continues with the thinning and vegetal migration of the blastoderm over the yolk cell until 50% of the yolk surface is covered (50% epiboly) (40–42), at which time the deep cells begin the second phase of gastrulation involving dorsal convergence and involution movements which form the germ cell layers. Concomitant with this, each of the three cell layers continues to spread over the yolk in the epiboly process until the yolk cell is completely covered and internalized (41–44). Reducing the levels of cholesterol metabolites in zebrafish embryos results in an epiboly-delay phenotype (36).
In this study we have identified and cloned the zebrafish npc1 gene and found that it is widely present during early embryonic development. Using targeted morpholino (MO) antisense oligonucleotides, we have demonstrated that loss of npc1 leads to sterol localization defects in early embryos, similar to defects observed in fly and mammalian cells lacking Npc1. Our gene knockdown studies further revealed that npc1 is required for normal epiboly movement. Epiboly defects may be in part due to abnormal cytoskeletal structures, as we observed disruptions in the actin cytoskeleton in npc1morphants. Unlike some zebrafish mutants and morphants that have epiboly delay, Npc1-morphant zebrafish did not show a reduction in EVL integrity (45, 46) or any decline to the ability of the leading cells to undergo necessary shape changes during epiboly (47). The phenotype cannot be attributed to cell death during early developmental stages as we did not detect any increases in apoptosis in these morphant embryos. The epiboly defect in morphants was rescued by the co-injection of mouse Npc1 mRNA at the 1-cell stage or into the yolk cell of a 1,000-cell stage embryo, showing conservation of function between the fish and mammalian orthologs. Moreover, two downstream components of steroid synthesis, including the cholesterol derivative pregnenolone (P5) and glucocorticoid dexamethasone (Dex), could partially rescue the epiboly defects, demonstrating that such deficits are likely due, at least in part, to a steroid deficiency. Embryos that make it through epiboly continue to show deficiencies in gastrulation movements and also demonstrate cell death in the developing central nervous system. In full, this study indicates that zebrafish possess a functional ortholog of the mammalian Npc1 gene, which functions in sterol trafficking and is required for early epiboly movements in developing embryos. Furthermore, the morphant studies reported herein demonstrate that the zebrafish is an excellent vertebrate model system to further study the in vivo mechanisms related to altered cholesterol trafficking and steroid biosynthesis, as well as a potential model for future therapeutic and genetic interaction studies for Npc1.
EXPERIMENTAL PROCEDURES
Animals
Zebrafish (Danio rerio) embryos were obtained from natural crosses and staged as previously described (48). The Tübingen strain of fish was originally obtained from the International Zebrafish Resource Center (Eugene, OR), and Ekkwill wild-type fish were obtained from EkkWill Waterlife Resources (Gibsonton, FL). Zebrafish were maintained according to IACUC regulations and standard practices (49).
Cloning and RT-PCR
To obtain the npc1 gene, data mining revealed a partial clone (NCBI Accession number BG799923). An oligonucleotide sequence recognizing this partial clone was designed and used for 5′ RACE and 3′ RACE (SMART Race kit, Clontech, Mountain View, CA). cDNA was synthesized (SuperScript III RT First-Strand Synthesis System, Invitrogen, Carlsbad, CA) using RNA isolated and purified (PerfectPure RNA Tissue Kit, 5 Prime Inc., Gaithersburg, MD) from 24 h post fertilization (hpf) Ekkwill zebrafish.
The npc1 nucleotide sequences were analyzed, and a putative open reading frame was designed using MacVector software that was blasted against the zebrafish genome at the Sanger Institute. Putative protein sequence was obtained using MacVector software. Phylogenetic software analysis was accessed at Expasy (www.expasy.ch/tools). GenBank accession numbers for Npc1 and Niemann-Pick 1, like 1 (Npc1L1) proteins used in comparative analyses were Homo sapiens (human) NPC1 (NP_000262), Mus musculus (mouse) Npc1 (NP_032746), Oryctolagus cuniculus (rabbit) NPC1 (NP_001075540), Sus scrofa (pig) NPC1 (NP_999487), Drosophila melanogaster (fly) Npc1 (AAf52874), Saccharomyces cerevisiae (yeast) NCR1 (Niemann-Pick related protein, NP_508771), human Npc1L1 (EAL23753), mouse Npc1L1 (NP_997125), and rabbit Npc1L1 (NP_001075697).
To determine npc1 gene expression, total RNA was extracted, and cDNA was synthesized as described above from approximately 30 embryos at developmental stages from 1 hpf to 7 days post fertilization (dpf). Oligonucleotides were designed to amplify a ∼300 basepair npc1 fragment. As a PCR loading control, gapdh gene specific primers were used. Additional control samples, without reverse transcriptase, were performed for each sample. Primers used to amplify npc1: Npc1_F: 5′ CAATGACAACTGCACCATCC 3′, Npc1_R: 5′ GTGTCATTCAGGCAGTTGGTGAC 3′. Primers used to amplify gapdh: Gapdh_F: 5′ GAAGGTGGCAAACTGGTCAT 3′ and Gapdh_R: 5′ TTGCACCACCCTTAATGTGA 3′.
Morpholino design and testing
Anti-sense MOs were developed (GeneTools LLC, Philomath, OR) to target the translational start site of Npc1 (MO1, TGTGGTTTCTCCCCAGCAGAAGCAT) or the splice site acceptor for Npc1 exon 13 (MO2, GAGTCCACCTGTACAACATTTACAG). MO1 can disrupt translation from both zygotic and maternal mature mRNAs; however, some maternal mRNAs may become translated prior to injection of MO1. MO2 disrupts an mRNA processing event, and thus, it can only inhibit the proper translation of zygotic transcripts. Two 5-basepair-mismatch morpholinos were also generated to serve as controls for MO1 and MO2. A p53 MO (GCGCCATTGCTTTGCAAGAATTG), reported to suppress apoptotic effects induced by MO, was also occasionally used (50). Co-injection of an Npc1 MO and p53 MO did not alter any of the phenotypes examined, suggesting that they were not due to off-target activation of p53 activity. MOs were diluted to working concentrations of 0.5-3 mg/ml in Danieau solution (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca (NO3)2, 5 mM HEPES, pH 7.6).
For detecting spliced products in MO2-injected embryos, total RNA extraction, cDNA synthesis, and RT-PCR were performed as described above. Oligonucleotides were designed to flank exon 13, which is removed due to a MO2-induced RNA splicing deficit. The forward primer is in exon 12, 5′ GGCATGGGAGAAAGAGTTTATTAGG 3′, and the reverse primer is in exon 14, 5′ CATGTTCCTCTCTTCCTTCTCTGAG 3′. The wild type product, which includes exon 13 (178 nucleotides), is 480 nucleotides in length. Mis-splicing produced by MO2 will result in a fragment that is 302 nucleotides in length, as well as the wild-type fragments. To determine the identity of npc1 fragments in MO2-injected fish, PCR was performed with the primers above, and products were separated on an agarose gel. The DNA bands were extracted and purified using the Qiaquick Gel Extraction kit (Qiagen, Valencia, CA), and DNA sequencing was performed (Genewiz, Inc., South Plainfield, NJ).
Capped mRNAs, of either mouse Npc1 or synthetic transpose (for control injections), were synthesized using the mMESSAGE mMACHINE kit (Ambion, Inc., Austin, TX) according to manufacturer's instructions. Nanoliter quantities of 100 pg/nl mRNA solutions, suspended in 0.2 M KCl, were pressure-injected at the 1-cell stage, alone or in conjunction with MO1, or at the 1,000-cell stage, depending on the experimental design. Diluted MOs were injected into the yolk of 1-cell stage embryos at a concentration of 2.5-7.5 ng per embryo or into the yolk syncytial layer of a 1,000-cell stage embryo at a concentration of 5.0 ng per embryo, along with 0.1% rhodamine dextran solution (Sigma-Aldrich, St. Louis, MO).
Whole mount in situ hybridization and embryo staining
Embryo fixation and whole-mount in situ hybridization was conducted essentially as previously described (51, 52). Antisense riboprobe for npc1 was made by PCR amplifying ∼1 kb fragment from 24 hpf Ekkwill cDNA using oligonucleotides that recognize the npc1 gene sequence. The oligonucleotides were as follows: npc1 forward: 5′ GCGATACACAACAGCTCAAC 3′, npc1 reverse: 5′ TTCCAACTGCAGATCCAGCAC 3′. The fragment was cloned into pGemTEasy (Promega, Madison, WI). To obtain sectioned tissues, npc1-stained embryos were cryo-protected, embedded in gelatin, and sectioned 10 µm on a Leica cryostat.
We constructed riboprobes from plasmids containing a selected region of ntl (53), fkd6 (54), papc (55), gsc (56), krt4 (57), krt18 (58), cldnE (59), krt8 (45, 46, 60, 61), and sox32 (62) zebrafish genes. Embryos were visualized on a Leica MZ16F microscope equipped with a Leica DFC 490 digital camera. Images were processed using Adobe Photoshop CS2.
Actin staining was performed essentially as previously described (63). Actin was visualized by incubating fixed embryos with Alexa Fluor 568 phalloidin (Invitrogen, Carlsbad, CA). Alexa Fluor 568 phalloidin was diluted to 1:1000 in 1% goat serum. Embryos were blocked in blocking buffer [1% goat serum, 10% BSA in PBT (phosphate buffered saline + 0.1% Tween)], then incubated in phalloidin at room temperature for 1-2 h, followed by multiple washes in PBT. Images were acquired using the Zeiss 510 META confocal laser scanning microscope.
Filipin complex from Streptomyces filipinensis (F9765; Sigma-Aldrich) was diluted to a stock concentration of 25 mg/ml in DMSO and stored at 4.0°C. The stock concentration was further diluted to 0.05 mg/ml in 1% goat serum (filipin staining solution). Embryos were fixed at 12 hpf in 4% paraformaldehyde for 1 h at room temperature, followed by multiple washes in PBT. After the last wash, PBT was removed and replaced with filipin staining solution. Fixed embryos were incubated in the dark, at room temperature, in the filipin staining solution for 2 h. After incubation, embryos were rinsed in PBT and gradually move into 80% glycerol/20% PBT. Embryos were visualized in PBT:glycerol by confocal microscopy with a Zeiss 510 META confocal laser scanning microscope.
DAPI staining to visualize cell nuclei was performed essentially as previously described (63). Fixed embryos were incubated in a DAPI solution (Sigma-Aldrich), which was diluted 1:50,000 in equal parts PBT and glycerol. Embryos were blocked in blocking buffer for 2 h at room temperature prior to incubation in the DAPI solution for 30 min, followed by multiple washes in PBT. Images were acquired using a Zeiss 700 META confocal laser scanning microscope.
Cell death was visualized in live embryos by incubation in 5 mg/ml Acridine Orange (Sigma-Aldrich) for 1 h at room temperature, followed by three rinses in PBS. Images were acquired using the Zeiss 700 META confocal laser scanning microscope.
Steroid treatments
Pregnenolone (P5) and Dexamethasone (Dex) (Sigma-Aldrich) were solubilized in DMSO to obtain stock solutions of 20 mM that were stored at −20°C. Stock solutions were further diluted at 1:500-1,000 in zebrafish embryo water to obtain a final working concentration of 20-40 μM. Manually dechorionated embryos were bathed in the P5 or Dex solutions from the 16-cell stage until the time they were visually analyzed or fixed (generally between 50-95% epiboly) in 4% paraformaldehyde, and then analyzed by in situ hybridization. Vehicle-only controls were included for each steroid treatment experiment by diluting DMSO alone at 1:500-1,000 in zebrafish embryo water.
Measurements of epiboly
Epiboly progression was measured as previously described (37). The percentage of epiboly was calculated by determining the distance between the animal pole to the blastoderm margin (which was marked using in situ hybridization staining with an ntl riboprobe) divided by the distance between the animal pole and the vegetal pole. Measurements were acquired using Adobe Photoshop software. Statistical significance between variant groups was determined using either a two-tailed Student's t-test or multivariate ANOVA followed by Tukey's posthoc analysis (Excel; PASW 18/SPSS).
