Abstract
Shiga toxin–producing Escherichia coli O157 is a leading cause of foodborne illness worldwide. To evaluate better methods to rapidly detect and genotype E. coli O157 strains, the present study evaluated the use of ampliPHOX, a novel colorimetric detection method based on photopolymerization, for pathogen identification with DNA microarrays. A low-density DNA oligonucleotide microarray was designed to target stx1 and stx2 genes encoding Shiga toxin production, the eae gene coding for adherence membrane protein, and the per gene encoding the O157-antigen perosamine synthetase. Results from the validation experiments demonstrated that the use of ampliPHOX allowed the accurate genotyping of the tested E. coli strains, and positive hybridization signals were observed for only probes targeting virulence genes present in the reference strains. Quantification showed that the average signal-to-noise ratio values ranged from 47.73 ± 7.12 to 76.71 ± 8.33, whereas average signal-to-noise ratio values below 2.5 were determined for probes where no polymer was formed due to lack of specific hybridization. Sensitivity tests demonstrated that the sensitivity threshold for E. coli O157 detection was 100–1000 CFU/mL. Thus, the use of DNA microarrays in combination with photopolymerization allowed the rapid and accurate genotyping of E. coli O157 strains.
Introduction
Shiga toxin–producing Escherichia coli O157:H7 (E. coli O157) is an enteric pathogen known to cause human gastrointestinal illnesses with clinical spectra, ranging from bloody diarrhea and hemorrhagic colitis to life-threatening hemolytic-uremic syndrome (Rangel et al., 2005; Karmali, 2009). In recent years, most risk management efforts have focused on identifying the presence and distribution of E. coli O157 in foods as part of imperative constituents of food safety programs (Karmali, 2009; Buchanan and Appel, 2010). Thus, the O157 serotype is considered to be the most commonly reported serotype linked to outbreaks in North America.
The rise in foodborne-related outbreaks of E. coli O157 has heightened the importance of developing improved methods to rapidly detect and characterize virulent strains (Bettelheim and Beutin, 2003; Bettelheim, 2007; Buchanan and Appel, 2010). Established culturing methods for the identification of pathogenic E. coli can be labor intensive and time consuming and assess phenotypic and genotypic markers by screening only a small number of determinants. Molecular-based technologies, such as DNA microarrays, offer a viable alternative to screen multiple markers simultaneously (Uttamchandani et al., 2009; Call, 2005).
One challenge of using the DNA microarray platforms for pathogen detection has been developing cost effective as well as sensitive procedures that would indicate the positive signals on the array (Call, 2005; Kuck and Taylor, 2008; Vora et al., 2008). Fluorescent assays are commonly used methods for microarray-based detection of pathogens. However, limitations of these assays are that labeling of target DNA can be inconsistent and highly variable, and they utilize expensive and nonportable scanners for data acquisition and analysis (Call, 2005; Kuck and Taylor, 2008; Vora et al., 2008). Other nonfluorescent, colorimetric assays employ the use of unstable reagents that require temperature-controlled environments and that have variable development times, leading to an increase in nonspecific backgrounds (Kuck and Taylor, 2008). Improved procedures for pathogen surveillance are thus needed with sufficient sensitivity, cost effectiveness, and suitability for routine testing. To implement the use of better detection methods for categorizing E. coli O157 strains, the present study evaluated ampliPHOX, a novel and innovative colorimetric procedure that is based on light-initiated signal amplification through polymerization (Kuck and Taylor, 2008; Sikes et al., 2008; Dawson et al., 2009). Our findings demonstrate that photopolymerization is a simple and rapid method for the accurate genotyping of E. coli O157 strains with DNA microarrays.
Materials and Methods
Bacterial strains and growth conditions
E. coli O157:H7 strains RM1600 (feedlot strain 8) and RM1697 (feedlot strain 42) were isolated from beef cattle feed yards in Kansas (Kimura et al., 2000). E. coli O157:H7 strains RM4876 and RM6011 were isolated from watershed samples collected in the Salinas Valley in California (Cooley et al., 2007; Quiñones et al., 2009). E. coli O55:H7 strain RM3654 (CPK100) and E. coli O113:H21 strain RM3655 (CPK117) were obtained from the Canadian Pediatric Kidney Disease Research Center in Ottawa, Ontario, Canada. Nonpathogenic E. coli strain RM5034 (strain K12; ATCC-29425) was obtained from the American Type Culture Collection (Manassas, VA). The genotypic and phenotypic characteristics of the E. coli reference strains were described previously (Kimura et al., 2000; Cooley et al., 2007; Quiñones et al., 2009). Bacterial cultures were propagated on Luria-Bertani (LB) agar (Difco, Detroit, MI) or were grown aerobically with constant shaking (200 rpm) in LB broth (Difco) at 37°C.
