Abstract
Previously, we have shown that Pck1 expression in mammary gland adipocytes and white adipose tissue maintains triglyceride stores through glyceroneogenesis, and these lipids were used for synthesis of milk triglycerides during lactation. Reduced milk triglycerides during lactation resulted in patterning of the newborn for insulin resistance. In this study, the role of Pck1 in mammary gland epithelial cells was analyzed. The developmental expression of Pck1 decreased in isolated mouse mammary gland epithelial cells through development and during lactation. Using HC11, a clonal mammary epithelial cell line, we found that both Janus kinase 2 signal transducers and activators of transcription 5 and the AKT pathways contributed to the repression of Pck1 mRNA by prolactin. These pathways necessitate three accessory factor regions of the Pck1 promoter for repression by prolactin. Using [U-13C6]glucose, [U-13C3]pyruvate, and [U-13C3]glycerol in HC11 cells, we determined that Pck1 functions in the pathway for the conversion of gluconeogenic precursors to glucose and contributes to glycerol-3-phosphate synthesis through glyceroneogenesis. Therefore, Pck1 plays an important role in both the mammary gland adipocytes and epithelial cells during lactation.
Keywords: adipocytes, gluconeogenesis, glyceroneogenesis
During lactation, there are profound modifications in the intermediary metabolism of different tissues to adequately supply nutrients for milk production (1, 2). There are decreases in both lipogenic capacity and activities of several lipogenic enzymes in adipose tissue (3–7). Prolactin is one of the key signals that integrates metabolism of tissues during lactation, promoting lipogenesis over esterification in the adipose tissue (2, 3, 7, 8). These changes cause mobilization of lipid stores in adipose tissues to support milk production in the mammary gland.
One gene that is present in both adipocytes and epithelial cells of the mammary gland is the phosphoenolpyruvate carboxykinase gene Pck1. The role of Pck1 in adipocytes of white adipose tissue (WAT) has been clearly defined as glyceroneogenic. Over 30 years ago, Ballard, Hanson, and Leveille (9) and Reshef, Niv, and Shapiro (10, 11) demonstrated that in vitro incubation of rat epididymal fat pad with pyruvate reduced the amount of FFAs released by 65% (10, 11). However, the rate of lipolysis remained unaffected. In WAT, glycerol is released during lipolysis, but it cannot be phosphorylated in preparation for triglyceride synthesis because the tissue manifests negligible glycerol kinase activity. Ballard and Hanson (12) and Reshef, Hanson, and Ballard (13) determined that during fasting, gluconeogenic precursors such as pyruvate and alanine are converted into the glycerol backbone of triglycerides through the glyceroneogenic pathway. Support for a glyceroneogenic role for Pck1 in the mammary gland was established by Jimenez et al. (14), who established incorporation of labeled oleate into glycerol-3-phosphate in isolated acini from lactating Wistar rats. However, they suggested that the last steps of gluconeogenesis between triose-phosphate and glucose-6-phosphate are not operative in rat mammary gland acinar cells (14).
Previously, we have shown that the role for Pck1 in mammary gland adipocytes is the formation of glycerol-3-phosphate through glyceroneogenesis (15). We analyzed mice in which the binding site for peroxisome proliferator-activated receptor γ (PPARγ) was deleted from the promoter of Pck1 (PPARE−/−). Pck1 expression and triglyceride content in the milk were measured. The Pck1 mRNA expression was lowered to 2.2% that of wild-type (WT) mice in mammary gland adipocytes in PPARE−/− mice; however, the expression levels of Pck1 mRNA were similar in epithelial cells from PPARE−/−, PPARE+/−, and WT mice. The female PPARE−/− mice presented reduced lipid storage in mammary gland adipocytes and in WAT, resulting in a 40% reduction of milk triglycerides during lactation. However, because the PPARE−/− females had normal expression of Pck1 in mammary gland epithelial cells, we wanted to determine the role of the Pck1 gene in mammary gland epithelial cells during lactation. We hypothesized that Pck1 provides glucose through gluconeogenesis in mammary epithelial cells and may contribute to lipid synthesis by the formation of glycerol-3-phosphate through glyceroneogenesis. In this study, we investigated the role of Pck1 in mammary gland epithelial cells during lactation in mice and in HC11 cells.
EXPERIMENTAL PROCEDURES
Prolactin injection
Wild-type mice that were used previously for published studies on the role of Pck1 in mammary gland (15) were further characterized in this study. The mice had free access to water and were fed a normal mouse diet ad libitum (LabDiet, St. Louis, MO; Diet # 5P76). The approximate composition of the diet was 1.08 kJ protein, 0.58 kJ fat, and 14.33 kJ carbohydrate. The diet consisted of 4,100 kJ × kg−1 gross energy (16). Four week-old virgin female mice were injected intraperitoneally with prolactin (1 µg/g body weight) (7). Thirty minutes after the prolactin injection, the mice were euthanized, and thoracic mammary gland tissues were collected. We chose this time point because we found in a time course study that 30 minutes was adequate for repression of Pck1 gene expression by prolactin (see supplementary ). All experimental protocols were approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Isolation of adipocytes and mammary epithelial cells
Isolation of adipocytes and epithelial cells was performed as previously described (17, 18). Briefly, thoracic mammary glands were dissected from virgin female mice (three glands for each preparation). The lymph node was removed, and they were minced with a razor blade and incubated at 37°C with gentle shaking for 1 h in DMEM:F12 medium containing 0·2% collagenase. Three batches of isolated epithelial cells and three batches of isolated adipocytes were prepared. After the dissociation was complete, the cell suspension was separated at 176.08 g for 10 min. The adipocytes and the epithelial cells were collected separately and washed extensively, and any endothelial contaminants were removed by sedimentation. The adipocytes and epithelial cells were collected and stored in RNAlater solubilization buffer (Qiagen; Valencia, CA) for isolation of RNA.
Plasmid constructs
The plasmid 490-Luc was generated by ligating the XbaI/Pst1 fragment of the rat Pck1 promoter into pGL3-basic (which contains the luciferase structural gene) that had been digested with restriction enzymes PstI and BglII. The plasmids were isolated with Plasmid Mini Kits (Qiagen). Truncations of the gene promoter were created using restriction enzymes to delete segments of the promoter within p490-Luc to generate 402-Luc and 206-Luc. Heterologous constructs were made with the −490 bp to −402 bp fragment, and a −490 bp to −206 bp fragment of the rat Pck1 promoter was ligated before the SV40 promoter in the pGL3 promoter vector (Promega; Madison,WI).
Site-directed mutagenesis
The AF1 (−451 bp to −439 bp), AF2 (−416 bp to −407 bp), GR1 (−391 bp to −371 bp), GR2 (−370 bp to −350 bp), and AF3 (−326 bp to −321 bp) were mutated using the following oligonucleotide (the mutated bases are in bold) primers for mutated AF1: forward: 5′GCTGAATTCCCTTCTCACTACGATTGGCCGTGGGAGTGAC-3′, reverse: 5′ GTCACTCCCACGGCCAATCGTAGTGAGAAGGGAATTCAGC-3′; primers for mutated AF2: forward, 5′-GACACCTCACAGCTGTAGCATACTGACAACCAGCAGCCAC-3′; reverse, 5′-GTGGCTGCTGGTTGTCAGTATGCTACAGCTGTGAGGTGTC-3′; primers for mutated GR1: forward, 5′-CAGCAGCCACTGGAATTCAAGATCTAATGCCAGCAGCATA-3′; reverse, 5′-TATGCTGCTGGCATTAGATCTTGAATTCCAGTGGCTGCTG-3′; primers for mutated GR2, forward: 5′- ATGTGCAGCCAGCAGCTGGTAATCCGTAAGAGGCGTCCCG-3′; reverse: 5′-CGGGACGCCTCTTACGGATTACCAGCTGCTGGCTGCACAT-3′; primers for mutated AF3: forward, 5′-CCGGCCAGCCCTGTCCTCGGTACCCACCTGACAATTAGGG-3′; reverse, 5′-CCTTAATTGTCAGGTGGGTACCGAGGACAGGGCTGGCCGG-3′. These sites were mutated in combination or individually to generate vectors using the QuickChange mutagenesis kit (Stratagene; La Jolla, CA). The plasmids resulting from the above mutations were isolated, and their sequence was confirmed by restriction digestion and DNA sequencing performed by the Case Western Reserve University Molecular Biology Core Laboratory.
Cell culture
HC11 mouse mammary gland epithelial cells were routinely maintained in RPMI 1640 medium supplemented with 10% FBS, 5 µg/ml insulin, and 10 ng/ml epidermal growth factor (EGF) (19). For induction of differentiation, cells were grown to confluence and then kept in medium without EGF for 48 h to induce competence. Competent cells were incubated with differentiation medium (serum-free RPMI 1640 medium containing 5 μg/ml insulin, 1 mM dexamethasone, and 1 μg/ml ovine prolactin) for 72 h.