RESULTS
Zebrafish Npc1 has a conserved protein identity and function in sterol trafficking
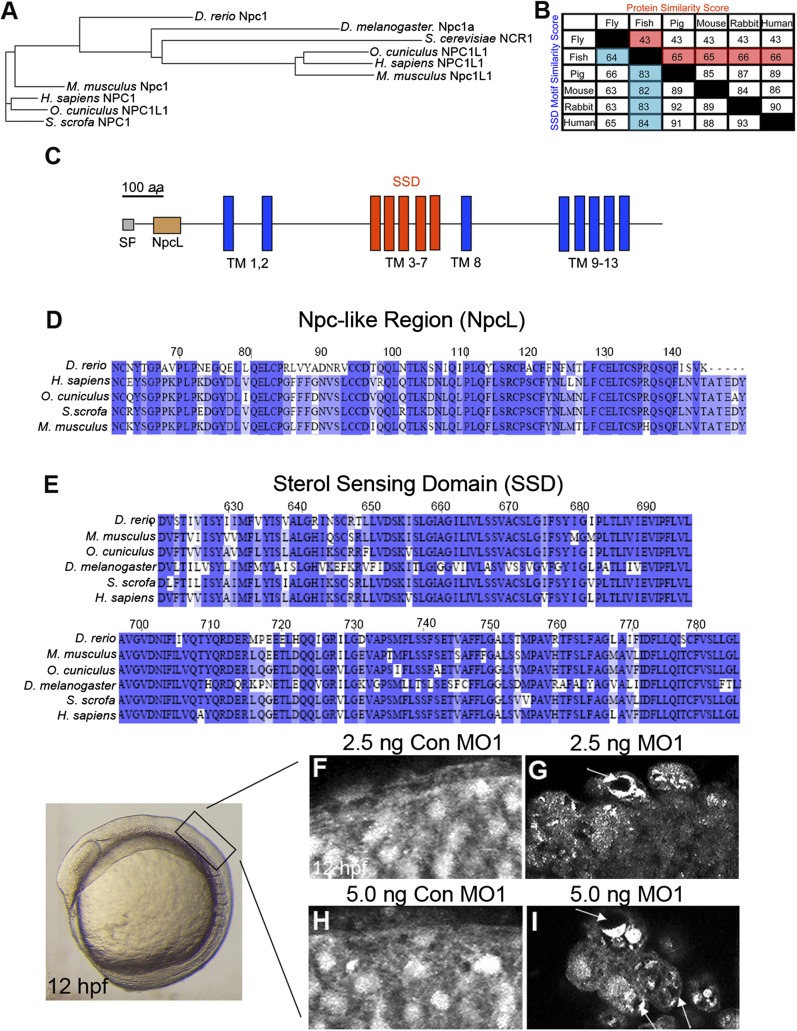
Zebrafish npc1 was identified by homology searches using human NPC1 to screen genomic and cDNA zebrafish databases, followed by 3′ and 5′ RACE PCRs to obtain the putative mRNA sequence [deposited in the National Center for Biotechnology Information (NCBI) as JF951427]. Zebrafish npc1 is predicted to encode a 1,278 amino acid protein with 60% identity and 66% similarity to human NPC1, and 59% identity and 65% similarity to mouse Npc1 (Fig. 1A, B). Peptide analysis software predicted that zebrafish Npc1 is a 13-transmembrane-spanning domain protein, harboring a putative leucine zipper motif with surrounding sequences that collectively form the so-called Npc-like region (NpcL) and the 5-transmembrane-spanning SSD motif (Fig. 1C). These protein characteristics have been described in all NPC1 orthologs studied to-date (64). Previous genetic analysis has shown that the NpcL region and the SSD motif are both necessary for protein function (65, 66). Protein sequence alignments of these two domains between zebrafish and other species revealed a high level of conservation (Fig. 1D, E), strongly implying that the ascribed role for human NPC1 in lipid homeostasis and trafficking may be conserved in the zebrafish Npc1 protein. In further support of this prediction, the presence of a dileucine target motif (LLSY) in the carboxy terminus of the zebrafish Npc1 protein (data not shown) is highly reminiscent of similar motifs identified in mammalian Npc1 that have been suggested to promote protein localization to the endosomal and lysosomal pathways (64, 67, 68).
Fig.1.
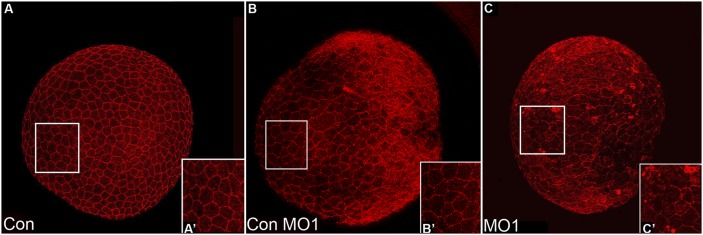
Zebrafish Npc1 protein and function is highly conserved. (A) Phylogenetic tree of D. rerio (zebrafish), D. melanogaster (fly), S. cerevisiae (yeast), H. sapiens (human), M. musculus (mouse), and S. scrofa (pig) NPC1 proteins, and O. cuniculus (rabbit), human, and mouse NPC1-like1 (Npc1L1) proteins. (B) Similarity score (the percentage of identical amino acids) for whole protein and the SSD motif among Npc1 amino acid sequences for a variety of species. Zebrafish column and row are highlighted. Common names are used for space considerations. (C) Zebrafish Npc1 protein contains a putative signal peptide (SP) at the N-terminus, 13 transmembrane (TM) spanning domains, a Npc-like region (NpcL), and a sterol sensing domain (SSD) motif. A line corresponding to the length of 100 amino acids is provided for reference. (D–E) Protein sequence alignments for the NpcL (D) and the SSD (E) are compared between zebrafish Npc1 and known NPC1 orthologs. Identical residues are shaded. The numbers above the aligned sequences in D and E correspond to the amino acid sequence of zebrafish Npc1. (F–I) Sterol localization defects in npc1 morphants, images taken from the region of a 12 hpf embryo detailed on the left. Lateral views, anterior to the left, of confocal stack projections obtained by imaging flat-mounted trunk tissue from 12 hpf embryos injected with 2.5-5.0 ng Con MO1 (F, H) or their stage-matched siblings injected with 2.5-5.0 ng MO1 (G, I). (F, H) In controls, filipin staining was diffuse and consistent throughout the field of cells. (G, I) In npc1 morphants, filipin staining appeared punctuate and uneven in many cells, often revealing darker areas within a single cell where the fluorescent signal was not equally dispersed (arrows).
To determine if zebrafish npc1 is required for sterol trafficking, we performed sterol localization studies. To do this, we employed a whole-embryo, filipin-staining protocol following the reduction of Npc1 protein levels by MO injection. Filipin stains free 3-β-hydroxysterols, which include unesterified cholesterol (69). Npc1 protein levels were reduced by injecting a translation-blocking morpholino (MO1) into one-cell-stage embryos. MO1-injected embryos exhibited early embryonic defects (discussed in detail below); however, these defects did not prohibit us from determining the distribution of endogenous cholesterol in npc1 morphants. We found that cells within the dorsal trunk of 12 hpf noninjected wild-type or control embryos injected with 2.5-5.0 ng/embryo of a 5-basepair-mismatch MO1 (Con MO1) at the one-cell stage displayed a diffuse staining pattern for filipin, revealing that sterols were evenly distributed throughout the cell (Fig. 1F, H). In contrast, one-cell-stage MO1 injections produced embryos that displayed uneven, patchy sterol distribution in cells (Fig. 1G, I). Fluorescent filipin staining in npc1 morphants was punctate within individual cells, suggesting that sterols were accumulating in subcellular locations. Moreover, while dark areas (free of fluorescent filipin signal) were not apparent in cells of control embryos (Fig. 1F, H), these areas were commonly seen in npc1-morphant cells, further reflecting the uneven distribution of sterols (Fig. 1G, I; arrows). The sterol localization deficit in npc1-morphant cells is similar to what has been shown in NPC1 mutant fly and mammalian cells stained with filipin (23, 70), suggesting that the zebrafish npc1 gene shares a conserved function in cholesterol trafficking.
Expression of zebrafish npc1
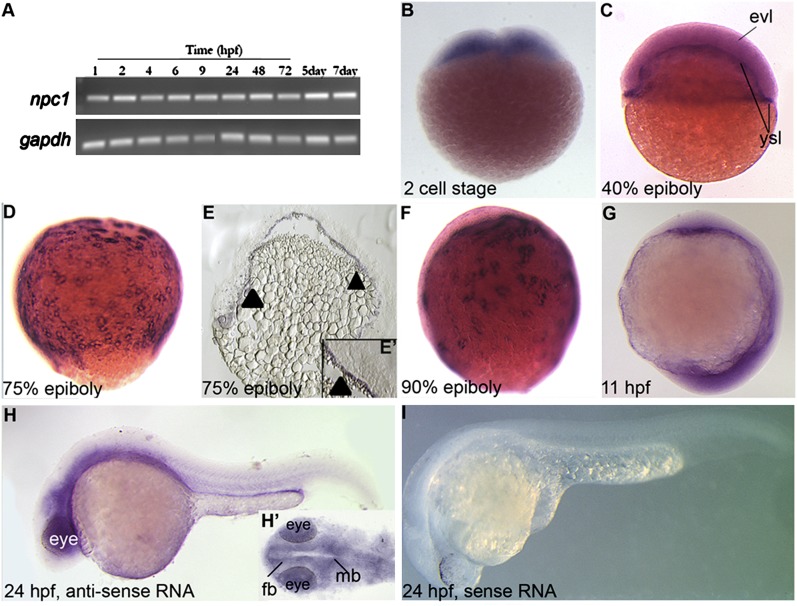
npc1 could be detected throughout embryogenesis and early larval development, from 1 hpf up to 7 dpf, by RT-PCR (Fig. 2A). No signal was seen in minus RT control samples (data not shown). npc1 antisense riboprobes were generated to examine the spatiotemporal expression of npc1 by in situ hybridization during embryogenesis. Consistent with RT-PCR data, we found that npc1 was present in the blastodiscs at the two-cell stage, ∼1 hpf (Fig. 2B), revealing that the earliest npc1 transcripts are maternally deposited. During epiboly stages, npc1 was ubiquitously expressed throughout the blastomeres extending from the animal to the vegetal pole (Fig. 2CndashF). Sectioning stained embryos revealed that npc1 was strongly expressed in the extra-embryonic YSL (Fig. 2E, E′; arrowheads). As embryos ended epiboly and began somitogenesis, npc1 expression was expressed ubiquitously at a low level, yet the expression levels were more intense in tissues bordering the yolk (Fig. 2G). npc1 expression became intense in anterior regions of the embryo proper by 24 hpf (Fig. 2H), wherein the staining was strongest in neural tissues (Fig. 2H′). Importantly, background signal with npc1 sense RNA probe was minimal at all stages tested, which included 24 hpf (Fig. 2I). This expression data revealed that npc1 is expressed dynamically throughout early development in embryonic and extra-embryonic tissues.
Fig.2.
Expression of zebrafish npc1 during embryogenesis. (A) RT-PCR analysis of zebrafish npc1 from embryos aged from 1 hpf to 7 dpf (RT samples showed no signal, not shown). (B–H) Lateral (B–I) or dorsal (H′) views of embryos stained with a RNA probe for npc1, (B) in blastodiscs at the 2 cell stage, (C–F) during epiboly stages in the blastomeres extending from the animal to vegetal poles and in the YSL. Expression in the YSL tissue was confirmed in embryo sections (arrowheads in E) and at higher magnifications of 75% epiboly sectioned embryo (E′). (G) During early somitogenesis npc1 was ubiquitously expressed at low levels. (H, H′) By 24 hpf, npc1 became mostly localized to the anterior tissues, with strong staining in the neural tissues, as visible in ventral views of stained heads (H′). (I) Embryos stained at 24 hpf with a npc1 sense RNA probe had no detectable signal. fb, forebrain; mb, midbrain.