Construction of the DNA microarray
The 30-mer oligonucleotide probes targeting the intimin adherence protein (eae) (Zhang et al., 2002; Ramachandran et al., 2003), O157 O-antigen perosamine synthetase (per) (Bilge et al., 1996; Reeves et al., 1996; Shimizu et al., 1999), Shiga toxin 1 (stx1), and Shiga toxin 2 (stx2) (Nataro and Kaper, 1998; Bettelheim, 2007; Serna and Boedeker, 2008) in pathogenic E. coli are shown in Table 1. The E. coli O157–specific probes were designed by using PRIMER3 software (Rozen and Skaletsky, 2000) and were purchased from Eurofins MWG Operon (Huntsville, AL) with a 5′-amino-C6 modification for covalent binding to the slide surface. The probes were spotted in triplicate at a final concentration of 20 μM on aldehydesilane-coated glass slides (Nexterion® Slide AL; SCHOTT, Inc., Elmsford, NY). As a control for the photopolymerization detection process, a synthetic, 24-mer oligonucleotide probe with 5′-amino-C6 and 3′-biotin modifications (InDevR, Inc., Boulder, CO) was spotted at a final concentration of 500 nM. This biotinylated control oligonucleotide did not have any sequence homology to any E. coli genes. The microarrays were manufactured by Applied Microarrays, Inc. (Tempe, AZ) with a spot diameter size of 250 μm and a center-to-center spacing of 700 μm. After printing, an adhesive silicone well (0.5 mm thickness, 9 mm diameter; Grace Bio-Labs, Bend, OR) was placed in the center of the printed array, and the microarray slides were stored in a desiccator until further use.
Table 1.
List of DNA Oligonucleotides Used in the DNA Microarray
| |
|
Primer sequences for PCR |
|
|
|
|
|---|---|---|---|---|---|---|
| Target gene | Probe sequences (5′ → 3′) | Forward (5′ → 3′) | Reverse (5′ → 3′) | Amplicon size (bp) | Accession no. | Reference |
| stx1A | GTTCTTATGTAATGACTGCTGAAGATGTTG | CTGTGGCAAGAGCGATGTTA | CTCAACCTTCCCCAGTTCAA | 163 | AAG57228 | This study |
| stx2A | GTTAATGGAGTTCAGTGGTAATACAATGAC | AATCAGTCGTCACTCACTGGTT | CGGCGTCATCGTATACACAG | 193 | AAG55587 | This study |
| per | TAGGCTACAATTATAGGATGACAAATATCT | GGTGAAGGTGGAATGGTTGTC | TCAGCAATTTCACGTTTTCGTG | 206 | AAG57096 | This study |
| eae | GTTAATCTGCAGAGTGGTAATAACTTTGAC | CCGCTCTTGGTATCGCTGGT | CGCTGACCCGCACCTAAA | 223 | AAG58823 | This study |
PCR, polymerase chain reaction.
Polymerase chain reaction
Polymerase chain reaction (PCR) reagents were supplied by New England Biolabs (Ipswich, MA). PCR primers were purchased from Eurofins MWG Operon with a 5′-phosphorylated modification for the forward primers and a 5′-biotin modification for the reverse primers (Table 1). For preparing bacterial crude lysates, cultures of the reference strains were grown aerobically in LB broth for 24 h, and 100 μL of the bacterial culture was collected by centrifugation at 2000 g for 5 min. Cell pellets were resuspended in 100 μL of HyPure™ molecular biology-grade water (HyClone Laboratories, Inc., Logan, UT) and incubated at 100°C for 20 min. The lysates were centrifuged at 2000 g for 5 min, and the supernatants were collected and frozen until further use. For experiments that assessed the sensitivity threshold of the detection assay, cell lysates were prepared from tenfold dilutions of a culture from E. coli O157 strain RM1697. Multiplex PCR amplifications were performed in a 50 μL reaction mixture, each containing 5 μL of the bacterial crude lysate, 0.5 μM of eae primers, 0.1 μM (each) of per, stx1, and stx2 primers, 1 × ThermoPol Taq buffer with 2 mM MgSO4, 200 μM deoxynucleoside triphosphates, and 2.5 U of Taq polymerase. The reaction mixtures were placed in a Dyad Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA) with the following settings: 2 min at 94°C, followed by 30 cycles of 30 sec at 94°C, 1 min at 60°C, 45 sec at 72°C, and a final extension time of 5 min at 72°C.