Transient transfection and luciferase assay
All plasmids were purified using the HiSpeed Plasmid Midi Kit (Qiagen). Cells were seeded in 12-well plates and transfected with plasmid DNA using FuGENE-HD transfection reagent (Roche Applied Science; Indianapolis, IN). Forty-eight hours after transfection, cells were incubated with serum-free RPMI 1640 medium containing lactogenic hormone mix [dexamethasone, insulin and prolactin (DIP)]: 1 mM dexamethasone, 5 mg/ml insulin, and 1 µg/ml ovine prolactin (Sigma; St. Louis, MO) for 72 h. Controls were treated with dexamethasone and insulin only (DI). After 72 h treatment, the cells were washed with ice-cold 1× PBS, pH7.4, and lysed by the addition of 250 µl of 1× passive lysis buffer. The insoluble material was pelleted by centrifugation in 1.5 ml eppendorf tubes for 6 min at 12,000 g, and the supernatant was separated from the pellet for the measurement of luciferase activity. For luciferase activity, 20 µl of the cell lysate was used to measure the integrated light units over 10 s. Firefly and Renilla luciferase activities were measured using dual-luciferase assay reagents (Promega) on a luminometer. Relative luciferase activity was calculated by dividing firefly luciferase activity by Renilla luciferase activity and expressed as relative Pck1-luciferase activity (relative activity). For the inhibition of signaling pathways in a separate experiment, cells were grown and transfected as described above. At 48 h after treatment with DI or DIP, the cells were treated with specific inhibitors Akt1/2 inhibitor (1 μM) (Akt inhibitor VIII, Calbiochem/EMD Chemicals, Inc.; San Diego, CA) (20), MAPK/ERK inhibitor (1 μM) (U0126, EMD Chemicals, Inc.) (21) and the STAT5 inhibitor (1 μM) (Calbiochem/EMD Chemicals, Inc.) (22). After 24 h, relative activity was determined as described above. The assay was carried out in quadruplicate.
Real-time quantitative reverse-transcription PCR
Total RNA was prepared from 100 mg of liver, WAT, brown adipose tissue (BAT), and mammary gland from mice or HC11 cells with the RNeasy Lipid Tissue Mini Kit (Qiagen). The single-strand cDNAs were synthesized from 2 µg total RNA with random hexamer primers and mouse mammary tumor virus reverse transcription (Ambion; Austin, TX). The cDNAs were amplified using Syber Green PCR Core Reagent Mix (Applied Biosystems; Foster City, CA). The real-time quantitative PCR was performed in the Chromo4 Cycler (MJ Research; Waltham, MA). The relative amounts of the mRNAs were determined by linear regression from standard curves derivative maximum method, with Opticon Monitor 3 software (MJ Research). RNA expression data were normalized to levels of 18S rRNA. The utilized primer sequences (Integrated DNA Technologies; San Diego, CA) are presented in supplementary Table I.
Stable isotope incubation conditions
Cells were incubated in glucose-free and 10% stripped FBS RPMI 1640 media [contains ∼0.1–0.2 mM amino acid, 2 mM glutamine, without sodium pyruvate and without glucose (Invitrogen; Carlsbad, CA)] that contained 5 mM of U13C-labeled substrates: Condition 1, [U-13C6]glucose-enriched 100% mass percent enrichment (MPE); Condition 2, [U-13C3]glycerol-enriched 100% MPE; Condition 3, [U-13C3]pyruvate-enriched 100% MPE. 13C label incorporation into cells at 80–90% confluence was measured after switching the cells into fresh glucose-free medium containing 100% of one of the labeled substrates outlined in the above conditions, and cultured at 37°C for 6 h. The media were then removed and saved for measurement of MPE of the 13C-labeled precursors and 13C label incorporation into the respective “products.” The isolated cells were stored at −80°C and then analyzed for MPE (13C label incorporation) of triglyceride-bound glycerol and palmitate. Measurement of MPE of glucose was also performed. All stable isotopomer analysis was completed at the Case Mouse Phenotyping Center. Briefly, total triglycerides were hydrolyzed and extracted from the cells, and the 13C enrichments of glycerol and palmitate were determined by using gas chromatography mass spectrometry (GCMS) (23). The media were assayed for precursor MPE for the purpose of normalizing the product enrichments to the precursor enrichments.
13C labeling of glucose, glycerol, and palmitate
Cells were saponified by performing complete hydrolysis using ethanol-hydroxide incubation at 70°C for 1 h and then extracted using the Folch procedure. Briefly, 3 ml of chloroform-methanol mix (2:1, v/v) was added, and cells were homogenized. Layers were separated by water, and the chloroform layer was removed following centrifugation. Both organic and aqueous layers were evaporated to dryness. The aqueous layer from the previous step was reconstituted in 100 μl water and divided into two equal parts. Total glycerol and palmitate MPEs were measured as the triglyceride-bound glycerol and palmitate and assayed from an aliquot containing either the organic extract or total palmitate. Also, the cell media (20 μl aliquot) was sampled to determine precursor enrichment of glucose, glycerol, or pyruvate.
To determine 13C labeling of glucose and glycerol in the media and/or cell extracts, aliquots containing the aqueous fraction were converted to their respective penta-acetate derivatives (24). This was done by derivatization accomplished by reacting it with 50 μl of acetic anhydride solvent and further heated for 2 h at 70°C. To determine 13C labeling by pyruvate or glycerol, an aliquot taken from the aqueous fraction (for media or cells) was first evaporated, and then pyruvate was reacted with 70 μl of bis(trimethylsilyl) trifluoroacetamide + 10% trimethylchlorosilane and incubation for 1 h at room temperature. Using the GCMS system (Agilent 5973N-MSD equipped with an Agilent 6890 GC system and a DB-17 MS capillary column) all samples were analyzed for the respective ions corresponding to MO, M+1, M+2, and M+3 through M+6 for each of the analytes (25).
MPE calculations
The fractional contribution of the precursor to the product was based on the MPEs derived from the 13C label incorporation into the total pool of each of the products (glycerol, palmitate, and glucose). Mass isotopomer analysis enables the measurements of unlabeled analyte (MO) to the labeled analyte (M+1, 2, or 3, etc.). The measured mass isotopomer distribution was corrected for natural enrichment of each mass (26, 27) by a technique that takes into account the skew of natural 13C enrichment of molecules as increasing numbers of carbon atoms become fully labeled (26, 28). For example, the MPE calculated for glycerol following incubation with the precursor of [U-13C6]glucose was calculated as the ratios of [(M+3/(MO+M3)·100)]glycerol to precursor MPE [(M+6/(MO+ M+6)·100)][U-13C6]glucose in the cells or media × 100. The MPE calculated for glycerol following incubation with the precursor of [U-13C3]pyruvate was calculated as the ratios of [(M+2/(M0+M+2)·100)]glycerol to precursor MPE [(M+3/(MO+ M+3)·100)][U-13C3]pyruvate in the media × 100 to account for the loss of label in the tricarboxylic acid (TCA) cycle (26, 29). We did not detect M+3 in our studies. Theoretically, since half of the phophoeonolpyruvate that emerges from the TCA cycle should have been randomized in the malate/fumarate exchange, we should have detected M+3. The lack of M+3 detection may be due to small sample size (too few cells) and thus M+3 was below the level of detection. The calculation of glycerol-bound triglyceride is a modified calculation of pyruvate to glucose as described (26).
Glycerol kinase activity
Glycerol kinase activity was measured as previously described (30, 31). Briefly, HC11 cells treated with or without prolactin were homogenized in extraction buffer (50 mM HEPES, pH 7.8, 40 mM KCl, 11 mM MgCl2, 1 mM EDTA, and 1 mM DTT) on ice. The samples were centrifuged at 15,000 g for 15 min at 4°C, and 10 µg of protein was used for the enzymatic assay. The protein samples were incubated with 50 µl of assay buffer (50 mM Tris-HCl, pH 7.2, 5 mM ATP, 10 mM MgCl2, 100 mM KCl, 2.5 mM DTT, 4 mM glycerol, and 500 µmol/l 3H-glycerol) for 90 min at 37°C. The reaction was terminated with 100 µl stop solution (ethanol-methanol, 97:3). Equal amounts of samples (20 µl) were spotted onto DE81 Whatman filters (Whatman, Inc.; Piscataway, NJ). The filters were air-dried and washed in water overnight. Radioactivity on the filters was measured by liquid scintillation counting.
Statistical analysis
The results were expressed as the mean ± SEM. For three or more comparisons, we analyzed the parameters by one-way ANOVA with Bonferoni's correction. For analysis of three or more means compared with HC11 cells without prolactin, we analyzed the data by one-way ANOVA. For analysis of two means, we used Student's t-test, with P < 0.05.
RESULTS
Expression of Pck1 in mammary gland
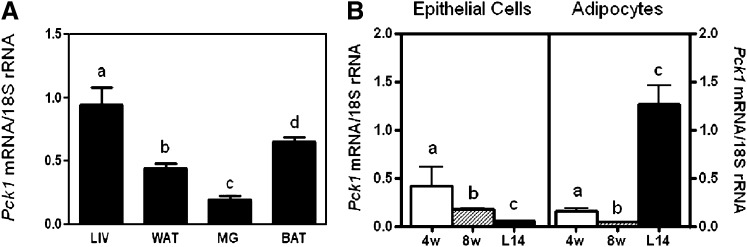
Pck1 mRNA is expressed in a variety of tissues. We compared the expression of Pck1 mRNA in the mammary gland to its expression in the liver, WAT, and BAT in wild-type mice (Fig. 1A). The mammary gland had 25% the expression of Pck1 mRNA compared with liver tissue. Previously, we established expression of Pck1 protein in mammary gland adipocytes and epithelial cells by immunohistochemistry (15). To determine developmental expression of Pck1 in mammary gland adipocytes and epithelial cells, we analyzed Pck1 mRNA expression during development at 4 weeks and 8 weeks and during lactation (Fig. 1B). We found that the expression of Pck1 in isolated mammary epithelial cells decreased over time in mice. However, in isolated adipocytes, we found increased expression of Pck1 mRNA during lactation.
Fig. 1.