Morpholino knockdown of npc1 causes epiboly delay
To examine the function of npc1 during embryonic development, we continued our gene knockdown studies using morpholino oligonucleotides. We injected two separate function-blocking MOs, one designed to interrupt protein translation (MO1) and the other to reduce Npc1 protein levels by altering RNA splicing, specifically to remove the SSD (MO2) (see Experimental Procedures for further details). Injection of either MO at the one- or two-cell stage of embryonic development led to a reduced progression of epiboly.
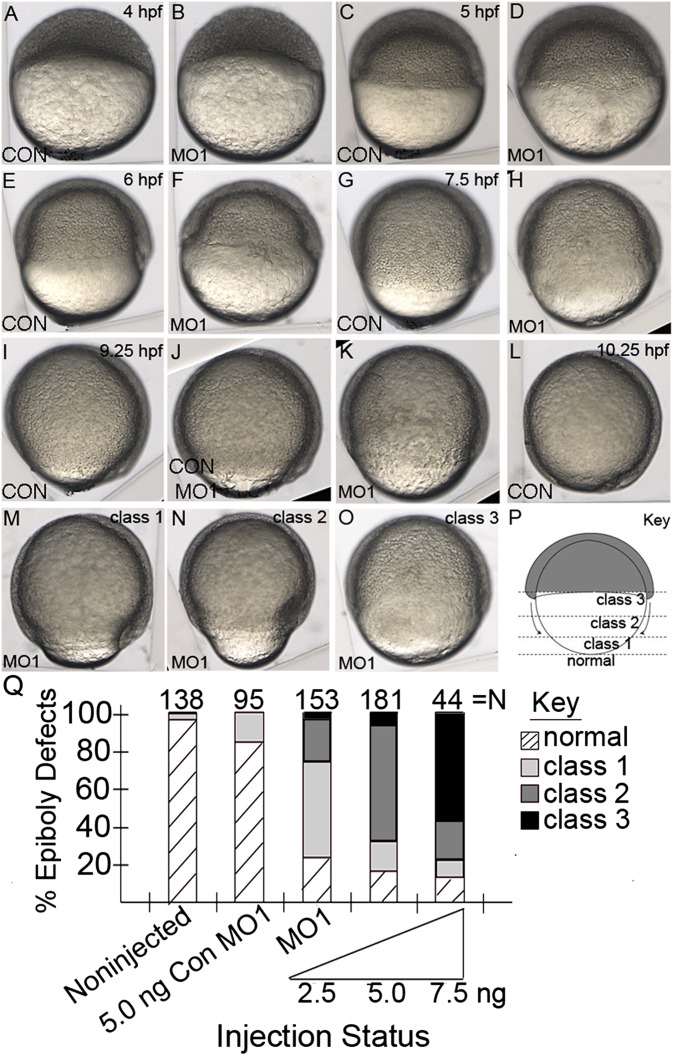
To identify when development delay first becomes apparent and to examine the subsequent epiboly delay outcomes in npc1 morphants, we carefully compared control embryos with morphants during epiboly stages (4-10 hpf). Early in development, MO1-injected embryos were phenotypically undetectable from control embryos at sphere stage (∼4 hpf, Fig. 3A, B) and 40% epiboly (∼5 hpf, Fig. 3C, D), suggesting that npc1 is dispensable for initiation of epiboly. However, by the time control embryos had reached shield stage (∼60% epiboly, 6 hpf), npc1-morphant embryos had only progressed to 45-50% epiboly (Fig. 3E, F), and when control embryos were at 75% epiboly, npc1 morphants lagged behind at 50-60% epiboly (Fig. 3G, H). Likewise, at 9.25 hpf, when controls (noninjected or Con MO1-injected embryos) had progressed to 95% epiboly, MO1-injected embryos lagged behind considerably in their epiboly progression (Fig. 3IndashK). Finally, at 10.25 hpf when the yolk was completely covered in control embryos, indicating the end of epiboly (Fig. 3L), npc1 morphants had not yet completed epiboly and were significantly delayed to varying degrees, depending on the dose of MO1 injected into the embryos (Fig. 3MndashO).
Fig.3.
Reduced npc1 expression results in epiboly delay. (A–O) Lateral views of live control or MO1-injected embryos 4-10.25 hpf. (A–D) Both control and time-matched MO1-injected embryos progressed to sphere stage (A, B) and 40% epiboly (C, D) at similar rates. (E, F) Control embryos were at shield stage at 6 hpf (E) and time-matched morphant embryo were at 45% epiboly (F). (G, H) Control embryos were at 75% epiboly at 7.5 hpf (G) while time-matched MO1-injected embryos were at approximately 50% epiboly (H). (I–K) Noninjected or Con MO1-injected controls were at 95% epiboly at 9.25 hpf (I, J), while MO1-injected embryos were at 60-75% epiboly (K). (L) Control embryos finished epiboly by 10.25 hpf (L), while time-matched MO1-injected embryos had delayed epiboly to varying degrees; class 1 epiboly-delay phenotype embryos were delayed at 90-95% epiboly (M), class 2 phenotypes were delayed to 70-89% epiboly (N), and class 3 phenotypes were delayed to 69% epiboly or earlier (O). (P) Schematic of embryo during epiboly showing blastoderm cells (gray) migrating downward over the vegetal yolk (white). Dotted lines designate different migration points of the blastoderm cells in morpholino-injected embryos when noninjected controls complete epiboly. (Q) The effect of injecting different concentrations of MO1 or Con MO1 on epiboly.
To describe the dose-dependent epiboly-delay effects in npc1 morphants, MO1- and Con MO1-injected embryos were collected, and their epiboly delay was scored at the point at which noninjected controls had completed epiboly (approximately 10.25 hpf). If injected embryos had completed epiboly at the same time as noninjected controls, they were scored as normal; however, if the embryo had delayed epiboly progression, it was characterized as a class 1, 2, or 3 depending on the severity (Fig. 3P). At higher doses (7.5 ng/embryo), well over half of the embryos (25/44, 57%) receiving MO1 exhibited a class 3 phenotype (Fig. 3O), meaning that embryos were still in the earliest stages of epiboly when control embryos had reached 100% epiboly. The majority of embryos receiving a moderate dose (5.0 ng/embryo) of MO1 exhibited class 2 phenotypes (109/180, 60%) (Fig. 3N). Embryos were assigned a class 2 phenotype if their epiboly progression lagged behind at 70-89% epiboly when control embryos had reached 100% epiboly. Finally, we found that low doses (2.5 ng/embryo) of MO1 was a sufficient amount of morpholino to delay epiboly in 79% of embryos (120/153), with the majority of embryos exhibiting a class 1 phenotype (78/153, 51%) (Fig. 3M), characterized by an embryo that had only progressed to 90-95% epiboly when wild-type embryos had reached 100% epiboly. In contrast to this, the vast majority of embryos injected with a moderate dose of Con MO1 displayed normal epiboly phenotypes, as did noninjected controls (77/95, 82% and 130/136, 96%, respectively), suggesting that MO injection itself does not lead to adverse epiboly effects (Fig. 3L). Those rare embryos in these two groups that showed an epiboly-delay phenotype could invariably be characterized as a class 1 phenotype (Fig. 3M). Overall, the severity of the epiboly-delay phenotype was proportional to the amount of MO1 injected (Fig. 3Q).
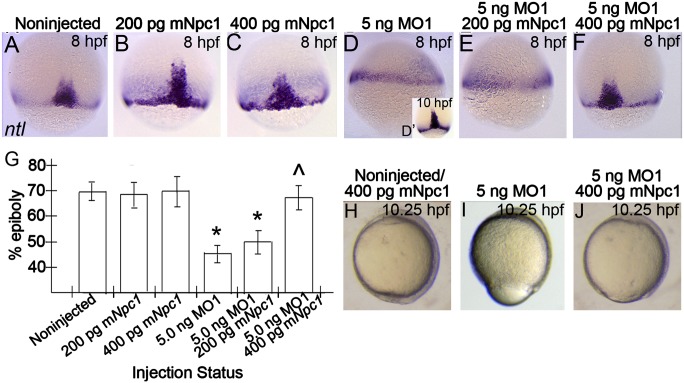
To confirm that MO1 was targeting Npc1 and to examine the conservation of action of Npc1 across species, RNA rescue experiments were performed using mouse Npc1 (mNpc1) RNA. mNpc1 mRNA cannot be recognized by MO1 due to sequence variation between the first exon of zebrafish and mouse Npc1 gene transcripts. Embryos injected with 200-400 pg mNpc1 mRNA alone were indistinguishable from controls throughout all stages of epiboly, indicating that overexpression of Npc1 had no apparent impact on epiboly progression (Fig. 4H). However, when 400 pg of mNpc1 mRNA was co-injected with 5 ng MO1, the negative effects of MO1 on epiboly progression were greatly reduced (Fig. 4I, J). To quantify the extent of epiboly rescue in npc1 morphants following mNpc1 mRNA injection, we stained embryos with no-tail (ntl), which is localized within the blastoderm margin. By marking the blastoderm margin we could measure the extent of blastoderm cell migration over the ventral yolk (see Experimental Procedures about how epiboly was measured in this study). To initiate the study, noninjected controls or sibling embryos that were injected with either MO1, mNpc1 mRNA, or both were fixed uniformly (time-matched) once the control siblings had progressed to ∼65-85% epiboly and the percentage of epiboly progression for each group was measured. We found that epiboly progression was not significantly altered between controls (epiboly progression of 69.1% ± 4.8%, n = 22) and their siblings injected with 200 pg of mNpc1 mRNA (epiboly progression of 67.1% ± 8.2%, n = 12) or 400 pg of mNpc1 mRNA (epiboly progression of 69.4% ± 4.5%, n = 10) (Fig. 4AndashC). In contrast, siblings receiving a moderate dose of MO1 had significantly delayed epiboly progression (epiboly progression of 47.5% ± 7.6, n = 19) compared with controls (Fig. 4D), which could be rescued upon co-injection with 400 pg mNpc1 (68.4% ± 7.4%, n = 11) (Fig. 4F), but not by 200 pg of mNpc1 (52.1% ± 7.2%, n = 15) (Fig. 4E). These results are summarized in Fig. 4G and represent the results of a single experiment, which was replicated twice (Table 1). Taken together, these studies strongly suggest that MO1 produces defects, specifically by reducing Npc1, and that zebrafish Npc1 and mouse Npc1 share functionality. Furthermore, they show that Npc1 functions to influence epiboly progression.
Fig.4.
Epiboly-delay phenotypes in npc1 morphants are rescued by co-injection with mouse Npc1 mRNA. (A–F) Lateral views, animal pole facing upward of fixed noninjected or injected embryos stained with ntl riboprobe at 8 hpf, or MO1-injected embryos at 10 hpf (D′). (A–D) Embryos injected with mNpc1 mRNA alone were indistinguishable from noninjected controls (A–C), while the blastoderm margin of time-matched embryos injected with MO1 had progressed at a lower rate by 8 hpf (D), but continued to progress and looked similar to 8 hpf noninjected controls by 10 hpf (D′). (E, F) Co-injection of 400 pg mNpc1, but not 200 pg mNpc1, rescued MO1-associated defects. (G) The effect of expressing mNpc1 RNA in wild-type and MO1-injected embryos. Graph represents the % of epiboly from averages obtained on analyzing the epiboly progression in each group from a single clutch of siblings. Error bars show standard deviation. Statistics derived from posthoc analysis after ANOVA. *P < 0.001 compared with control embryos; ^P < 0.001 compared with MO1 injection alone. (H–J) Lateral views, anterior structures facing upward of live controls or mRNA injected embryos (H), MO1-injected embryos (I), or embryos co-injected with RNA and MO1 (J) at 10.25 hpf.