Microarray hybridization
For each hybridization reaction, 45 μL of PCR amplicons was purified by using the MinElute® PCR purification kit (Qiagen, Valencia, CA), and 20 μL of the eluate was digested with 10 U of lambda exonuclease and 1 × lambda exonuclease reaction buffer (Epicenter Biotechnologies, Madison, WI) in a final volume of 23 μL for 5 min at 37°C, followed by immediate addition of an equal volume of InDevR 2 × Hyb Buffer (InDevR, Inc.). The hybridization mixture was applied to each microarray that was previously rinsed with distilled water for 5 min, and the microarray slides were further incubated for 1 h at room temperature. After hybridization, the slides were transferred to a microarray wash bin (InDevR, Inc.) and were rinsed briefly with InDevR Wash Buffer D, Wash Buffer A for 1 min, Wash Buffer D briefly, Wash Buffer B for 5 min, and Wash Buffer C for 5 min and then dried.
Microarray labeling, signal amplification, and analysis
The specific colorimetric detection of signals on the microarray were detected with the ampliPHOX™ Detection System (InDevR, Inc.), a light-initiated signal amplification through polymerization (Kuck and Taylor, 2008; Sikes et al., 2008; Dawson et al., 2009). The hybridized microarrays were labeled subsequently with ampliTAG™ labeling solution (InDevR, Inc.) for 5 min at room temperature in a dark humidity chamber, and immediately after labeling, microarrays were rinsed briefly with Wash Buffer D, Wash Buffer C for 5 min, and then dried. For the detection of positive signals on the microarray, ampliTAG-labeled microarrays were covered with 40 μL ampliPHY™ reagent and photoactivated for 220 sec by using the ampliPHOX Reader (InDevR, Inc.). Immediately after photoactivation, arrays were rinsed with water, stained with 2 drops of ampliRED™ for 5 min and finally rinsed again with water to observe polymer formation. Color digital images of the stained arrays were collected by using the ampliPHOX Reader and the associated ampliVIEW™ software 1.0 (InDevR, Inc.). For each spot, the quantification of signal and background intensities was quantified by using ampliVIEW software and expressed as signal-to-noise ratio (SNR). SNR values were calculated as the difference between the mean pixel intensities of the signal minus the mean pixel intensities of the background divided by the standard deviation of the background (He and Zhou, 2008).
Results
A novel colorimetric method for detection of pathogenic E. coli on DNA microarrays
To implement the use of improved detection methods with DNA microarrays, the present study evaluated photopolymerization, a novel colorimetric detection method (Kuck and Taylor, 2008; Sikes et al., 2008; Dawson et al., 2009), for genotyping E. coli O157 strains. To provide a proof of concept for testing photopolymerization, a low-density 30-mer DNA oligonucleotide array was constructed to target the E. coli O157 virulence genes eae, per, stx1, and stx2 (Table 1). To determine that a gene target was present in the sample, a multiplex PCR amplification was performed to generate biotinylated fragments, ranging from 163 to 223 base pairs (Table 1). The PCR-amplified fragments were subjected to lambda exonuclease digestion to produce single-stranded DNA targets for achieving a rapid microarray hybridization (Boissinot et al., 2007). For photopolymerization to occur, a streptavidin-conjugated photoinitiator is used to specifically label the microarrays that have been hybridized with the biotin-labeled, single-stranded DNA targets (Kuck and Taylor, 2008). After irradiating at a wavelength absorbed by the photoinitiator (λ = 404 nm), polymer forms exclusively in only a few minutes where the probe sequences were spotted on the microarray (Kuck and Taylor, 2008; Dawson et al., 2009); polymer formation can be observed after staining (Kuck and Taylor, 2008).