Pck1 expression in mammary gland. A: Expression of Pck1 mRNA compared with liver (LIV), white adipose tissue (WAT), mammary gland (MG), and brown adipose tissue (BAT). Values are the mean ± SEM, n = 4; subscripts with a different letter are statistically significant, P < 0.05. B: Mammary gland epithelial cells and mammary adipocytes were isolated from mice at 4 weeks (4w), 8 weeks (8w), and after 14 days of lactation (L14), and Pck1 mRNA expression was measured by RT-PCR and normalized by 18S rRNA. Values are the mean ± SEM, n = 5; subscripts with a different letter are statistically significant, P < 0.05.
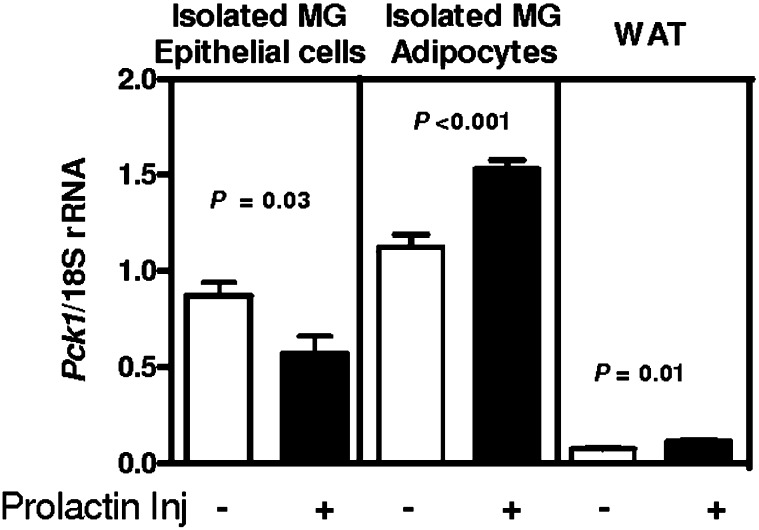
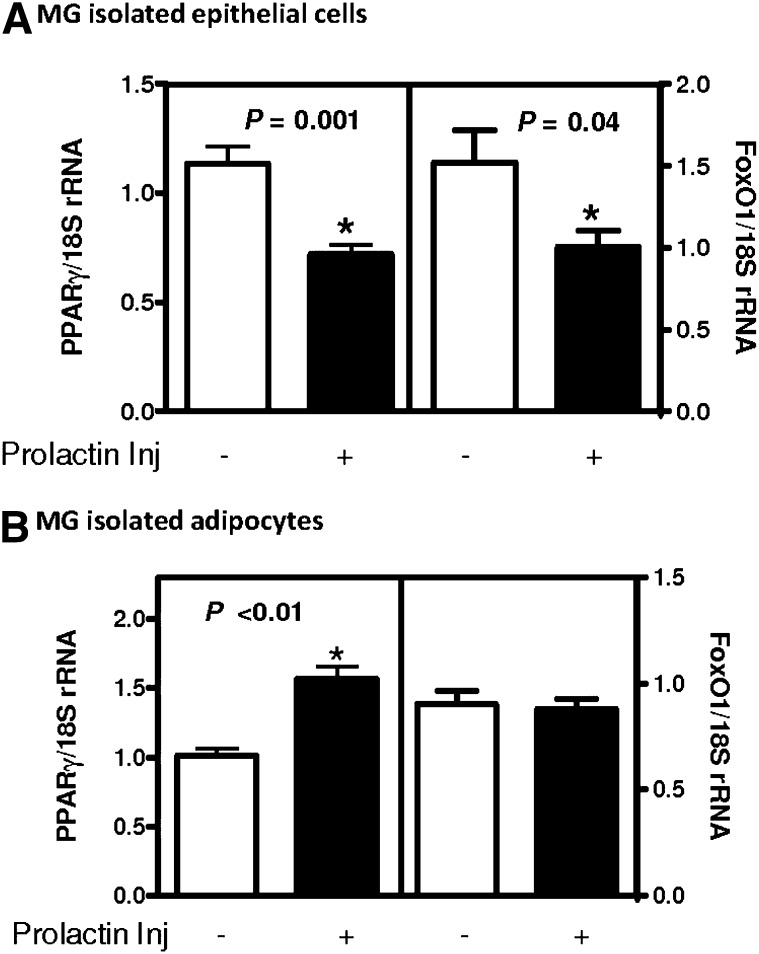
Because Pck1 was greatly induced in the adipose tissue, whereas it was reduced in the epithelial cells of the mammary gland during lactation, we wanted to determine whether this was a direct response to prolactin, one of the key hormones present during lactation. In female mice, epithelial cells and adipose cells were isolated from the mammary gland 30 min after injection of prolactin. We found a reciprocal response to prolactin in epithelial cells compared with adipocytes. Pck1 mRNA was reduced in mammary epithelial cells (Fig. 2), whereas mammary gland adipocytes and WAT have increased Pck1 mRNA expression (Fig. 2). Two transcription factors that are known regulators of Pck1 expression are PPARγ and forkhead box protein O1(FoxO1) (32–36). We measured these in the same samples and found reduced levels of PPARγ and FoxO1 after prolactin injection in isolated mammary gland epithelial cells (Fig. 3A). However, in isolated adipocytes, PPARγ expression was induced, yet there was no effect on FoxO1 expression (Fig. 3B).
Fig. 2.
2. The effect of prolactin injection on Pck1 mRNA expression. Mice were treated with saline (−) or with 1 µg/g body weight of prolactin (+), and after 30 min, tissues were isolated. Pck1 mRNA expression was measured by RT-PCR and normalized with 18S rRNA in isolated mammary gland (MG) epithelial cells, isolated mammary gland adipocytes, or isolated white adipose tissue (WAT). Values are the mean ± SEM; n = 5 mice per time point.
Fig. 3.
3. PPARγ and FoxO1 expression in mammary adipocytes. Mice were treated with saline (−) or with 1 µg/g body weight of prolactin (+), and after 30 min, tissues were isolated. RNA was isolated, and gene expression was measured by RT-PCR and normalized by 18S rRNA. Expression of (A) PPARγ and FoxO1 in isolated mammary epithelium and (B) PPARγ and FoxO1 in isolated mammary gland adipocytes. Values are the mean ± SEM; n = 3–5 mice per time point, *P < 0.05.
Mammary epithelial cell line HC11
To elucidate the mechanism for regulation of Pck1 in mammary gland epithelial cells during lactation, we chose to study HC11 cells. HC11 cells are a mammary epithelial cell line derived from spontaneously immortalized COMMA-D epithelial cells, isolated from the mammary gland of pregnant BALB/c mice (37). HC11 cells can functionally differentiate and express milk proteins in vitro and can reconstitute ductal epithelium (37). HC11 cells have been used extensively to study the hormonal regulation of mammary epithelial cell differentiation and casein gene expression (38–40). When grown at confluence for several days after exposure to certain growth factors, such as EGF, and then treated with the lactogenic hormones prolactin, dexamethasone, and insulin, the cells produce β-casein milk protein.
We analyzed HC11 cells treated with or without prolactin. For a control and to ensure that the cells were responding to the lactogenic hormones, we measured β-casein gene expression. We found that β-casein gene expression was induced with prolactin treatment (see supplementary ). However, the expression of Pck1 mRNA was reduced in response to prolactin (see supplementary ). Thus, these cells are responding to the lactogenic hormones and Pck1 expression is responding in a similar manner as in vivo. We measured phosphorylation of FoxO1 in HC11 cells treated with prolactin (see supplementary ). We found increased phosphorylated FoxO1 in the cytoplasm of prolactin-treated cells. AKT (Akt/PKB) phosphorylates FoxO1, thus allowing it to migrate to the cytoplasm. Therefore, we tested whether AKT phosphorylation was altered. We found slightly increased levels of AKT phosphorylation in the prolactin-treated cells. The total amount of FoxO1 and AKT were not changed with prolactin treatment. To confirm that the cytoplasmic and nuclear fractions were not contaminated with other cell fractions, we measured cAMP response element binding (CREB), a marker for nuclear proteins, and AKT, a marker for cytosolic proteins (see supplementary ). We found CREB in the nuclear cell fractions and AKT in the cytosolic cell fractions.