TABLE 1.
Epiboly Percentage Following MO1 Injections and Mouse Npc1 (mNpc1) mRNA Injections
| Clutch Number | One-way ANOVA Results | Control | 200 pg mNpc1 | 400 pg mNpc1 | 5 ng MO1 | 5 ng MO1+ 200 pg mNpc1 | 5 ng MO1+ 400 pg mNpc1 |
|---|---|---|---|---|---|---|---|
| 1 | F (5, 88) = 26.8 P < 0.001 | 69.1 ± 4.8(N = 22) | 67.2 ± 8.2d(N = 12) | 69.5 ± 4.5d(N = 10) | 47.6 ± 7.6a(N = 19) | 52.1 ± 7.2a(N = 15) | 68.4 ± 7.4b,c(N = 11) |
| 2 | F (5, 79) = 10.2 P < 0.001 | 66.4 ± 6.0(N = 16) | 65.7 ± 6.1d(N = 16) | 65.9 ± 7.0d(N = 10) | 55.7 ± 5.0a(N = 7) | 56.5 ± 4.8a(N = 21) | 67.8 ± 6.4b,c(N = 10) |
| 3 | F (5, 88) = 304.5 P < 0.001 | 84.9 ± 3.3(N = 26) | 81.3 ± 4.7d(N = 12) | 80.0 ± 4.9d(N = 7) | 39.7 ± 5.6a(N = 15) | 42.5 ± 3.8a(N = 13) | 77.6 ± 6.0b,c(N = 15) |
Top number indicates the mean level of epiboly achieved for each group (see Results for technique) ± standard deviation. Each clutch demonstrated significance at the levels shown. Data from C1 are shown in graphical form in Fig. 4G. C, clutch; N, number of embryos analyzed per group.
Posthoc analysis using Tukey's HSD indicated that these groups had significantly reduced (P < 0.001 in all cases) epiboly progression compared with control embryos.
Posthoc analysis using Tukey's HSD indicated that these groups had no significant difference when compared with controls (C1: P = 1.00; C2: P = 0.993; C3: P = 0.309).
Posthoc analysis using Tukey's HSD indicated that these groups were significantly different to embryos injected with MO1 alone (C1: P < 0.001; C2: P = 0.002; C3: P < 0.001).
Posthoc analysis using Tukey's HSD indicated that these groups were not different from control embryos (P values between 0.116 and 1.0), indicating that injection alone did not result in a change in epiboly progression.
The SSD domain of Npc1 is required for protein function during epiboly
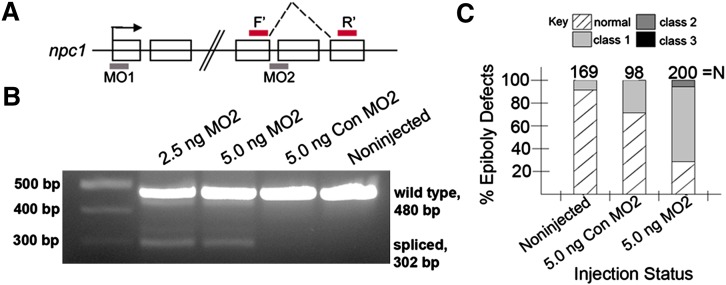
Since npc1 is expressed as both maternal and zygotic transcripts (see Fig. 2), both may be involved in epiboly. Zygotic RNA is subject to processing events, including RNA splicing, while maternally deposited RNA transcripts are already spliced, allowing exclusive reduction of zygotic npc1 expression by utilizing a morpholino designed to perturb npc1 RNA splicing (MO2). MO2 was designed to target the deletion of exon 13 (Fig. 5A), chosen because it contains SSD amino acids, a motif required for Npc1 function in higher vertebrates. While the exact function of the SSD motif in Npc1 is not fully understood, biochemical evidence has revealed that the domain is necessary for sterol trafficking, possibly by directly binding cholesterol (65, 66) Thus, this MO also tests the necessity of the SSD domain in zygotic transcripts of the Npc1 protein.
Fig.5.
Zygotic npc1 transcripts containing SSD amino acids are essential for normal epiboly. (A) Genomic structure of zebrafish npc1. Open boxes indicate coding regions of exons. Exons shown in structure include exons 1, 2, 12, 13, and 14. Arrow indicates translational start site in exon 1. Gray shaded regions below the boxes indicate the recognition site for morpholinos used in this study. Dotted lines indicate the consequence of exon disorganization due to redirected splicing upon utilization of MO2. Red lines above the boxes denoting exons 12 and 14 indicate site of PCR primers (B) Smaller DNA fragments, due to abnormal splicing, were only visible in lanes loaded with cDNA from embryos receiving MO2. (C) The effect on epiboly following MO2 and Con MO2 injections at the one cell stage.
To determine the extent of MO2 activity, we developed primers complimentary to sequences contained in exon 12 and exon 14 that amplify 480 bp of the npc1 gene. PCR with these primers revealed the presence of smaller (∼300 bp) fragments in MO2-injected embryos (Fig. 5B) resulting from abnormal slicing. DNA sequencing of the gel bands confirmed that the loss of exon 13 nucleotides (supplementary Fig. I), ∼180 bp in length, was the specific consequence of MO2. Perturbing normal RNA splicing could be achieved with a concentration of 2.5 ng MO2; increasing the concentration seemingly did not lead to an increased amount of mis-spliced product (Fig. 5B). Importantly, a 5-basepair-mismatch control morpholino for MO2 (Con MO2) had no effect on splicing when it was injected into fish at low or moderate concentrations (Fig. 5B).
In a manner similar to MO1, animals injected with 5.0 ng/embryo MO2 displayed a higher incidence of epiboly delay (142/200, 71% with epiboly-delay phenotypes), compared with sibling embryos injected with Con MO2 (28/98, 29% with epiboly-delay phenotypes) or noninjected controls (17/169, 10% with epiboly-delay phenotypes) (Fig. 5C). Epiboly delay in MO2-injected embryos was most commonly characterized as a class 1 phenotype (132/200, 66%), with a smaller percentage showing a class 2 phenotype (10/200, 5%) (refer to Fig. 3MndashO for representative images of class 1-3 phenotypes). Moreover, epiboly-delay phenotypes in control embryos were invariably class 1 (Figs. 3M and 5C). These data suggest that zygotic npc1 is essential for normal epiboly movements and points to an important role for the SSD, which is specifically disrupted by MO2, in protein function during epiboly.
The role of npc1 in the YSL
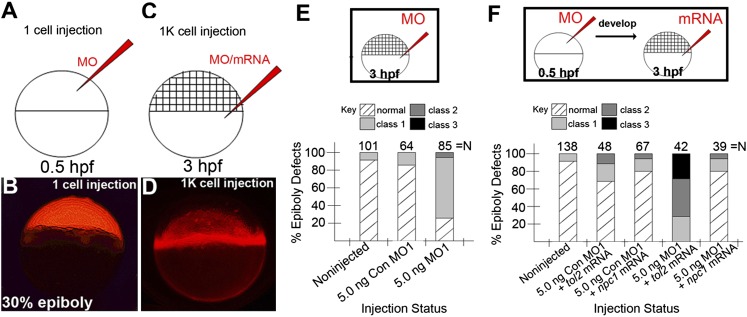
The YSL is one critical location of early npc1 expression (Fig. 2). It has long been recognized that the YSL is required for teleost epiboly movements (38), although the molecular mechanisms behind this requirement is poorly understood. To examine the functional contributions of YSL-localized npc1 to epiboly progression, we specifically reduced YSL-localized Npc1 by co-injecting 5.0 ng/embryo MO1 and 0.1% rhodamine dextran directly into the YSL at the 1,000-cell stage. When viewing these embryos at 30% epiboly, the distribution of the dextran solution was limited to the yolk and YSL, and we only selected those embryos with YSL-limited injections for our analysis (Fig. 6C, D). This is in contrast to all other injections in this study, which were performed at the 1- or 2-cell stage, which allows the injected material to be ubiquitously distributed in the embryo, including the YSL (Fig. 6A, B). When examined at 10.25 hpf, embryos receiving YSL-limited injections of MO1 were more often delayed (61/85, 72% with epiboly-delay phenotypes), compared with their siblings receiving a similar injection of Con MO1 (11/64, 18% with epiboly-delay phenotypes) or noninjected controls (5/101, 5% with epiboly-delay phenotypes) (Fig. 6E). Epiboly delays following YSL-specific Npc1 knockdown were most commonly characterized as class 1 phenotypes (57/85, 67%) (Fig. 3M), while the remaining epiboly-delay embryos showed class 2 phenotypes (4/85, 5%) (Figs. 3N and 6E). This result indicates that npc1 expressed in the YSL is critical for normal epiboly movements.
Fig.6.
Npc1 action in the YSL is critical for normal epiboly progression. (A, C) Schematic of injection strategies performed in this study. Injected material was either injected at the 1- or 2-cell stage (A) or at 3 hpf to target expression into the yolk and YSL nuclei (C). (B, D) Lateral views, animal pole facing upward displaying fluorescent signals at 30% epiboly in embryos co-injected with MO1 and rhodamine dextran at the 1-cell (B) or 1,000-cell stage (D). (E) The effect on epiboly following MO1 and Con MO1 injections into the YSL at the 1,000-cell stage. (F) The effect on epiboly following npc1 or tol2 mRNA injection into the YSL following Npc1 knockdown by MO1 injection at the 1- or 2-cell stage.
Since directed injection of MO into the yolk cell resulted in epiboly delay, we next sought to determine if epiboly delay in npc1 morphants injected with MO1 at the one-cell stage could be rescued by microinjection of mNpc1 mRNA into the YSL. Sibling embryos were injected with MO1 or Con MO1 at the one-cell stage and then allowed to develop normally until 3 hpf/1,000-cell stage, at which point mNpc1 mRNA or control mRNA encoding the Tol2 transposase was injected into the YSL (Fig. 6F). Control mRNA injections had little impact on epiboly progression; however, YSL-targeted mNpc1 mRNA microinjections were capable of rescuing a majority of the epiboly-delay phenotypes evident in embryos injected at the one-cell stage with MO1 (Fig. 6F). In contrast, MO1-injected embryos receiving control mRNA into the YSL showed high incidence of epiboly delay, consistent with the epiboly delay seen in nonrescued MO1-injected embryos (Fig. 6F, compare with Fig. 3Q). In full, these data strongly imply that the YSL is a critical site of action for Npc1 to promote normal epiboly progression during early embryonic development.
Deep cell fates form normally in npc1 morphants
As YSL-deposited genes are known to be involved in mesendoderm induction and reduced epiboly cell movements could negatively influence proper germ layer induction (71), we next assessed whether deep cell fates and mesendoderm induction was altered in npc1-morphant embryos. Since any changes to cell and germ layer induction would be apparent by 70-80% epiboly, we allowed npc1-morphant embryos to reach this developmental stage prior to fixation and analysis. At 75% epiboly, ntl normally marks the mesoderm and dorsal forerunner cells (Fig. 4A). We found that expression of ntl was identical between embryos injected with MO1 at the one-cell stage and their stage-matched, noninjected siblings (Fig. 4D, D′). Next, we found the expression of the mesendodermal marker gsc to be indistinguishable between MO1-injected embryos and their stage matched controls (supplementary Fig. IIA–C). Moreover, we found that expression of the early endodermal marker sox32/cas was indistinguishable between npc1 morphants and stage-matched controls (supplementary Fig. IID–F). These results strongly suggest that mesendoderm initiation and deep cell fates become specified normally in npc1 morphants. During epiboly, the cell layers have characteristic movement behaviors, and these movements can be affected differently by different mutations or disruptions. In some cases, the EVL integrity is altered, which contributes to abnormal epiboly phenotypes (45, 46, 60). To examine the EVL in npc1 morphants and controls, a number of EVL differentiation markers were used. We found that the expression of EVL markers krt4 and krt18 were well maintained in MO1-injected embryos (supplementary Fig. IIG–N), as were the markers cldnE and krt8 (data not shown). Moreover, YSL nuclei marked by dapi staining were associated appropriately with the epiboly leading edge (data not shown). These results suggest that, in the case of npc1 morphants, the EVL integrity is maintained and all cell layers show roughly equivalent epiboly delay.