Specificity and sensitivity for pathogenic E. coli detection with photopolymerization
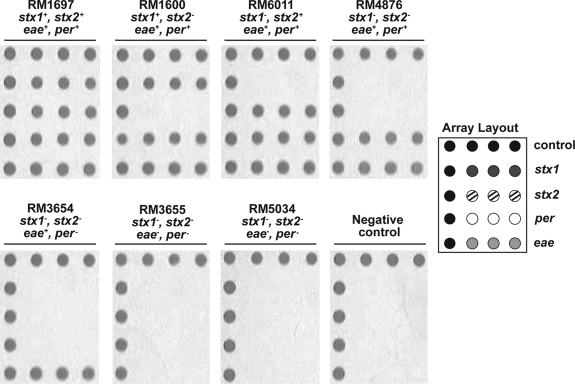
To validate the specificity of using photopolymerization with a low-density microarray for detecting pathogenic E. coli O157, several reference strains with different genotypes were examined (see Materials and Methods section). Bacterial crude lysates, prepared from the tested strains, were subjected to a multiplex PCR amplification of eae, per, stx1, and stx2 loci, and the amplified gene targets were hybridized to the DNA microarray. The labeling and subsequent microarray signal amplification was achieved using photopolymerization. The results demonstrated polymer formation for all spots targeting the genes represented on the microarray when E. coli O157 strain RM1697, a strain that harbors all four genes, was examined (Fig. 1). Polymer formation on microarray probes targeting stx1, eae, and per but not for stx2 was observed for the stx2-negative E. coli O157 strain RM1600 (Fig. 1). Further, specificity in photopolymerization was observed also when analyzing the E. coli O157 strains RM6011 and RM4876 and the E. coli O55 strain RM3654 (Fig. 2). Polymer formation was only observed on control probes for the photopolymerization reaction for the E. coli O113 strain RM3655 (Fig. 1), a strain that tested negative for the genes represented on the microarray, as determined in a PCR assay (data not shown). Similar results to those obtained for strain RM3655 were also observed for the nonpathogenic E. coli strain RM5034 and for a negative control reaction lacking any DNA template (Fig. 1). These findings demonstrated a high level of probe specificity, and the patterns of photopolymerization correlated with the genotype of the strain.
FIG. 1.
Specific identification of pathogenic Escherichia coli. E. coli reference strains with different genotypes were examined by DNA microarrays and photopolymerization (see Materials and Methods section). Each panel demonstrates polymer formation on probes spotted on the DNA microarrays. The array layout shows the construction of the microarray. Probes targeting stx1 (dark gray), stx2 (hatched), per (white), and eae (light gray) were spotted in triplicate, and positive controls for polymer formation (black) were spotted along the edges.
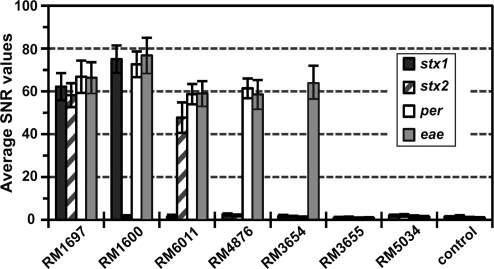
FIG. 2.
Quantification of signals on the DNA microarray. E. coli reference strains were tested by using DNA microarray and ampliPHOX detection (see Materials and Methods section). For each spot on the microarray, the signals for each spot on the microarray were quantified and expressed as signal-to-noise ratio (SNR). Each bar represents the average SNR values ± standard deviations of three independent experiments with triplicate measurements for each probe.
To quantitate the specificity of the photopolymerization reaction, the SNR values were measured for each probe on the microarray when testing the E. coli reference strains. Image quantification of E. coli O157 strain RM1697 showed that positive signals detected for all probes targeting virulence genes had SNR values, ranging from 58.2 ± 5.58 to 66.83 ± 7.55 (Fig. 2). Similarly, high SNR values, ranging from 47.73 ± 7.12 to 76.71 ± 8.33, were measured after analysis of positive signals for the other E. coli O157 strains RM1600, RM6011, and RM4876 as well as for E. coli O55 strain RM3654 (Fig. 2). Probes that had no polymer formation due to a lack of specific hybridization had significantly lower SNR values, ranging from 0.69 ± 0.39 to 2.48 ± 0.50 (Fig. 2). The results from the specificity experiments indicated a clear difference between signal intensities and background noise for probes with polymer formation.
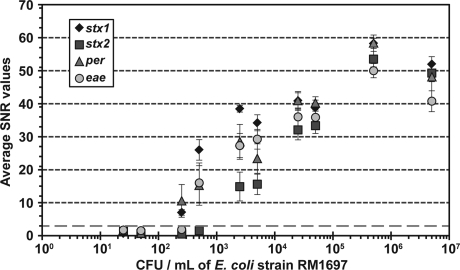
The sensitivity threshold for identifying pathogenic E. coli by using DNA microarrays with photopolymerization was further investigated. Various cell concentrations of E. coli O157 strain RM1697 were tested on the microarray, and the SNR values were quantified after performing the photopolymerization reaction. The limit of detection was defined by the findings from validation experiments for specificity (Fig. 2), demonstrating that spots on the array without specific formation of polymer yielded SNR values < 2.5. In addition, previous studies have shown that an SNR value of 3 is considered to be the minimum signal intensity for accurate quantification (Verdnick et al., 2002; He and Zhou, 2008; Park et al., 2008). Based on these observations an SNR value of 3 was established as our detection limit for positive signals (Fig. 3), and the results from the sensitivity experiments demonstrated that the threshold for detecting strain RM1697 by using DNA microarrays with photopolymerization was determined to be between 100 and 1000 CFU/mL (Fig. 3).