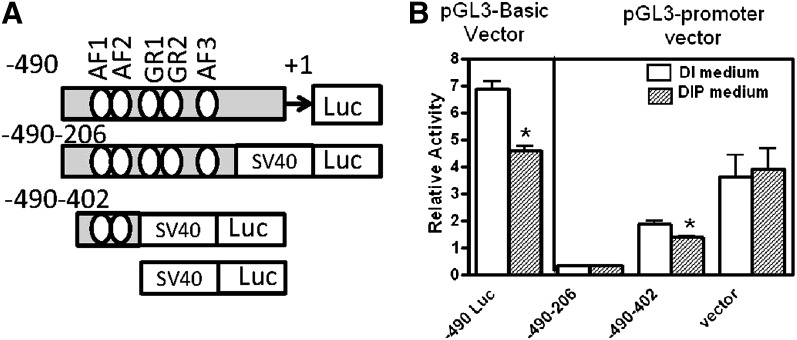
Region −490 bp to −402 bp in the Pck1 promoter is required for repression by prolactin
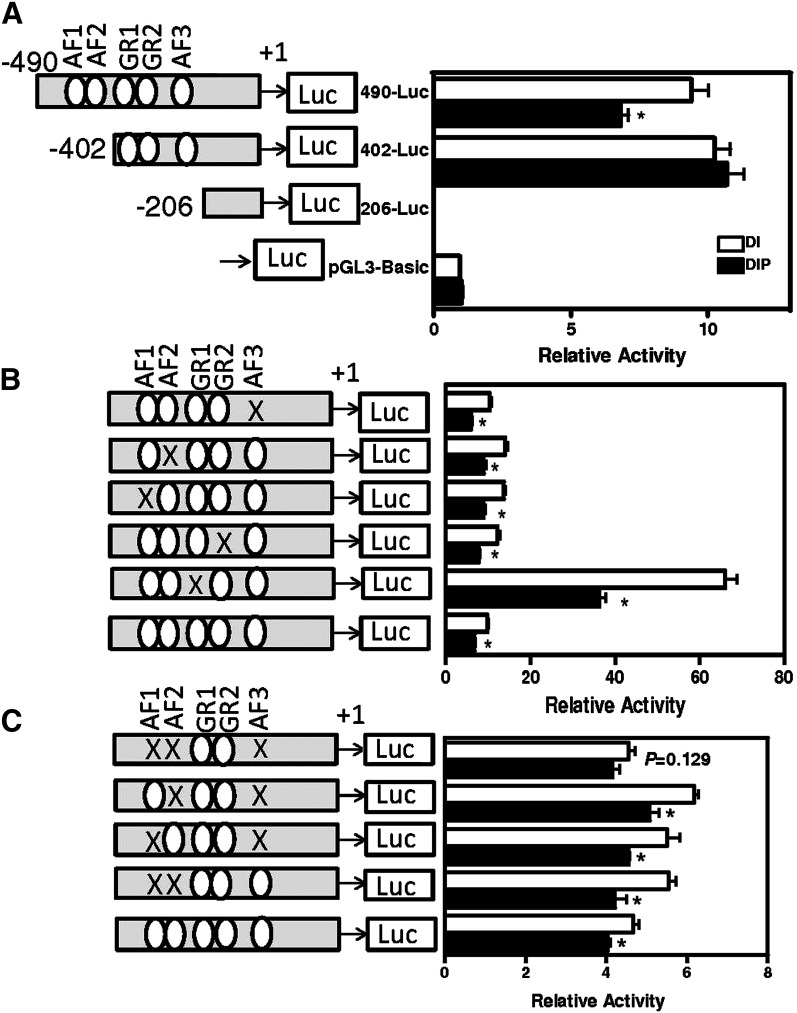
We wanted to determine which region(s) of the Pck1 promoter were required for repression by prolactin. Previous studies have shown that −490 bp of Pck1 promoter are adequate for regulated expression of Pck1 mRNA and is a minimal promoter construct (41–43). Thus we used −490-Luc (−490 bp to +72 bp Pck1 promoter ligated to luciferase reporter gene) as a control. First, from deletion analysis of the Pck1 promoter, we found that repression by prolactin was lost when the −402 bp-Luc construct was tested (Fig. 4A). However, the −490 bp-Luc construct had all of the necessary promoter elements for Pck1 mRNA repression. Hence the region −490 bp to −402 bp in the Pck1 promoter is required for repression by prolactin.
Fig. 4.
4. Deletions and mutation of GRE and AF sites in Pck1 promoter. A series of constructs were made that contained serial deletions of the Pck1 promoter-firefly luciferase or contained mutations of GRE and AF sites (represented by X) and transfected into HC11 cells. Cells were incubated with the lactogenic hormones dexamethasone, insulin, and prolactin (DIP) or lactogenic hormones minus prolactin [dexamethasone and insulin (DI)] as described in Experimental Procedures, and relative luciferase activity was measured in cell extracts. A: For serial deletion analysis, portions of the Pck1 promoter (−490 bp to +72 bp, −402 bp to +72 bp, −206 bp to +72 bp) were cloned into pGL3-basic promoter vector and analyzed, or (B) −490 bp to +72 bp of Pck1 promoter was cloned into pGL3 basic vector with single site mutations of GRE and/or AF sites or (C) multiple site mutations of GRE and/or AF sites as described in Experimental Procedures. The data are expressed as the means ± SEM, n = 6; *P < 0.05.
We mutated the regions within Pck1 promoter that are known regulators of Pck1 expression. Accessory factors (AFs) and glucocorticoid response elements (GREs) have been shown to regulate Pck1 mRNA expression by glucocorticoids (41–44). Using single mutations of each AF (Fig. 4B, mutAF1, mutAF2, mutAF3) and combinations of these mutations (Fig. 4C, mutAF1 + mutAF2, mutAF1+ mutAF3, mutAF2+ mutAF3 and mutAF1 + mutAF2 + mutAF3), we found that AF1, AF2, and AF3 were required for the prolactin repression of Pck1. We also analyzed constructs that were mutated for GRE, but loss of GRE in the promoter still repressed luciferase expression with prolactin treatment; thus it was not required for repression of Pck1 in HC11 cells.
To elucidate if AF1, AF2, and AF3 were acting as general repressors of gene expression or if this was specific to the Pck1 promoter, we made heterologous constructs with AF1+AF2 and AF1+AF2+AF3 before SV40 minimal luciferase promoter (Fig. 5). The promoter fragment −490 bp to −402 bp, which contains AF1 and AF2, recapitulated repression by prolactin. Because this repression was not complete, it suggested that AF1+AF2+AF3 were required for prolactin repression in the Pck1 promoter only and not acting as a general repressor of gene expression.
Fig. 5.
5. AF1, AF2, and AF3 regions in the Pck1 promoter. A: Portions of the Pck1 promoter (−490 to −402, −490 to −206) were cloned into pGL3-promoter vector or −490 to +72 of Pck1 promoter was cloned into pGL3 basic vector. B: The constructs and pGL3 promoter vector were transfected into HC11 cells. After transfection, cells were treated with the lactogenic hormones dexamethasone, insulin, and prolactin (DIP) or lactogenic hormones minus prolactin [dexamethasone and insulin (DI)] as described in Experimental Procedures, and luciferase relative activity was measured in cell extracts. The data are expressed as the means ± SEM; n = 6, *P < 0.05.
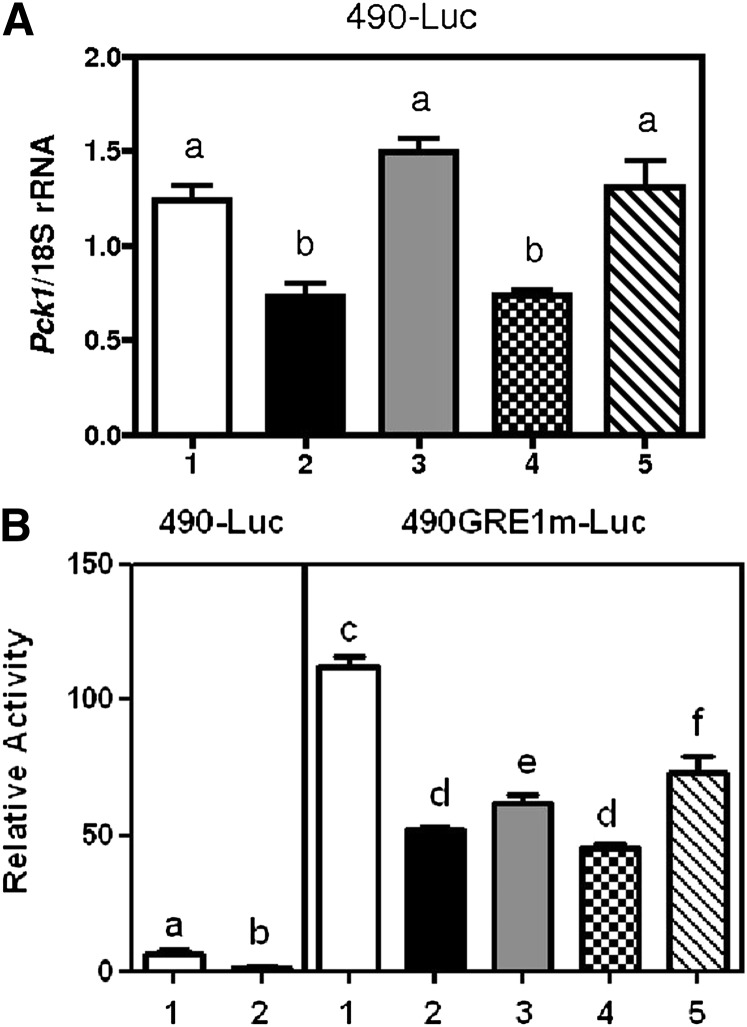
AKT and STAT5 pathways contribute to repression of Pck1 by prolactin
Upon prolactin binding to its receptor, three signaling pathways may mediate the activation of the receptor. These include the Janus kinase 2 (Jak2)/Signal transducers and activators of transcription 5 (STAT5) pathway, phosphatidylinositol-3-kinase (PI3K) pathway, and the mitogen-activated protein kinase (MAPK) pathway. To determine which pathway was necessary for the regulation of Pck1 expression in HC11 cells, we used inhibitors of each pathway and then assessed whether Pck1 mRNA expression was repressed after treatment with prolactin in −490 bp Pck1 promoter constructs and in GR1 mutated Pck1 promoter construct. We chose to test both constructs because the −490 bp Pck1 promoter only had 25% repression by prolactin. The GR1 mutated version had a more robust response. We used AKT inhibitor (AKT1/2) to test the PI3K pathway, since it is downstream of PI3K. To test the Jak2/STAT5 signaling pathway, we used Stat5 inhibitor. To test the importance of the MAPK signaling pathway, we used U0126 inhibitor, which is a highly selective inhibitor of MAPK/ERK kinase. We found that the AKT and Stat5 pathways, but not the MAPK pathway, were required for full repression of Pck1 expression in HC11 cells for both constructs (Fig. 6).