Disruption of Npc1 function results in actin cytoskeleton disorganization
Epiboly defects are often associated with disruption to the actin cytoskeleton (47, 72), and examination of the actin cytoskeleton can reveal alterations in cell shape that contribute to epiboly delay (47). To further investigate the loss of npc1 and its possible effect on epiboly, we visualized the actin cytoskeleton with Alexa Fluor-labeled phalloidin in control embryos for comparison with embryos injected with a low dose (3 ng/embryo) of MO1. In initial experiments, we collected both control and morphant embryos when they had reached 90-100% epiboly. To do this, we allowed our morphant embryos to continue to develop after collection of the control embryos, making them stage-matched. Upon examination of these embryos, we found that the actin cytoskeleton was disorganized in the npc1-morphant embryos, with both actin clumping as well as localized loss of the actin microfilaments in other cells (Fig. 7C, C′; n = 32). In contrast, this phenotype was not seen with control MO injections (Fig. 7B, B′). Moreover, the leading edge of the actin, which forms a contractile actin ring, was occasionally absent in the morphant embryos, a phenotype that was never evident in control embryos (data not shown). During normal epiboly, the leading cells undergo a distinctive cell shape change that can be seen when looking at the actin cytoskeleton. It has been demonstrated that in some cases where epiboly is disrupted, this cell shape change fails to occur (47). When we examined MO-injected embryos at different stages of epiboly, there was no difference in the cell shape of the cells on the leading edge of epiboly (data not shown). Actin clumping and disorganization is often a byproduct of cell death (73, 74). Since increased cell death has been reported in many mammalian tissues in response to loss of npc1 function, we examined epiboly-staged embryos for evidence of cell death. When examining the deep cells (data not shown) and leading edge of epiboly-staged embryos, the number of cells with pyknotic nuclei was not significantly increased between npc1 morphants and controls (P < 0.05; supplementary Fig. III). By these data, we could conclude that reducing Npc1 in zebrafish embryos did not impact cell survival during epiboly, but it did lead to perturbations to the actin cytoskeleton network.
Fig.7.
Disruption of Npc1 function results in actin cytoskeleton disorganization. Lateral views of stage-matched control and MO-injected embryos. Actin filaments were labeled with Alexa Fluor 568 Phalloidin in noninjected (A) and Con MO-injected controls (B) and in embryos injected with a low dose (3.0 ng) of MO1 (C). MO1-morphant embryos revealed disorganization, loss of continuity, or clumping of actin microfilaments (C′) compared with control embryos (A′, B′).
Treating npc1 morphants with steroid hormones P5 and Dex partially rescues epiboly delay
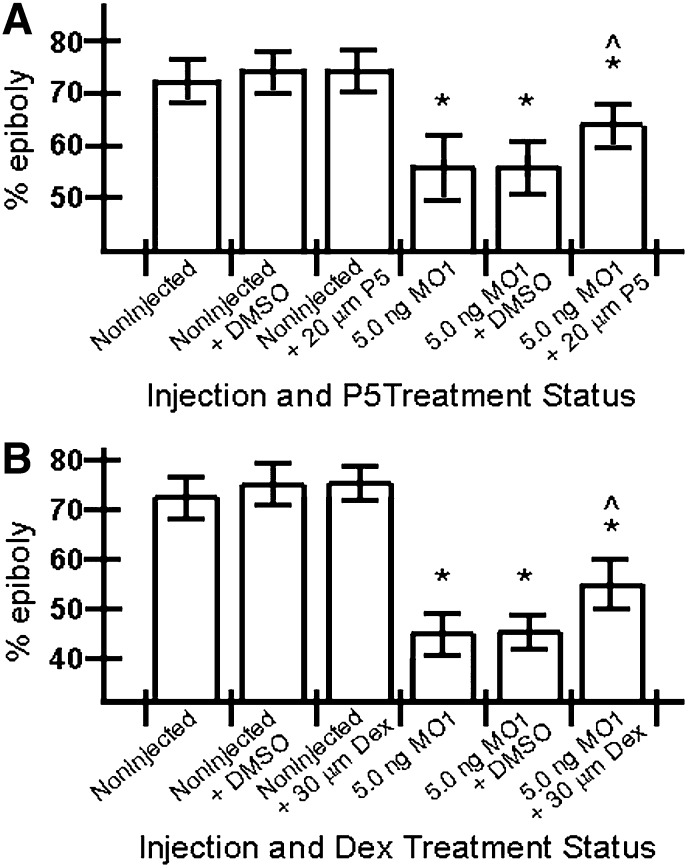
The synthesis of neurosteroids and steroid hormones is significantly reduced in vertebrate and invertebrate animals lacking a functional Npc1 (21, 23, 26), likely due to reduced amounts of cholesterol available for steroid metabolism (23). The precursor for all steroid hormones in the zebrafish is pregnenolone (P5), which is metabolized from cholesterol precursors by the mitochondrial biosynthesis enzyme cyp11a1 (25, 75). Knockdown of cyp11a by MO in the zebrafish results in an epiboly-delay defect that can be partially rescued by incubating cyp11a morphants in P5 at early developmental stages prior to the doming of the yolk (37). We reasoned that if epiboly delay in npc1 morphants is due to reduced steroid hormone production, then treating npc1 morphants with P5 would promote faster rates of epiboly. Indeed, we found that incubating npc1 morphants in 20 or 30 μM P5 solution, beginning at early cleavage stages (16-cell stage) and continuing throughout epiboly stages, partially rescued the epiboly-delay defect. Fig. 8A shows results from single experiment, and Table 2 summarizes data from three replicate experiments. Concurrently, incubations of control embryos with P5 alone under identical conditions had no impact on epiboly progression (Fig. 8A).
Fig.8.
P5 and Dex steroids partially rescue epiboly delay following npc1 gene knockdown. (A, B) The effects of P5 (A) and Dex (B) on epiboly progression in control and morphant embryos. Error bars show standard deviation. Statistics derived from posthoc analysis after ANOVA. *P < 0.001 compared with control embryos; ^P < 0.001 compared with MO1 alone.
TABLE 2.
Epiboly Percentage Following MO1 Injections and P5 treatments
| Clutch Number | One-way ANOVA Results | Control | Control + DMSO | Control + 20 µM P5 | 5 ng MO1 | 5 ng MO1 + DMSO | 5 ng MO1 + 20 µM P5 |
|---|---|---|---|---|---|---|---|
| 1 | F (5, 80) = 36.3 P < 0.001 | 86.6 ± 3.3(N = 11) | 85.9 ± 4.0(N = 11) | 86.21 ± 5.8(N = 8) | 70.7 ± 3.9a(N = 21) | 70.6 ± 5.8a(N = 12) | 77.3 ± 4.9ab(N = 19) |
| 2 | F (5, 124) = 53.6 P < 0.001 | 78.2 ± 5.0(N = 28) | 80.1± 3.9(N = 33) | 80.9 ± 5.3(N = 21) | 58.5 ± 9.2a(N = 22) | 63.3 ± 4.7a(N = 11) | 67.1 ± 8.0a,b(N = 10) |
| 3 | F (5, 114) = 58.4 P < 0.001 | 72.5 ± 3.7(N = 23) | 74.1 ± 4.7(N = 28) | 74.2 ± 4.1(N = 21) | 55.4 ± 7.8a(N = 13) | 55.9 ± 5.8a(N = 16) | 62.11 ± 4.3a,b(N = 12) |
Top number indicates the mean level of epiboly achieved for each group (see Results for technique) ± standard deviation. Each clutch demonstrated significance at the levels shown. Data from C3 are shown in graphical form in Fig. 8A. C, clutch; N, number of embryos analyzed per group.
Posthoc analysis using Tukey's HSD indicated that these groups had significantly reduced (P < 0.001 in all cases) epiboly progression compared with control embryos.
Posthoc analysis using Tukey's HSD indicated that these groups were significantly different to embryos injected with MO1 alone (C1: P = 0.001; C2: P = 0.004; C3: P = 0.013).
Following the production of P5 from cholesterol, P5 becomes the steroidogenic precursor for glucocorticoids (76), which play a central role in vertebrate development (77–79). Zebrafish embryos express a glucocorticoid receptor at early stages and readily process a synthetic glucocorticoid, Dex, when delivered in the embryo water (79, 80). This makes Dex an ideal candidate to determine if ectopic glucocorticoid treatment can reduce the epiboly delay seen in npc1 morphants. The npc1 morphants and control embryos were incubated in a 20 or 30 µM Dex solution in an identical fashion to the P5 treatments described above. Upon assessing epiboly progression among treated embryos, we found that 30 µM Dex solution led to a partial rescue of epiboly delay in npc1 morphants. Fig. 8B represents results from single experiment, and Table 3 summarizes three replicate experiments. Collectively, these findings suggest that a reduction in steroidogenesis is at least partly responsible for the epiboly-delay phenotype in npc1 morphants.
TABLE 3.
Epiboly Percentage Following MO1 Injections and Dex Treatments
| Clutch Number | One way ANOVA Results | Control | Control + DMSO | Control + 30 µM Dex | 5 ng MO1 | 5 ng MO1 + DMSO | 5 ng MO1 + 30 µM Dex |
|---|---|---|---|---|---|---|---|
| 1 | F (5, 62) = 24.7P < 0.001 | 75.4 ± 5.8(N = 7) | 74.8 ± 2.2(N = 8) | 74.6 ± 3.9(N = 8) | 53.0 ± 7.8a(N = 7) | 54.4 ± 6.0a(N = 7) | 59.7 ± 4.5a(N = 19) |
| 2 | F (5, 64) = 66.7P < 0.001 | 70.8 ± 3.9(N = 15) | 72.5 ± 5.8(N = 13) | 72.0 ± 4.0(N = 11) | 40.0 ± 4.5a(N = 9) | 41.7 ± 6.4a(N = 9) | 53.5 ± 11.8a,b(N = 8) |
| 3 | F (5, 92) = 116.5P < 0.001 | 72.1 ± 4.9(N = 11) | 74.0 ± 5.4(N = 20) | 74.9 ± 3.9(N = 17) | 46.1 ± 5.7a(N = 13) | 47.3 ± 4.0a(N = 18) | 53.2 ± 6.6a,b(N = 14) |
Top number indicates the mean level of epiboly achieved for each group (see Results for technique) ± standard deviation. Each clutch demonstrated significance at the levels shown. Data from C2 are shown in graphical form in Fig. 8B. C, clutch; N, number of embryos analyzed per group.
Posthoc analysis using Tukey's HSD indicated that these groups had significantly reduced (P < 0.001 in all cases) epiboly progression compared with control embryos.
Posthoc analysis using Tukey's HSD indicated that these groups were significantly different to embryos injected with MO1 alone (C2: P < 0.001; C3: P = 0.006).