Fig. 6.
6. Inhibitors of prolactin signaling pathways. A: −490 bp to +72 bp and (B) −490 GRE1m to +72 bp Pck1 promoter were ligated to firefly luciferase and were transfected into HC11 cells. HC11 cells were treated without (open white bar) and with (shaded bars) prolactin (1 µg/ml) and inhibitor for prolactin pathway for 72 h. 1) Cells treated with dexamethasone and insulin (DI); 2) cells treated with dexamethasone, insulin and prolactin (DPI); 3) cells treated with DIP + AKT inhibitor (1 µM); 4) cells treated with DIP + Stat5 inhibitor (1 µM); 5) cells treated with DIP + MAPK inhibitor (U0126, 1 µM). Total RNA was analyzed for Pck1 expression and normalized with 18S rRNA. Values are the mean ± SEM, n = 4; subscripts with a different letter are statistically significant, P < 0.05.
Function of Pck1 in mammary epithelial cells
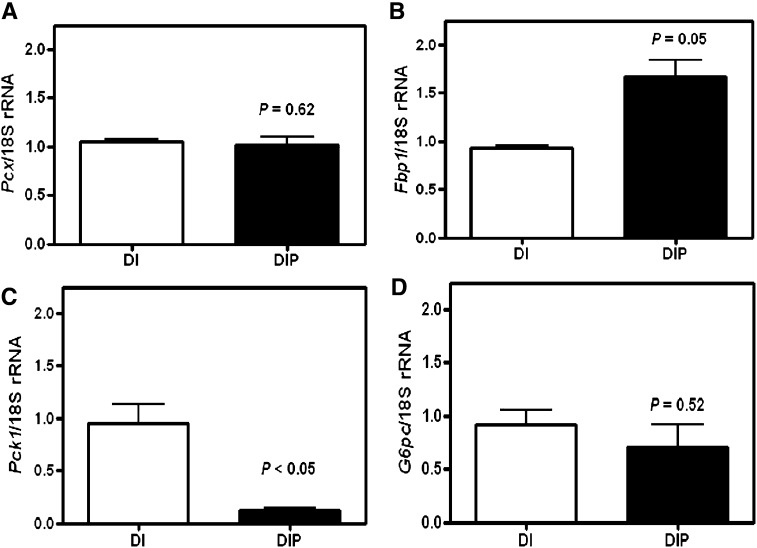
Previously, we have shown that the function of Pck1 in the adipocytes of the mammary gland was glyceroneogenesis (15). To determine the function of Pck1 in HC11 epithelial cells, we first measured the genes in the gluconeogenic pathway, pyruvate carboxylase (Pcx), fructose-6-bisphosphatase (Fbp), Pck1, and glucose-6-phosphatase (G6p) in HC11 cells treated with or without prolactin. We found that all of these gluconeogenic genes were expressed in HC11 cells. However, Pcx and G6p mRNA expression was not regulated by prolactin. Fbp had increased mRNA expression with prolactin treatment, whereas Pck1 expression was repressed (Fig. 7).
Fig. 7.
7. Expression of gluconeogenic genes. RNA was isolated from HC11 cells treated with the lactogenic hormones dexamethasone, insulin, and prolactin (DIP) or lactogenic hormones minus prolactin [dexamethasone and insulin (DI)] as described in Experimental Procedures. Gene expression was measured by RT-PCR and normalized with 18S rRNA for A: pyruvate carboxylase, Pcx, B: fructose bisphosphatase, Fbp1, C: phosphoenolpyruvate carboxykinase, Pck1 and D: glucose-6-phosphatase, G6P. Values are the mean ± SEM, n = 4–6.
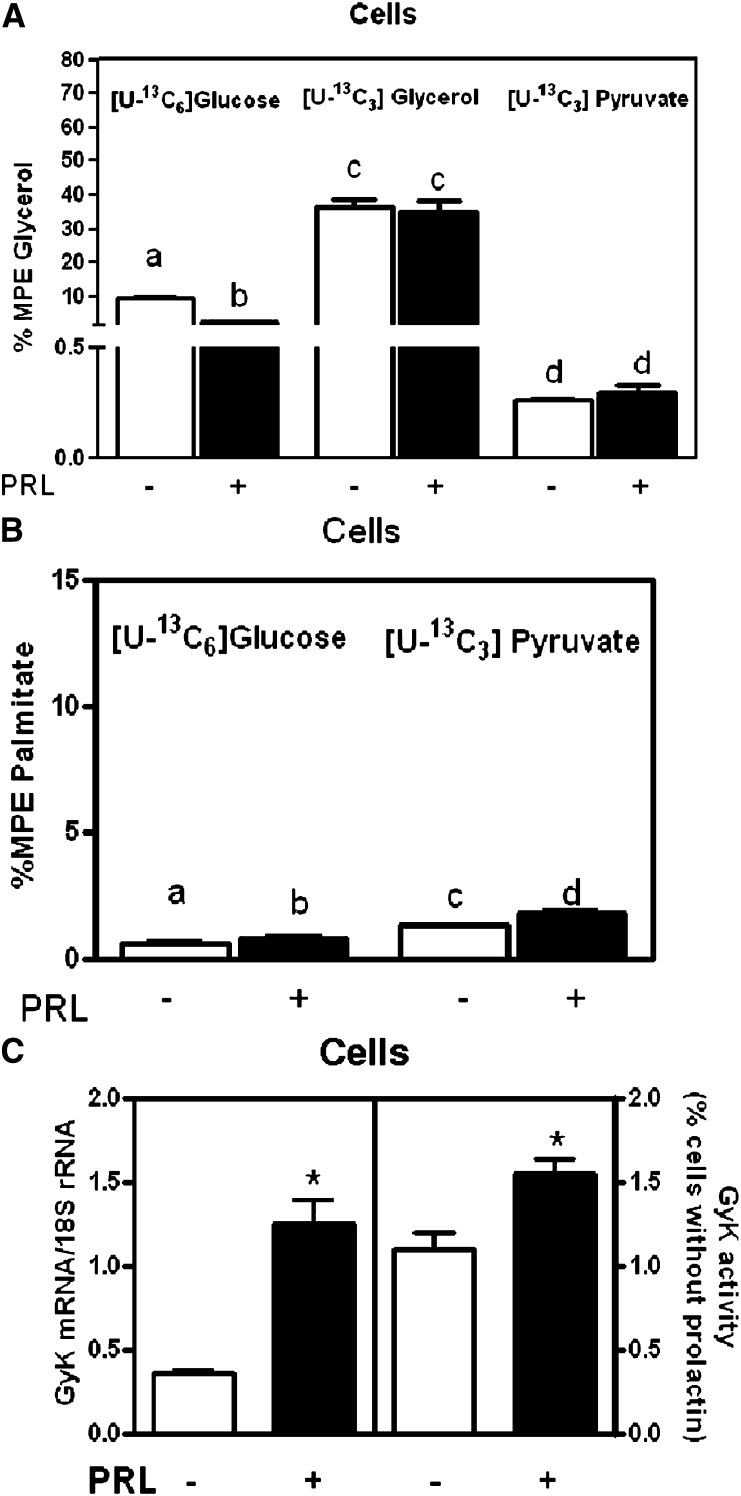
To analyze the metabolic function of these genes in HC11 cells, we measured synthesis of triglyceride (synthesized glycerol bound to triglyceride) from either [U-13C6]glucose, [U-13C3]glycerol or [U-13C3]pyruvate using MS analyses (Fig. 8A). We established incorporation of [U-13C 6]glucose into triglyceride-bound glycerol (Fig. 8A), suggesting that glycerol-3-phosphate was synthesized from glucose through glycolysis. We also found significant incorporation of label into triglyceride-bound glycerol using [U-13C3]glycerol (Fig. 8A), suggesting that glycerol is a precursor for glycerol-3-phosphate via glycerol kinase reaction. Finally, we found incorporation of [U-13C3]pyruvate into triglyceride-bound glycerol, supporting the role of glyceroneogenesis through Pck1. We also measured incorporation of either glucose or pyruvate into the newly synthesized FA, palmitate (Fig. 8B). We found that both can contribute to FA synthesis.
Fig. 8.
8. Synthesis of glycerol-bond triglyceride and palmitate in HC11 cells. A: Percent MPE glycerol synthesis from labeled [U-13C6]glucose, labeled [U-13C3]glycerol, or [U-13C3]pyruvate in HC11 cells treated with the lactogenic hormones dexamethasone, insulin, and prolactin [DIP (black bars)] or lactogenic hormones minus prolactin [dexamethasone and insulin (DI) open white bars] as described in Experimental Procedures. B: Percent MPE palmitate synthesis from labeled [U-13C3]pyruvate and [U-13C6]glucose in cells from HC11-treated cells. C: Glycerol kinase mRNA expression normalized to 18S rRNA and Gyk activity in HC11-treated cells. The data are expressed as the means ± SEM, n = 6; *P < 0.05. Subscripts that are different are statistically significant, P < 0.05.
Because the amount of newly made triglyceride-bound glycerol was significant from [U-13C3]glycerol, we tested the expression of glycerol kinase (GyK) and its activity in HC11 cells. We found a 70% increase in GyK mRNA expression and a 50% increase in GyK activity with prolactin administration. Therefore GyK also contributes to the formation of the glycerol-3-phosphate backbone (Fig. 8C).
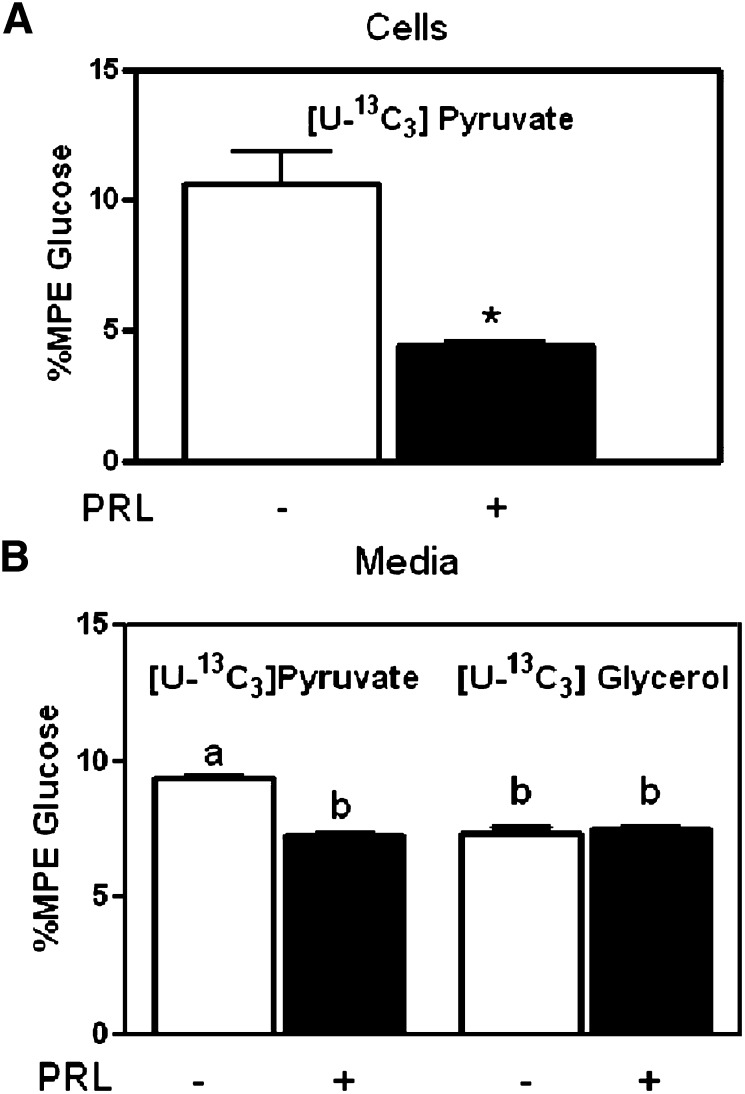
To further define the metabolic function of Pck1 in HC11 cells, we measured synthesis of glucose from [U-13C3]glycerol or [U-13C3]pyruvate using MS analyses (Fig. 8). We found significant incorporation of [U-13C3]pyruvate into glucose in cells and media that was reduced with prolactin treatment (Fig. 9A, B), suggesting that gluconeogenesis occurred through Pck1. We also found significant incorporation of [U-13C3]glycerol into glucose in the media that was not regulated by prolactin treatment (Fig. 9B), suggesting that gluconeogenesis was generating glucose through F6P and bypassing the Pcx and Pck1 steps in gluconeogenesis.
Fig. 9.
9. Glucose synthesis in HC11 cells. A: Percent MPE glucose synthesis from labeled [U-13C3]pyruvate in HC11 cells treated with the lactogenic hormones dexamethasone, insulin, and prolactin [(DIP) black bars] or lactogenic hormones minus prolactin [dexamethasone and insulin (DI) open white bars] as described in Experimental Procedures. B: Percent MPE glucose synthesis from labeled [U-13C3]pyruvate and [U-13C3]glycerol in media from HC11-treated cells. The data are expressed as the means ± SEM, n = 6; *P < 0.05. Subscripts that are different are statistically significant, P < 0.05.
DISCUSSION
In this study, we investigated the role of Pck1 in mammary gland epithelial cells during lactation in mice and in HC11 cells. Previously, mammary gland expression of Pck1 mRNA has been studied using transgenic mice containing a chimeric gene with the promoter regulatory regions of rat Pck1 linked to the bovine growth hormone structural gene. In these studies, expression of the transgene is detected in the mammary gland and requires the −355 bp to −460 bp region of the promoter for expression (19). However, our data indicated that the region from −490 bp to −202 bp in the promoter was required for Pck1 expression in the HC11 cells. The discrepancy may be explained by the use of a different reporter gene. Although we used the same rat promoter as McGrane et al. (19), we used a different reporter gene. McGrane et al. used a growth hormone reporter gene, whereas we used a luciferase reporter gene. In the McGrane et al. studies, they analyzed transgenic mice, which also may be influenced by site of insertion and the enhancers near the transgene.
The presence of Pck1 activity in the mammary gland of lactating rats and its coordinate regulation by prolactin, glucocorticoids, and insulin has been previously discussed by Lobato et al. (45). Lobato et al. administered 2-bromo-α-ergocryptine, which reduces serum prolactin levels in lactating rats. Prolactin was given back by injection of prolactin, and mammary gland tissue was isolated. Lobato et al. found a 50% increase of Pck1 activity with administration of prolactin (38). The mammary gland tissue is composed of several cell types, including epithelial cells and adipocytes. We suggest that the increase of Pck1 activity for the Lobato studies was due to changes in adipocyte expression of Pck1, because in our studies, we found prolactin increased Pck1 mRNA expression in isolated adipocytes from the mammary gland after prolactin administration. Prolactin has been shown not only to regulate lactation in the mammary gland, but also to regulate adipocyte metabolism (46). In WAT, Pck1 is increased during fasting to reesterify FA back to triglycerides after lipolysis. A key transcription factor that induces Pck1 expression in WAT is PPARγ. We found that prolactin induces PPARγ expression in mammary gland adipose cells. Thus, we propose that prolactin induces glyceroneogenesis in the adipocytes to reesterify excess FAs acids back to triglycerides after lipolysis. We hypothesize that this is necessary to maintain lipid stores in the mammary gland adipocytes for triglyceride synthesis during lactation.
Previously Jimenez et al. (14) established a role for glyceroneogenesis in mammary gland epithelial cells. They analyzed incorporation of labeled acetate and oleate into glycerol-3-phosphate in isolated acini from lactating Wistar rats. However, they suggest that the last steps of glyconeogenesis between triose-phosphate and glucose-6-phosphate are not operative in rat mammary gland acinar cells (14). In our HC11 studies, we found that all of the key gluconeogenic genes were expressed (Pcx, Pck1, F6p, and G6p). We showed in our stable isotope studies that glucose was synthesized from labeled pyruvate and that glycerol and was released into the media. Thus, the gluconeogenic pathways were active in the HC11 epithelial cell line. Our isotope data also suggest that glucose and glycerol are the main substrates for glycerol-3-phosphate formation for triglyceride synthesis in HC11 cells. This supports earlier findings by Rao and Abraham (47) that found glucose oxidation for NADPH production and glycerol-3-phosphate production is required for FA synthesis. Our data also suggest that the role of Pck1 in HC11 cells (epithelial cells) is glyceroneogenesis. We detected incorporation of labeled pyruvate into triglyceride-bound glycerol in HC11 cells.
The Pck1 promoter has been well characterized (41, 48–53). During fasting and diabetes, cAMP, as well as glucocorticoids, is elevated and induces hepatic Pck1 expression. The regions of the promoter responsible for glucocorticoid regulation span –455 bp to –321 bp in the Pck1 promoter and include AF1, AF2, and AF3. The proper positions of AF1, AF2, and AF3 in the Pck1 promoter are required for complete glucocorticoid response (44). In the current study, we found that these AF regions in the Pck1 promoter are necessary for regulation of mRNA expression by prolactin in HC11 cells. In the mammary gland, prolactin binds to the extracellular domain of the prolactin receptor and causes dimerization and activation of Jak2, a tyrosine kinase (54–57). Phosphorylation of the intracellular domain recruits Stat5 through its SH2 domain to the receptor and phosphorylates Stat5, resulting in dimerization and nuclear localization. Specific DNA binding of Stat5 response element (Stat5-RE) TTCNNNGAA results in induction of gene transcription. Glucocorticoids can bind to its receptor and translocate into the nucleus. The glucocorticoid receptor (GR) binds to its glucocorticoid response element (GRE) AGAACANNNTGTTCT in the promoter. In the presence of both hormones, Stat5 and the GR form a molecular complex that diminishes the glucocorticoid response of a GRE. Complex formation between Stat5 and the GR prevents binding to the consensus promoter sequence and therefore decreases transcription (58). The promoter of Pck1 does not have a consensus Stat5-RE, but does have glucocorticoid response unit (GRU), which consists of two GREs (GR1, GR2) and three AF (AF1, AF2, AF3) regions in the −490 bp upstream of the start site. In our model, Pck1 expression required all three AF sites but not the GRE sites for full repression by prolactin. From the inhibitor studies, we found that the Stat5 pathway is also required for Pck1. We propose that Stat5 is interacting with glucocorticoids bound to GR, thus inhibiting binding to the GRU and decreasing Pck1 gene expression.