In further experiments, we examined the potential of ectopic steroid hormone treatments to correct the actin cytoskeleton defects visualized in npc1 morphants. In these experiments, embryos were analyzed from a single clutch that was collected for analysis when the noninjected siblings reached 60-70% epiboly (supplementary Fig. IVA–C). Thus, unlike the previous actin figure (Fig. 7), these embryos were time-matched. The actin cytoskeleton defects associated with MO1 injection could be partially rescued by incubating npc1 morphants in 40 µM Dex (supplementary Fig. IVB and C, n = 38 and 20, respectively) or 20 µM P5 (data not shown, n = 13). Collectively, these data provide evidence that the epiboly delay in npc1 morphants is at least partly due to changes in the actin cytoskeleton network. Our data further suggest that steroid hormones function to stabilize the actin cytoskeleton during epiboly in addition to stabilizing yolk cell microtubules during epiboly stages (76).
npc1 morphants that complete epiboly display a shorter body axis and neural cell death phenotypes
Most embryos receiving 2.5-5.0 ng/embryo of MO1 eventually completed epiboly. When we allowed npc1 morphants to develop beyond epiboly, they consistently displayed a shorter and wider body axis. These morphant embryos invariably died prematurely, prior to 2 dpf. The severity of the phenotypes were morpholino dose-dependent, and embryos receiving low doses of MO1 (2.5 ng/embryo) were consistently healthier and more viable than siblings receiving moderate MO1 doses (5.0 ng/embryo). For this reason, the remaining analyses were performed on morphants receiving low doses of MO1.
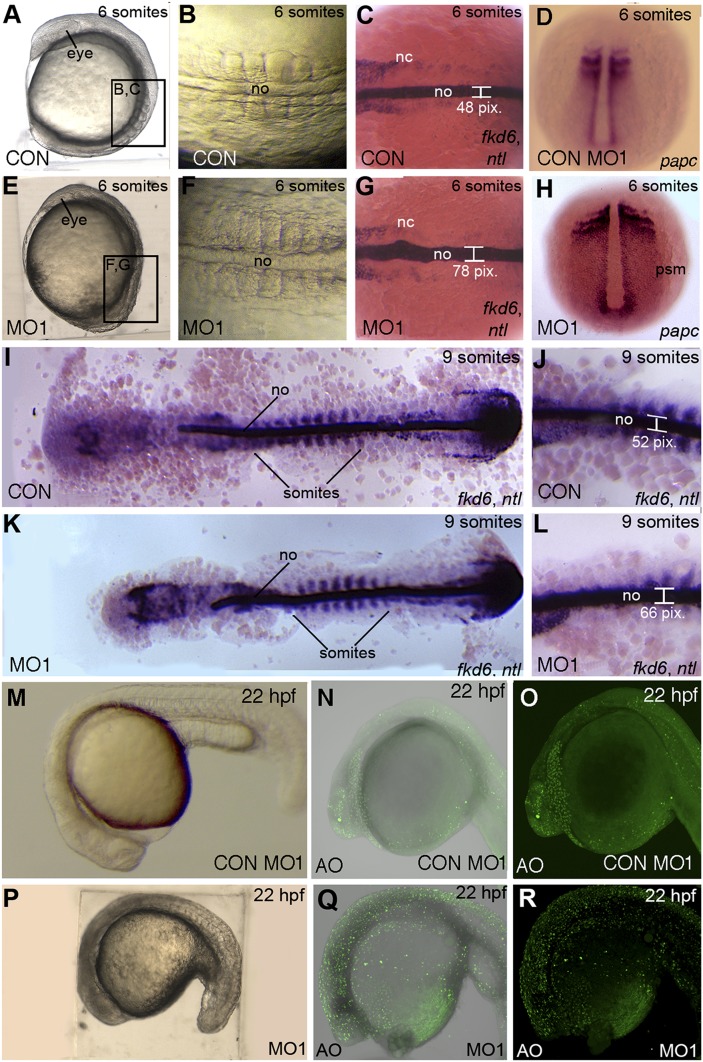
In npc1 morphants, cells in the anterior and posterior body axis migrated a shorter overall distance by the six-somite stage compared with stage-matched control embryos (Fig. 9A, E). Moreover, npc1 morphants displayed a wider notochord and somites than their stage-matched controls (Fig. 9B, F). Analysis with various cell markers along the anterioposterior (AP) axis throughout somitogenesis supported earlier evidence (supplementary Fig. II) that all germ layers became properly specified in npc1 morphants. However, comparing expression patterns between npc1 morphants and wild-type, noninjected siblings suggested that gastrulation cell movements were negatively affected in morphant embryos. At the six-somite stage, npc1 morphants displayed laterally expanded midline mesoderm and presomitic mesoderm tissue, as evidenced by ntl and papc staining, respectively (Fig. 9C, D, G, H). While the mediolateral (ML) expansion of the presomitic mesoderm in papc-stained npc1 morphants was clear by visual assessment, we quantified the width of the notochord in npc1 morphants to verify that these embryos had ML expansion. To determine the extent of notochord widening in morphants, the notochord width was quantified by measuring the span of the ntl-stained notochord. We found the notochord of npc1 morphants to be significantly wider compared with noninjected, stage-matched siblings at six somites. Later in development, at nine somites, npc1 morphants and their control siblings that were deyolked and physically flattened revealed the extent of the AP shortening and ML expansion in the morphant embryos (Fig. 9IndashL). In these embryos, fkd6 staining marks anterior structures while ntl staining marks both the posterior tailbud and the somites, thus allowing for proper stage matching based on the number of somites expressing ntl (Fig. 9I, K). Comparison of these embryos showed that npc1 morphants were relatively well developed; however, they continued to be shorter (Fig. 9I, K) than control embryos. Moreover, npc1 morphants continued to display a significantly widened notochord (Fig. 9J, L). These results suggest that npc1 may be required for proper gastrulation movements in cells undergoing convergence and extension during somitogenesis.
Fig.9.
The npc1 morphants display body axis defects and cell death. Lateral views of six-somite stage (A, E) and 22 hpf (M–R), or dorsal views of six-somite stage (B, C, D, F, G, H) and nine-somite stage (I–L) control (A–D, I, J, M) or npc1 morphants (E–H, K, L, N, O). (A, B, E, F) Nomarski optics reveal that npc1 morphants have a shorter and wider body axis than stage-matched controls at the six-somite stage. (C, D, G–L) Staining the midline mesoderm with an RNA probe for ntl revealed a widening of npc1-morphant midline (width 70 ± 5.1 pixels; n = 14) compared with controls (width: 56 ± 7.1 pixels; n = 11; P < 0.01) while papc staining revealed widening of the lateral mesodermal structures. (I, K) Deyolked and flat-mounted npc1 morphants are shorter in the anterior-posterior direction at the nine-somite stage. ntl staining marks nine somites in each embryo for proper stage matching. (J, L) ntl-stained npc1 morphants display a wider notochord at nine somites (64 ± 5.7 pixels; n = 12) compared with controls (52.6 ± 5.2 pixels; n = 10; P < 0.01. (M, P) By 22 hpf, MO1-injected embryos had darkened populations of cells in anterior regions, suggestive of cell death. (N, O, Q, R) Brightfield and fluorescent images of acridine orange (AO)-stained embryos. The npc1 morphants displayed increased levels of cell death, compared with Con MO-injected siblings. nc, neural crest; no, notochord; psm, presomitic mesoderm.
At later stages of somitogenesis (22 hpf), MO1-injected embryos remained shortened and exhibited dark staining in cells in anterior regions, characteristic of cell death (Fig. 9M, P). Cell death can be monitored in live fish using acridine orange (AO), a vital dye that stains dying cells, including apoptotic cells (81). AO staining indicated that knocking down npc1 led to increased cell death in embryos, becoming apparent near the end of the first day of development (Fig. 9N, O, Q, R). In contrast to these findings, control MO injections did not result in increased cell death (Fig. 9), nor was increased cellular death seen in npc1 morphants at earlier stages of development during epiboly (supplementary Fig. III). Cell death in npc1 morphants was followed by a variety of abnormal phenotypes during the second day of development, with morphant embryos rarely surviving beyond 48 hpf. Because npc1 knockdown was lethal, even at low doses, in the early embryonic development of zebrafish, further investigations at later embryonic or larval stages was precluded.
DISCUSSION
Several lines of evidence suggest that the zebrafish npc1 gene identified in this report is functionally similar to mammalian NPC1. First, the protein encoded by the zebrafish npc1 gene is a multi-transmembrane protein that is highly conserved when compared with NPC1 proteins from other species, including human. Analysis of the zebrafish Npc1 protein sequence revealed the strongest homology to other Npc1 species in known functional motifs of the protein, including the SSD motif, which encompasses transmembrane spanning domains 3-7, as well as the NpcL region that is present at the protein's N terminus and contains a leucine zipper motif (64). Second, zebrafish npc1 morphants exhibited phenotypes similar to those reported in other Npc1-deficient model systems, including mislocalization of endogenous cholesterol within cells, reduced cell mobility, increased cell death, decreased lifespan, and epiboly delay, a phenotype indicative of reduced steroid hormone production (37, 76). Thus, the severe phenotypic consequences seen in npc1 morphants likely reflect the dual requirements for Npc1 function in cholesterol trafficking and steroid production, reminiscent of findings reported in mouse and fly embryos that first suggested a connection between NP-C disease and reduced steroid production. Finally, protein function conservation between zebrafish Npc1 and mammalian NPC1 was confirmed by rescuing the primary developmental defect observed in zebrafish npc1-morphant embryos with mouse Npc1 RNA.
To gain insight into the requirement for npc1 during embryonic development, we studied its gene expression pattern and examined the consequences of reducing protein function in embryos injected with morpholinos. Zebrafish npc1 was present from fertilization up to 7 dpf. During gastrulation, we found that zebrafish npc1 was expressed abundantly, specifically in the extra-embryonic YSL cells, but it was also present in the embryonic cells. Reducing Npc1 protein levels in the whole embryo by a protein translation-blocking MO (MO1), as well as in YSL cells only, impacted the normal epiboly movements. In npc1 morphants, we observed that epiboly progression of the YSL and deep blastodermal cells was equally delayed during epiboly stages; however, involution of marginal cells occurred normally as evidenced by gsc staining during late epiboly stages. Importantly, adding mNpc1 mRNA to the YSL cells in embryos where Npc1 protein levels were reduced throughout the embryo restored epiboly progression, suggesting that YSL-localized Npc1 protein is critical for normal epiboly movements. Finally, a second MO (MO2), which acts by altering normal RNA splicing, also led to slower rates of epiboly upon microinjection at the one-cell stage, although its effect on epiboly progression was not as strong MO1. This may be due to maternal stores of npc1, which bypass splicing, as well as incomplete levels of splicing by MO2.
The npc1 gene expression pattern during epiboly stages is similar to the expression patterns of two crucial steroidogenic enzymes, cyp11a1 and hsd3b, which, respectively, catalyze cholesterol to P5 before converting P5 to subsequent steroid hormones (36, 76). Like npc1 morphants, the primary developmental defect in cyp11a1-depleted embryos is a slower rate of epiboly (37). In either morphant embryo, adding exogenous P5 was sufficient to partially rescue the epiboly delay, indicating that the epiboly deficit in npc1 morphants may be at least partly due to reduced steroid levels. Zebrafish epiboly is driven by yolk microtubules and the forces exerted by the actin cytoskeleton. Disrupting either actin or microtubule filaments in zebrafish embryos significantly delays or arrests epiboly progression (63, 82). Since hsd3b-depleted embryos, which cannot metabolize P5, display normal epiboly movements, it has been suggested that P5 alone influences proper epiboly function and its metabolites do not play a sufficient role in promoting epiboly (37). However, these findings do not exclude two further possibilities: i) that P5 metabolites share a redundant function with P5 in promoting epiboly movements and ii) that P5 and/or its metabolites may control additional cytoskeleton behaviors during epiboly. Along these lines, we found that Dex, a synthetic glucocorticoid, was capable of partially rescuing the epiboly-delay phenotype in npc1 morphants to a similar degree as was shown for phenotypic rescue with P5. Like humans, zebrafish express a single glucocorticoid receptor (83) likely to interact with endogenous cortisol, and in zebrafish this receptor is strongly expressed as a maternal and zygotic transcript during epiboly and gastrulation stages (84, 85). In fact, the glucocorticoid receptor is the most highly expressed steroid receptor in zebrafish embryos, suggesting that glucocorticoids have an important role in embryogenesis (85); however, such a requirement has not been examined in zebrafish. This is the first report, to our knowledge, that suggests a requirement for glucocorticoids or any P5 metabolite during epiboly. This is in contrast to progesterone, which was shown to worsen the epiboly-delay phenotype when added to zebrafish embryos with reduced steroidogenic enzyme activity (37). Considering that npc1 morphants would have reduced levels of both P5 and its metabolites (i.e., glucocorticoids), it is not surprising that these embryos experience epiboly-delay phenotypes as severe as cyp11a1 morphants.