FoxO transcription factors are widely expressed (59) and are important targets of insulin action (60). FoxO proteins form a subgroup within the family of Forkhead box transcription factors (61). FoxO1 is expressed in the liver and has been shown to stimulate expression of genes in the gluconeogenic pathway (Pck1 and glucose-6-phosphatase) (62, 63). FoxO1 interacts with insulin response sequences in the promoter of Pck1 (64, 65). Inactivation of FoxO1 is through the action of insulin binding to its receptor and activating the insulin signaling cascade. AKT phosphorylates FoxO1, and the transcription factor then translocates into the cytoplasm. As a result of this translocation, when insulin is present, Pck1 mRNA expression is reduced. In our study, we found reduced expression of FoxO1 mRNA in mammary epithelium cells after prolactin treatment. Using inhibitors for signaling pathways, we established that the AKT pathway is also required for Pck1 repression by prolactin. Therefore, we propose that prolactin requires both the AKT and Stat5 pathways for repression of Pck1 in HC11 cells. However, in the inhibitor studies, we did not fully restore 490GRE1m-Luc expression to untreated levels. This suggests that although AKT and Stat5 pathways contribute to regulation of Pck1 mRNA expression, other factors are also required
During lactation, why is Pck1 mRNA expression induced in mammary gland adipocytes while it is reduced in mammary gland epithelial cells? We propose that the increased Pck1 mRNA expression in mammary gland adipocytes during lactation maintains mammary gland adipocyte lipid stores through reesterification of glycerol back to glycerol-3-phosphate. Thus, the adipocytes surrounding the epithelial cells, as well as the diet, would be important sources of FAs and glycerol for milk triglycerides. In the epithelial cells, we found that Pck1 mRNA was reduced in response to prolactin and participated in gluconeogenesis and glyceroneogenesis. Lobato et al. (45) have shown that hepatic Pck1 mRNA is increased with prolactin administration. Thus, we hypothesize that the mammary gland relies on hepatic gluconeogenesis for glucose production and hepatic glyceroneogenesis for triglyceride synthesis for milk droplets during lactation. In addition GyK expression and activity within mammary gland epithelial cells also contributed to triglyceride production during lactation; we have shown an increase of Gyk activity in HC11 cells with prolactin administration. This supports that the mammary epithelial cells use glycerol from either the diet or hydrolyzed triglycerides from surrounding adipocytes or other fat stores for glycerol-3-phosphate synthesis and triglyceride formation. Does Pck1 have a different role in the mammary gland earlier in development? Because we found the highest expression of Pck1 mRNA at 4 weeks in isolated mammary epithelial cells, it is possible that Pck1 plays an important role in providing glucose and/or triglycerides during ductal elongation at puberty. Future studies will elucidate the metabolic role of Pck1 in the developing mammary gland at this earlier time.
Supplementary Material
Footnotes
Abbreviations:
- AF
- accessory factor
- BAT
- brown adipose tissue
- CREB
- cAMP response element binding
- DI
- dexamethasone and insulin
- DIP
- dexamethasone, insulin and prolactin
- EGF
- epidermal growth factor
- GCMS
- gas chromatography mass spectrometry
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element
- GRU
- glucocorticoid response unit
- MAPK
- mitogen-activated protein kinase
- MPE
- mass percent enrichment
- PPARγ
- peroxisome proliferator-activated receptor γ
- TCA
- tricarboxylic acid
- WAT
- white adipose tissue
- WT
- wild type
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK-075040 (CMC). The stable isotope studies were performed at the Case Western Reserve University (Cleveland, Ohio) Mouse Metabolic Phenotyping Center supported by NIDDK Grant DK-76769.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and one table.
REFERENCES
- 1.Vernon R. G., Flint D. J. 1983. Control of fatty acid synthesis in lactation. Proc. Nutr. Soc. 42: 315–331. [DOI] [PubMed] [Google Scholar]
- 2.Williamson D. H. 1980. Integration of metabolism in tissues of the lactating rat. FEBS Lett. 117 (Suppl.): K93–K105. [DOI] [PubMed] [Google Scholar]
- 3.Flint D. J., Clegg R. A., Vernon R. G. 1983. Adipose tissue metabolism during early pregnancy in the rat: temporal relationships of changes in the metabolic activity, number of insulin receptors, and serum hormone concentrations. Arch. Biochem. Biophys. 224: 677–681. [DOI] [PubMed] [Google Scholar]
- 4.Robinson A. M., Williamson D. H. 1978. Control of glucose metabolism in isolated acini of the lactating mammary gland of the rat. Effects of oleate on glucose utilization and lipogenesis. Biochem. J. 170: 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinnett-Smith P. A., Vernon R. G., Mayer R. J. 1980. Lipogenic enzymes in rat maternal adipose tissue in the perinatal period. Biochem. J. 186: 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubero A., Ros M., Lobato M. F., Carcia-Ruiz J. P., Moreno F. J. 1983. Coordination of glucose metabolism and NADPH formation in the adipose tissue and mammary gland during the lactation-weaning transition. Enzyme. 30: 38–47. [DOI] [PubMed] [Google Scholar]
- 7.Ros M., Lobato M. F., Garcia-Ruiz J. P., Moreno F. J. 1990. Integration of lipid metabolism in the mammary gland and adipose tissue by prolactin during lactation. Mol. Cell. Biochem. 93: 185–194. [DOI] [PubMed] [Google Scholar]
- 8.Ros M., Alonso G., Moreno F. J. 1992. Effects of litter removal on the lipolytic response and the regulatory components of the adenylate cyclase in adipocytes isolated from lactating rats. Biochem. J. 281: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballard F. J., Hanson R. W., Leveille G. A. 1967. Phosphoenolpyruvate carboxykinase and the synthesis of glyceride-glycerol from pyruvate in adipose tissue. J. Biol. Chem. 242: 2746–2750. [PubMed] [Google Scholar]
- 10.Reshef L., Niv J., Shapiro B. 1967. Effect of propionate on pyruvate metabolism in adipose tissue. J. Lipid Res. 8: 688–691. [PubMed] [Google Scholar]
- 11.Reshef L., Niv J., Shapiro B. 1967. Effect of propionate on lipogenesis in adipose tissue. J. Lipid Res. 8: 682–687. [PubMed] [Google Scholar]
- 12.Ballard F. J., Hanson R. W. 1967. Phosphoenolpyruvate carboxykinase and pyruvate carboxylase in developing rat liver. Biochem. J. 104: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reshef L., Hanson R. W., Ballard F. J. 1970. A possible physiological role for glyceroneogenesis in rat adipose tissue. J. Biol. Chem. 245: 5979–5984. [PubMed] [Google Scholar]
- 14.Jimenez J., Page-Penuelas A., Ros M., Garcia-Ruiz J. P., Moreno F. J. 1987. Glycerogenic pathway in the rat mammary gland. Int. J. Biochem. 19: 201–204. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh C. W., Millward C. A., DeSantis D., Pisano S., Machova J., Perales J. C., Croniger C. M. 2009. Reduced milk triglycerides in mice lacking phosphoenolpyruvate carboxykinase in mammary gland adipocytes and white adipose tissue contribute to the development of insulin resistance in pups. J. Nutr. 139: 2257–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardones P., Strobel P., Miranda S., Leighton F., Quinones V., Amigo L., Rozowski J., Krieger M., Rigotti A. 2002. Alpha-tocopherol metabolism is abnormal in scavenger receptor class B type I (SR-BI)-deficient mice. J. Nutr. 132: 443–449. [DOI] [PubMed] [Google Scholar]
- 17.Sebastian B. M., Kang L., Chen X., Nagy L. E. 2008. Methods to investigate the effects of chronic ethanol on adipocytes. Methods Mol. Biol. 447: 357–366. [DOI] [PubMed] [Google Scholar]
- 18.Smalley M. J., Titley J., Paterson H., Perusinghe N., Clarke C., O'Hare M. J. 1999. Differentiation of separated mouse mammary luminal epithelial and myoepithelial cells cultured on EHS matrix analyzed by indirect immunofluorescence of cytoskeletal antigens. J. Histochem. Cytochem. 47: 1513–1524. [DOI] [PubMed] [Google Scholar]
- 19.McGrane M. M., Yun J. S., Roesler W. J., Park E. A., Wagner T. E., Hanson R. W. 1990. Developmental regulation and tissue-specific expression of a chimaeric phosphoenolpyruvate carboxykinase/bovine growth hormone gene in transgenic animals. J Reprod Fertil Suppl 41: 17–23. [PubMed] [Google Scholar]
- 20.Logie L., Ruiz-Alcaraz A. J., Keane M., Woods Y. L., Bain J., Marquez R., Alessi D. R., Sutherland C. 2007. Characterization of a protein kinase B inhibitor in vitro and in insulin-treated liver cells. Diabetes. 56: 2218–2227. [DOI] [PubMed] [Google Scholar]
- 21.Duncia J. V., Santella III J. B., Higley C. A., Pitts W. J., Wityak J., Frietze W. E., Rankin F. W., Sun J. H., Earl R. A., Tabaka A. C., et al. 1998. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg. Med. Chem. Lett. 8: 2839–2844. [DOI] [PubMed] [Google Scholar]
- 22.Muller J., Sperl B., Reindl W., Kiessling A., Berg T. 2008. Discovery of chromone-based inhibitors of the transcription factor STAT5. ChemBioChem. 9: 723–727. [DOI] [PubMed] [Google Scholar]