Overall, npc1-depleted embryos had a disorganized actin cytoskeleton network within the blastoderm cells. P5 or Dex treatments were sufficient to partially rescue the changes to actin filament organization, leading us to suggest that npc1 likely regulates the cytoskeletal network either directly or indirectly in the early embryo. It is conceivable that P5 and/or its metabolites function to stabilize or maintain normal actin filaments during epiboly, similar to the ascribed function for P5 on microtubule stabilization during epiboly (37); however, more work will be required to determine the exact nature of steroid regulation on the actin cytoskeleton network. Importantly, the disordered actin skeleton in npc1 morphants was not associated with any increases in cell death, at least during early epiboly stages.
npc1-depleted embryos that received low to moderate doses (2.5 ng-5.0 ng/embryo) of MO1 often completed epiboly but were significantly shorter, with wider somites and notochord tissues and a high degree of cell death, especially in anterior neural tissues. In spite of this, these morphant embryos showed essentially normal expression levels and appropriate relative localization of a number of differentiation markers that we examined, including gsc, foxd3, sox32, and ntl. These findings suggest that Npc1 affects cell behavior but not cell specification during gastrula and segmentation stages. In the future it would be interesting to study the specific cellular behaviors of npc1-depleted cells to determine what role, if any, disrupting normal sterol-cellular biology has on in vivo cell behavior during early embryonic development. Moreover, it may be interesting to determine if earlier cell movement defects are present in other NP-C models.
This study demonstrates that zebrafish possess an ortholog of the mammalian Npc1 gene that encodes a transmembrane protein that displays functional conservation with higher vertebrate proteins. The npc1 gene is expressed in early embryonic development and is essential for normal morphogenetic movements at these stages. One hallmark of NP-C disease in human patients is a progressive degeneration of brain function due to cholesterol accumulation in neural tissues. Although early developmental defects made it harder to assess the role of npc1 in central nervous system (CNS) development, it was possible to see increased cell death in the CNS of embryos treated with a low dose of morpholino. While the CNS cell death phenotype in zebrafish is reminiscent of the human disease condition as well as mouse models for the disease, at present it is unclear how much of the early developmental defects in npc1 morphants contribute to later CNS defects. It would be interesting to study CNS development at later stages (1 dpf or older) in npc1 morphants that had received doses of fish or mammal Npc1 genes at early stages to bypass the initial defects associated with Npc1 functional loss during epiboly. The early defects found in the zebrafish happen in less than 12 h. We have established that drug treatment, in this case P5 and Dex, can reduce the severity of this early Npc1-related phenotype. Finally, since mouse Npc1 can functionally rescue the morphant embryos, taken together our findings suggest that the zebrafish model system might be an appropriate venue for rapid testing of potential therapeutics and for distinguishing between cholesterol accumulation and metabolism defects as casual agents in the debilitating outcomes associated with NP-C.
Supplementary Material
Acknowledgments
The authors thank members of Drs. Sara Ahlgren's and Jacek Topczewski's laboratories at Children's Memorial Research Center for discussions and comments on the manuscript. The authors are indebted to Drs. Barbara Sisson and Jolanta Topczewska for helpful tips on actin-staining of zebrafish embryos. The authors would also like to thank Dr. Jill A. Morris for helpful discussion and insight at the start of this project. The authors also thank members of the Topczewski laboratory, Drs. Zhibing Zhang, Axel Nohturfft, Robert Cornell, Daniel Wagner, and Jerome F. Strauss, for plasmids used in the study. The authors are indebted to the members of the Zebrafish International Resource Center (ZIRC) and Gene Tools, Inc., for fish and reagents used in the study.
Footnotes
Abbreviations:
- AO
- acridine orange
- AP
- anterioposterior
- CNS
- central nervous system
- Dex
- dexamethasone
- dpf
- days post fertilization
- ER/TGN
- endoplasmic reticulum/trans-Golgi network
- EVL
- enveloping layer
- hpf
- hours post fertilization
- ML
- mediolateral
- MO
- morpholino
- NP-C
- Niemann-Pick type C disease
- NP-C
- Niemann-Pick disease , type C1
- NPC2
- Niemann-Pick disease, type C2
- NpcL
- Npc-like region
- P5
- pregnenolone
- SSD
- sterol sensing domain
- YSL
- yolk syncytial layer
This work was supported by a Ruth L. Kirschtein predoctoral National Research Service Award (NRSA) F31DE019604 from the National Institute of Dental and Craniofacial Research of the National Institutes of Health (T.S); National Institutes of Health Grant AA-13596; and the State of Illinois (S.C.A.). Zebrafish International Resource Center (ZIRC) is supported by Grant P40 RR-012546 from the National Center for Research Resources of the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of nine figures and supplementary data and references.
REFERENCES
- 1.Vanier M. T., Millat G. 2003. Niemann-Pick disease type C. Clin. Genet. 64: 269–281. [DOI] [PubMed] [Google Scholar]
- 2.Wraith J. E., Imrie J. 2009. New therapies in the management of Niemann-Pick type C disease: clinical utility of miglustat. Ther. Clin. Risk Manag. 5: 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liscum L., Ruggiero R. M., Faust J. R. 1989. The intracellular transport of low density lipoprotein-derived cholesterol is defective in Niemann-Pick type C fibroblasts. J. Cell Biol. 108: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pentchev P. G., Comly M. E., Kruth H. S., Tokoro T., Butler J., Sokol J., Filling-Katz M., Quirk J. M., Marshall D. C., Patel S. 1987. Group C Niemann-Pick disease: faulty regulation of low-density lipoprotein uptake and cholesterol storage in cultured fibroblasts. FASEB J. 1: 40–45. [DOI] [PubMed] [Google Scholar]
- 5.Pentchev P. G., Comly M. E., Kruth H. S., Vanier M. T., Wenger D. A., Patel S., Brady R. O. 1985. A defect in cholesterol esterification in Niemann-Pick disease (type C) patients. Proc. Natl. Acad. Sci. USA. 82: 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol J., Blanchette-Mackie J., Kruth H. S., Dwyer N. K., Amende L. M., Butler J. D., Robinson E., Patel S., Brady R. O., Comly M. E. 1988. Type C Niemann-Pick disease. Lysosomal accumulation and defective intracellular mobilization of low density lipoprotein cholesterol. J. Biol. Chem. 263: 3411–3417. [PubMed] [Google Scholar]
- 7.Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B., et al. 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 277: 228–231. [DOI] [PubMed] [Google Scholar]
- 8.Millat G., Bailo N., Molinero S., Rodriguez C., Chikh K., Vanier M. T. 2005. Niemann-Pick C disease: use of denaturing high performance liquid chromatography for the detection of NPC1 and NPC2 genetic variations and impact on management of patients and families. Mol. Genet. Metab. 86: 220–232. [DOI] [PubMed] [Google Scholar]
- 9.Millat G., Chikh K., Naureckiene S., Sleat D. E., Fensom A. H., Higaki K., Elleder M., Lobel P., Vanier M. T. 2001. Niemann-Pick disease type C: spectrum of HE1 mutations and genotype/phenotype correlations in the NPC2 group. Am. J. Hum. Genet. 69: 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., Lobel P. 2000. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 290: 2298–2301. [DOI] [PubMed] [Google Scholar]
- 11.Cruz J. C., Sugii S., Yu C., Chang T. Y. 2000. Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J. Biol. Chem. 275: 4013–4021. [DOI] [PubMed] [Google Scholar]
- 12.Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., Infante R. E. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pentchev P. G., Brady R. O., Blanchette-Mackie E. J., Vanier M. T., Carstea E. D., Parker C. C., Goldin E., Roff C. F. 1994. The Niemann-Pick C lesion and its relationship to the intracellular distribution and utilization of LDL cholesterol. Biochim. Biophys. Acta. 1225: 235–243. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto Y., Ninomiya H., Ohsaki Y., Higaki K., Davies J. P., Ioannou Y. A., Ohno K. 2001. Accumulation of cholera toxin and GM1 ganglioside in the early endosome of Niemann-Pick C1-deficient cells. Proc. Natl. Acad. Sci. USA. 98: 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M., Dwyer N. K., Neufeld E. B., Love D. C., Cooney A., Comly M., Patel S., Watari H., Strauss J. F., 3rd, Pentchev P. G., et al. 2001. Sterol-modulated glycolipid sorting occurs in niemann-pick C1 late endosomes. J. Biol. Chem. 276: 3417–3425. [DOI] [PubMed] [Google Scholar]
- 16.Phillips S. E., Woodruff E. A., 3rd, Liang P., Patten M., Broadie K. 2008. Neuronal loss of Drosophila NPC1a causes cholesterol aggregation and age-progressive neurodegeneration. J. Neurosci. 28: 6569–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojtanik K. M., Liscum L. 2003. The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J. Biol. Chem. 278: 14850–14856. [DOI] [PubMed] [Google Scholar]
- 18.Loftus S. K., Morris J. A., Carstea E. D., Gu J. Z., Cummings C., Brown A., Ellison J., Ohno K., Rosenfeld M. A., Tagle D. A., et al. 1997. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 277: 232–235. [DOI] [PubMed] [Google Scholar]
- 19.Morris J. A., Carstea E. D. 1998. Niemann-Pick C disease: cholesterol handling gone awry. Mol. Med. Today. 4: 525–531. [DOI] [PubMed] [Google Scholar]
- 20.Zervas M., Dobrenis K., Walkley S. U. 2001. Neurons in Niemann-Pick disease type C accumulate gangliosides as well as unesterified cholesterol and undergo dendritic and axonal alterations. J. Neuropathol. Exp. Neurol. 60: 49–64. [DOI] [PubMed] [Google Scholar]
- 21.Griffin L. D., Gong W., Verot L., Mellon S. H. 2004. Niemann-Pick type C disease involves disrupted neurosteroidogenesis and responds to allopregnanolone. Nat. Med. 10: 704–711. [DOI] [PubMed] [Google Scholar]
- 22.Huang X., Warren J. T., Buchanan J., Gilbert L. I., Scott M. P. 2007. Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development. 134: 3733–3742. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Suyama K., Buchanan J., Zhu A. J., Scott M. P. 2005. A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development. 132: 5115–5124. [DOI] [PubMed] [Google Scholar]
- 24.Miller W. L. 1988. Molecular biology of steroid hormone synthesis. Endocr. Rev. 9: 295–318. [DOI] [PubMed] [Google Scholar]
- 25.Chung B. C., Guo I. C., Chou S. J. 1997. Transcriptional regulation of the CYP11A1 and ferredoxin genes. Steroids. 62: 37–42. [DOI] [PubMed] [Google Scholar]
- 26.Fluegel M. L., Parker T. J., Pallanck L. J. 2006. Mutations of a Drosophila NPC1 gene confer sterol and ecdysone metabolic defects. Genetics. 172: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellon S. H., Gong W., Schonemann M. D. 2008. Endogenous and synthetic neurosteroids in treatment of Niemann-Pick Type C disease. Brain Res. Rev. 57: 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zampieri S., Mellon S. H., Butters T. D., Nevyjel M., Covey D. F., Bembi B., Dardis A. 2009. Oxidative stress in NPC1 deficient cells: protective effect of allopregnanolone. J. Cell. Mol. Med. 13: 3786–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busso D., Onate-Alvarado M. J., Balboa E., Zanlungo S., Moreno R. D. 2010. Female infertility due to anovulation and defective steroidogenesis in NPC2 deficient mice. Mol. Cell. Endocrinol. 315: 299–307. [DOI] [PubMed] [Google Scholar]
- 30.Gevry N. Y., Lopes F. L., Ledoux S., Murphy B. D. 2004. Aberrant intracellular cholesterol transport disrupts pituitary and ovarian function. Mol. Endocrinol. 18: 1778–1786. [DOI] [PubMed] [Google Scholar]
- 31.Higaki K., Almanzar-Paramio D., Sturley S. L. 2004. Metazoan and microbial models of Niemann-Pick Type C disease. Biochim. Biophys. Acta. 1685: 38–47. [DOI] [PubMed] [Google Scholar]
- 32.Davies J. P., Ioannou Y. A. 2000. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 275: 24367–24374. [DOI] [PubMed] [Google Scholar]
- 33.Kuwabara P. E., Labouesse M. 2002. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18: 193–201. [DOI] [PubMed] [Google Scholar]
- 34.Schwend T., Ahlgren S. C. 2009. Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development. BMC Dev. Biol. 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtta-Vuori M., Salo V. T., Nyberg L., Brackmann C., Enejder A., Panula P., Ikonen E. 2010. Zebrafish: gaining popularity in lipid research. Biochem. J. 429: 235–242. [DOI] [PubMed] [Google Scholar]
- 36.Hsu H. J., Hsiao P., Kuo M. W., Chung B. C. 2002. Expression of zebrafish cyp11a1 as a maternal transcript and in yolk syncytial layer. Gene Expr. Patterns. 2: 219–222. [DOI] [PubMed] [Google Scholar]
- 37.Hsu H. J., Liang M. R., Chen C. T., Chung B. C. 2006. Pregnenolone stabilizes microtubules and promotes zebrafish embryonic cell movement. Nature. 439: 480–483. [DOI] [PubMed] [Google Scholar]
- 38.Trinkaus J. 1951. A study of the mechanism of epiboly in the egg of Fundulus heteroclitus. J. Exp. Zool. 118: 269–319. [Google Scholar]
- 39.Strahle U., Jesuthasan S. 1993. Ultraviolet irradiation impairs epiboly in zebrafish embryos: evidence for a microtubule-dependent mechanism of epiboly. Development. 119: 909–919. [DOI] [PubMed] [Google Scholar]
- 40.Solnica-Krezel L. 2006. Gastrulation in zebrafish - all just about adhesion? Curr. Opin. Genet. Dev. 16: 433–441. [DOI] [PubMed] [Google Scholar]
- 41.Rohde L. A., Heisenberg C. P. 2007. Zebrafish gastrulation: cell movements, signals, and mechanisms. Int. Rev. Cytol. 261: 159–192. [DOI] [PubMed] [Google Scholar]
- 42.Warga R. M., Kimmel C. B. 1990. Cell movements during epiboly and gastrulation in zebrafish. Development. 108: 569–580. [DOI] [PubMed] [Google Scholar]
- 43.Holloway B. A., Gomez de la Torre Canny S., Ye Y., Slusarski D. C., Freisinger C. M., Dosch R., Chou M. M., Wagner D. S., Mullins M. C. 2009. A novel role for MAPKAPK2 in morphogenesis during zebrafish development. PLoS Genet. 5: e1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solnica-Krezel L. 2005. Conserved patterns of cell movements during vertebrate gastrulation. Curr. Biol. 15: R213–R228. [DOI] [PubMed] [Google Scholar]
- 45.Fukazawa C., Santiago C., Park K. M., Deery W. J., Gomez de la Torre Canny S., Holterhoff C. K., Wagner D. S. 2010. Poky/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev. Biol. 346: 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabel J. L., d'Alencon C., O'Brien E. K., Van Otterloo E., Lutz K., Cuykendall T. N., Schutte B. C., Houston D. W., Cornell R. A. 2009. Maternal interferon regulatory factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev. Biol. 325: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koppen M., Fernandez B. G., Carvalho L., Jacinto A., Heisenberg C. P. 2006. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 133: 2671–2681. [DOI] [PubMed] [Google Scholar]
- 48.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253–310. [DOI] [PubMed] [Google Scholar]
- 49.Westerfield M. 2000. 4th edition The Zebrafish Book. A Guide for the Labratory Use of Zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- 50.Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. 2007. P53 activation by knockdown technologies. PLoS Genet. 3: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thisse C., Thisse B., Schilling T. F., Postlethwait J. H. 1993. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 119: 1203–1215. [DOI] [PubMed] [Google Scholar]
- 52.Jowett T. 1997. Tissue In Situ Hybridization: Methods in Animal Development. John Wiley & Sons, NY. [Google Scholar]
- 53.Schulte-Merker S., van Eeden F. J., Halpern M. E., Kimmel C. B., Nusslein-Volhard C. 1994. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 120: 1009–1015. [DOI] [PubMed] [Google Scholar]
- 54.Odenthal J., Nusslein-Volhard C. 1998. Fork head domain genes in zebrafish. Dev. Genes Evol. 208: 245–258. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto A., Amacher S. L., Kim S. H., Geissert D., Kimmel C. B., De Robertis E. M. 1998. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 125: 3389–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blader P., Strahle U. 1998. Ethanol impairs migration of the prechordal plate in the zebrafish embryo. Dev. Biol. 201: 185–201. [DOI] [PubMed] [Google Scholar]
- 57.Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X., et al. 2001. Expression of the zebrafish genome during embryogenesis (NIH R01-RR15402). ZFIN Direct Data Submission (http://zfin.org). [Google Scholar]
- 58.Wang Y. H., Chen Y. H., Lin Y. J., Tsai H. J. 2006. Spatiotemporal expression of zebrafish keratin 18 during early embryogenesis and the establishment of a keratin 18:RFP transgenic line. Gene Expr. Patterns. 6: 335–339. [DOI] [PubMed] [Google Scholar]
- 59.Siddiqui M., Sheikh H., Tran C., Bruce A. E. 2010. The tight junction component Claudin E is required for zebrafish epiboly. Dev. Dyn. 239: 715–722. [DOI] [PubMed] [Google Scholar]
- 60.Pei W., Noushmehr H., Costa J., Ouspenskaia M. V., Elkahloun A. G., Feldman B. 2007. An early requirement for maternal FoxH1 during zebrafish gastrulation. Dev. Biol. 310: 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imboden M., Goblet C., Korn H., Vriz S. 1997. Cytokeratin 8 is a suitable epidermal marker during zebrafish development. C. R. Acad. Sci. III. 320: 689–700. [DOI] [PubMed] [Google Scholar]
- 62.Stafford D., White R. J., Kinkel M. D., Linville A., Schilling T. F., Prince V. E. 2006. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 133: 949–956. [DOI] [PubMed] [Google Scholar]
- 63.Cheng J. C., Miller A. L., Webb S. E. 2004. Organization and function of microfilaments during late epiboly in zebrafish embryos. Dev. Dyn. 231: 313–323. [DOI] [PubMed] [Google Scholar]
- 64.Ioannou Y. A. 2000. The structure and function of the Niemann-Pick C1 protein. Mol. Genet. Metab. 71: 175–181. [DOI] [PubMed] [Google Scholar]
- 65.Millard E. E., Gale S. E., Dudley N., Zhang J., Schaffer J. E., Ory D. S. 2005. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 280: 28581–28590. [DOI] [PubMed] [Google Scholar]
- 66.Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C., Chang T. Y. 2004. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 101: 12473–12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higgins M. E., Davies J. P., Chen F. W., Ioannou Y. A. 1999. Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol. Genet. Metab. 68: 1–13. [DOI] [PubMed] [Google Scholar]
- 68.Neufeld E. B., Wastney M., Patel S., Suresh S., Cooney A. M., Dwyer N. K., Roff C. F., Ohno K., Morris J. A., Carstea E. D., et al. 1999. The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J. Biol. Chem. 274: 9627–9635. [DOI] [PubMed] [Google Scholar]
- 69.Friend D. S., Bearer E. L. 1981. beta-Hydroxysterol distribution as determined by freeze-fracture cytochemistry. Histochem. J. 13: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gondre-Lewis M. C., McGlynn R., Walkley S. U. 2003. Cholesterol accumulation in NPC1-deficient neurons is ganglioside dependent. Curr. Biol. 13: 1324–1329. [DOI] [PubMed] [Google Scholar]
- 71.Chen S., Kimelman D. 2000. The role of the yolk syncytial layer in germ layer patterning in zebrafish. Development. 127: 4681–4689. [DOI] [PubMed] [Google Scholar]
- 72.Carreira-Barbosa F., Kajita M., Morel V., Wada H., Okamoto H., Martinez Arias A., Fujita Y., Wilson S. W., Tada M. 2009. Flamingo regulates epiboly and convergence/extension movements through cell cohesive and signalling functions during zebrafish gastrulation. Development. 136: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bursch W., Hochegger K., Torok L., Marian B., Ellinger A., Hermann R. S. 2000. Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J. Cell Sci. 113: 1189–1198. [DOI] [PubMed] [Google Scholar]
- 74.Chen G. C., Lee J. Y., Tang H. W., Debnath J., Thomas S. M., Settleman J. 2008. Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy. 4: 37–45. [DOI] [PubMed] [Google Scholar]
- 75.Chung B. C., Matteson K. J., Voutilainen R., Mohandas T. K., Miller W. L. 1986. Human cholesterol side-chain cleavage enzyme, P450scc: cDNA cloning, assignment of the gene to chromosome 15, and expression in the placenta. Proc. Natl. Acad. Sci. USA. 83: 8962–8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu H. J., Hsu N. C., Hu M. C., Chung B. C. 2006. Steroidogenesis in zebrafish and mouse models. Mol. Cell. Endocrinol. 248: 160–163. [DOI] [PubMed] [Google Scholar]
- 77.Canalis E., Pereira R. C., Delany A. M. 2002. Effects of glucocorticoids on the skeleton. J. Pediatr. Endocrinol. Metab. 15Suppl.5 1341–1345. [PubMed] [Google Scholar]
- 78.Dickmeis T., Lahiri K., Nica G., Vallone D., Santoriello C., Neumann C. J., Hammerschmidt M., Foulkes N. S. 2007. Glucocorticoids play a key role in circadian cell cycle rhythms. PLoS Biol. 5: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hillegass J. M., Villano C. M., Cooper K. R., White L. A. 2008. Glucocorticoids alter craniofacial development and increase expression and activity of matrix metalloproteinases in developing zebrafish (Danio rerio). Toxicol. Sci. 102: 413–424. [DOI] [PubMed] [Google Scholar]
- 80.de Graaf M., Zivkovic D., Joore J. 1998. Hormone-inducible expression of secreted factors in zebrafish embryos. Dev. Growth Differ. 40: 577–582. [DOI] [PubMed] [Google Scholar]
- 81.Furutani-Seiki M., Jiang Y. J., Brand M., Heisenberg C. P., Houart C., Beuchle D., van Eeden F. J., Granato M., Haffter P., Hammerschmidt M., et al. 1996. Neural degeneration mutants in the zebrafish, Danio rerio. Development. 123: 229–239. [DOI] [PubMed] [Google Scholar]
- 82.Solnica-Krezel L., Driever W. 1994. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 120: 2443–2455. [DOI] [PubMed] [Google Scholar]
- 83.Prunet P., Sturm A., Milla S. 2006. Multiple corticosteroid receptors in fish: from old ideas to new concepts. Gen. Comp. Endocrinol. 147: 17–23. [DOI] [PubMed] [Google Scholar]
- 84.Schaaf M. J., Champagne D., van Laanen I. H., van Wijk D. C., Meijer A. H., Meijer O. C., Spaink H. P., Richardson M. K. 2008. Discovery of a functional glucocorticoid receptor beta-isoform in zebrafish. Endocrinology. 149: 1591–1599. [DOI] [PubMed] [Google Scholar]
- 85.Pikulkaew S., De Nadai A., Belvedere P., Colombo L., Dalla Valle L. 2010. Expression analysis of steroid hormone receptor mRNAs during zebrafish embryogenesis. Gen. Comp. Endocrinol. 165: 215–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.