- 23.Bederman I. R., Foy S., Chandramouli V., Alexander J. C., Previs S. F. 2009. Triglyceride synthesis in epididymal adipose tissue: contribution of glucose and non-glucose carbon sources. J. Biol. Chem. 284: 6101–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu L., Zhang G. F., Kombu R. S., Allen F., Kutz G., Brewer W. U., Roe C. R., Brunengraber H. 2010. Parenteral and enteral metabolism of anaplerotic triheptanoin in normal rats. II. Effects on lipolysis, glucose production, and liver acyl-CoA profile. Am. J. Physiol. Endocrinol. Metab. 298: E362–E371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., Kombu R. S., Kasumov T., Zhu S. H., Cendrowski A. V., David F., Anderson V. E., Kelleher J. K., Brunengraber H. 2008. Metabolomic and mass isotopomer analysis of liver gluconeogenesis and citric acid cycle. I. Interrelation between gluconeogenesis and cataplerosis; formation of methoxamates from aminooxyacetate and ketoacids. J. Biol. Chem. 283: 21978–21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Previs S. F., Fernandez C. A., Yang D., Soloviev M. V., David F., Brunengraber H. 1995. Limitations of the mass isotopomer distribution analysis of glucose to study gluconeogenesis. Substrate cycling between glycerol and triose phosphates in liver. J. Biol. Chem. 270: 19806–19815. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez C. A., Des Rosiers C., Previs S. F., David F., Brunengraber H. 1996. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31: 255–262. [DOI] [PubMed] [Google Scholar]
- 28.McMenamy R. H., Kleineke J., Roil W., Soling H. D. 1981. Perifusion system for isolated cells. Anal. Biochem. 112: 117–127. [DOI] [PubMed] [Google Scholar]
- 29.Millward C. A., Desantis D., Hsieh C. W., Heaney J. D., Pisano S., Olswang Y., Reshef L., Beidelschies M., Puchowicz M., Croniger C. M. 2010. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/fatty acid cycle and development of insulin resistance in mice. J. Lipid Res. 51: 1452–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hibuse T., Maeda N., Funahashi T., Yamamoto K., Nagasawa A., Mizunoya W., Kishida K., Inoue K., Kuriyama H., Nakamura T., et al. 2005. Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc. Natl. Acad. Sci. USA. 102: 10993–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan H. P., Li Y., Jensen M. V., Newgard C. B., Steppan C. M., Lazar M. A. 2002. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat. Med. 8: 1122–1128. [DOI] [PubMed] [Google Scholar]
- 32.Olswang Y., Cohen H., Papo O., Cassuto H., Croniger C. M., Hakimi P., Tilghman S. M., Hanson R. W., Reshef L. 2002. A mutation in the peroxisome proliferator-activated receptor gamma-binding site in the gene for the cytosolic form of phosphoenolpyruvate carboxykinase reduces adipose tissue size and fat content in mice. Proc. Natl. Acad. Sci. USA. 99: 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono H., Shimano H., Katagiri H., Yahagi N., Sakoda H., Onishi Y., Anai M., Ogihara T., Fujishiro M., Viana A. Y., et al. 2003. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 52: 2905–2913. [DOI] [PubMed] [Google Scholar]
- 34.Whiteman E. L., Cho H., Birnbaum M. J. 2002. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 13: 444–451. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto M., Pocai A., Rossetti L., Depinho R. A., Accili D. 2007. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6: 208–216. [DOI] [PubMed] [Google Scholar]
- 36.Gross D. N., Wan M., Birnbaum M. J. 2009. The role of FOXO in the regulation of metabolism. Curr. Diab. Rep. 9: 208–214. [DOI] [PubMed] [Google Scholar]
- 37.Ball R. K., Friis R. R., Schoenenberger C. A., Doppler W., Groner B. 1988. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 7: 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chammas R., Taverna D., Cella N., Santos C., Hynes N. E. 1994. Laminin and tenascin assembly and expression regulate HC11 mouse mammary cell differentiation. J. Cell Sci. 107: 1031–1040. [DOI] [PubMed] [Google Scholar]
- 39.Doppler W., Hock W., Hofer P., Groner B., Ball R. K. 1990. Prolactin and glucocorticoid hormones control transcription of the beta-casein gene by kinetically distinct mechanisms. Mol. Endocrinol. 4: 912–919. [DOI] [PubMed] [Google Scholar]
- 40.Doppler W., Groner B., Ball R. K. 1989. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc. Natl. Acad. Sci. USA. 86: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott D. K., Stromstedt P. E., Wang J. C., Granner D. K. 1998. Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol. Endocrinol. 12: 482–491. [DOI] [PubMed] [Google Scholar]
- 42.Stafford J. M., Waltner-Law M., Granner D. K. 2001. Role of accessory factors and steroid receptor coactivator 1 in the regulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. J. Biol. Chem. 276: 3811–3819. [DOI] [PubMed] [Google Scholar]
- 43.Stafford J. M., Wilkinson J. C., Beechem J. M., Granner D. K. 2001. Accessory factors facilitate the binding of glucocorticoid receptor to the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 276: 39885–39891. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama T., Scott D. K., Wang J. C., Granner D. K. 1998. Structural requirements of the glucocorticoid and retinoic acid response units in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 12: 1487–1498. [DOI] [PubMed] [Google Scholar]
- 45.Lobato M. F., Careche M., Ros M., Moreno F. J., Garcia-Ruiz J. P. 1985. Effect of prolactin and glucocorticoids on P-enolpyruvate carboxykinase activity in liver and mammary gland from diabetic and lactating rats. Mol. Cell. Biochem. 67: 19–23. [DOI] [PubMed] [Google Scholar]
- 46.Brandebourg T., Hugo E., Ben-Jonathan N. 2007. Adipocyte prolactin: regulation of release and putative functions. Diabetes Obes. Metab. 9: 464–476. [DOI] [PubMed] [Google Scholar]
- 47.Rao G. A., Abraham S. 1975. Stimulatory effect of glucose upon triglyceride synthesis from acetate, decanoate, and palmitate by mammary gland slices from lactating mice. Lipids. 10: 409–412. [DOI] [PubMed] [Google Scholar]
- 48.Yamada K., Duong D. T., Scott D. K., Wang J. C., Granner D. K. 1999. CCAAT/enhancer-binding protein beta is an accessory factor for the glucocorticoid response from the cAMP response element in the rat phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 274: 5880–5887. [DOI] [PubMed] [Google Scholar]
- 49.Hall R. K., Yamasaki T., Kucera T., Waltner-Law M., O'Brien R., Granner D. K. 2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 275: 30169–30175. [DOI] [PubMed] [Google Scholar]
- 50.Cassuto H., Olswang Y., Heinemann S., Sabbagh K., Hanson R. W., Reshef L. 2003. The transcriptional regulation of phosphoenolpyruvate carboxykinase gene in the kidney requires the HNF-1 binding site of the gene. Gene. 318: 177–184. [DOI] [PubMed] [Google Scholar]
- 51.Waltner-Law M., Duong D. T., Daniels M. C., Herzog B., Wang X. L., Prasad R., Granner D. K. 2003. Elements of the glucocorticoid and retinoic acid response units are involved in cAMP-mediated expression of the PEPCK gene. J. Biol. Chem. 278: 10427–10435. [DOI] [PubMed] [Google Scholar]
- 52.Olswang Y., Blum B., Cassuto H., Cohen H., Biberman Y., Hanson R. W., Reshef L. 2003. Glucocorticoids repress transcription of phosphoenolpyruvate carboxykinase (GTP) gene in adipocytes by inhibiting its C/EBP-mediated activation. J. Biol. Chem. 278: 12929–12936. [DOI] [PubMed] [Google Scholar]
- 53.Tontonoz P., Hu E., Devine J., Beale E. G., Spiegelman B. M. 1995. PPAR gamma 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol. Cell. Biol. 15: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doppler W. 1994. Regulation of gene expression by prolactin. Rev. Physiol. Biochem. Pharmacol. 124: 93–130. [DOI] [PubMed] [Google Scholar]
- 55.Groner B., Gouilleux F. 1995. Prolactin-mediated gene activation in mammary epithelial cells. Curr. Opin. Genet. Dev. 5: 587–594. [DOI] [PubMed] [Google Scholar]
- 56.Groner B. 2002. Transcription factor regulation in mammary epithelial cells. Domest. Anim. Endocrinol. 23: 25–32. [DOI] [PubMed] [Google Scholar]
- 57.Hynes N. E., Cella N., Wartmann M. 1997. Prolactin mediated intracellular signaling in mammary epithelial cells. J. Mammary Gland Biol. Neoplasia. 2: 19–27. [DOI] [PubMed] [Google Scholar]
- 58.Groner B., Shemanko C. 2002. Cooperation of nuclear transcription factors regulated by steroid and peptide hormones. Ernst Schering Res. Found. Workshop. 213–231. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W., Patil S., Chauhan B., Guo S., Powell D. R., Le J., Klotsas A., Matika R., Xiao X., Franks R., et al. 2006. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281: 10105–10117. [DOI] [PubMed] [Google Scholar]
- 60.Barthel A., Schmoll D., Unterman T. G. 2005. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 16: 183–189. [DOI] [PubMed] [Google Scholar]
- 61.Kaestner K. H., Knochel W., Martinez D. E. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14: 142–146. [PubMed] [Google Scholar]
- 62.Nakae J., Cao Y., Oki M., Orba Y., Sawa H., Kiyonari H., Iskandar K., Suga K., Lombes M., Hayashi Y. 2008. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 57: 563–576. [DOI] [PubMed] [Google Scholar]
- 63.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 423: 550–555. [DOI] [PubMed] [Google Scholar]
- 64.Unterman T. G., Fareeduddin A., Harris M. A., Goswami R. G., Porcella A., Costa R. H., Lacson R. G. 1994. Hepatocyte nuclear factor-3 (HNF-3) binds to the insulin response sequence in the IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem. Biophys. Res. Commun. 203: 1835–1841. [DOI] [PubMed] [Google Scholar]
- 65.O'Brien R. M., Noisin E. L., Suwanichkul A., Yamasaki T., Lucas P. C., Wang J. C., Powell D. R., Granner D. K. 1995. Hepatic nuclear factor 3- and hormone-regulated expression of the phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein 1 genes. Mol. Cell. Biol. 15: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.