FIG. 3.
Sensitivity of E. coli O157 detection with DNA microarrays with ampliPHOX detection. Various cell concentrations of the reference E. coli O157 strain RM1697 were tested on the DNA microarray, and the SNR values were quantified after photopolymerization. The dashed line delineates the detection limit for positive signals, and each symbol represents the average SNR values ± standard deviations from three independent experiments with triplicate measurements for each probe.
Discussion
Outbreaks of foodborne disease caused by E. coli O157 continue to cause concern among consumers, regulatory agencies, and the food industry (Mandrell, 2009; Buchanan and Appel, 2010). To address this issue, risk management efforts have focused on identifying the presence and distribution of E. coli O157 in foods due to large outbreaks and severe illnesses associated with this particular serotype (Karmali, 2009; Buchanan and Appel, 2010). For use in surveillance and outbreak epidemiology, there is a need for the development of rapid, simple, and inexpensive assays for pathogen detection. Molecular-based technologies, such as DNA microarrays, offer an alternative to screen simultaneously for multiple virulence markers and to assist in the identification of the virulence types of foodborne pathogens (Call, 2005; Uttamchandani et al., 2009).
For pathogen detection with DNA microarrays, procedures that enzymatically incorporate a fluorescent dye during DNA amplification are most commonly used due to the availability of reagents and instrumentation (Call, 2005; Vora et al., 2008; Uttamchandani et al., 2009). However, these fluorescent assays can lack consistency in the labeling of the DNA and may require procedures that are time consuming and use expensive scanners for data analysis (Kuck and Taylor, 2008; Vora et al., 2008). Alternative methods that are colorimetric show more uniformity in DNA labeling. A limitation of these colorimetric assays, measuring alkaline phosphatase activity or silver precipitation, is that they employ posthybridization reagents that result in high variability in signal detection (Kuck and Taylor, 2008). Given the limitations of current procedures, the present study evaluated photopolymerization for identification of pathogenic E. coli, using low-cost reagents and instrumentation that are amenable to high-throughput sampling and routine pathogen surveillance (Kuck and Taylor, 2008). Previously, photopolymerization was employed successfully for molecular diagnostic assays detecting Streptococcus species (Dawson et al., 2009) and for genotyping influenza viruses (Kuck and Taylor, 2008). Real-time PCR methods have been developed for the identification of virulent strains of E. coli O157 (Kawasaki et al., 2010; Suo et al., 2010); however, the cost for the reagents and instrumentation required for these analyses with real-time PCR impede the use of this molecular method for routine strain characterization in smaller research facilities. To provide a proof of concept for testing photopolymerization for foodborne pathogen detection with DNA microarrays, the present study constructed a low-density DNA microarray targeting stx1, stx2, eae, and per, genes encoding virulence factors that are considered to be good indicators of pathogenic E. coli O157.
Validation experiments demonstrated that the use of DNA microarrays with photopolymerization for detection of E. coli O157 resulted in an accurate genotyping of the tested strains. Polymer formation was clearly evident only for those probes targeting virulence genes known to be present in the reference strains. Quantification of the specificity by measuring the SNR values for each probe on the array demonstrated that there was a clear distinction between signal intensities and background noise, an important aspect for data analysis and interpretation of results with DNA microarrays (Verdnick et al., 2002; He and Zhou, 2008). These results were achieved by using a 30-mer oligonucleotide probe allowing a more specific detection of the target gene. Previous studies that assessed optimal conditions for fluorescent detection on DNA microarrays demonstrated that the use of short, 30-mer probes were equivalent to background noise, whereas optimal hybridization signals required 70-mer or 100-mer oligonucleotide probes (Letowski et al., 2004). However, the use of longer probes resulted also in lower specificity due to cross-hybridization with nontarget genes in some species (Letowski et al., 2004). In the present study, the use of 30-mer probes on the microarray in combination with photopolymerization facilitated the detection of specific target regions in the genes selected for identifying E. coli O157. Our quantification analysis showed that positive signal intensities had SNR values that were at least 20 times higher than spots without any specific hybridization.
Further, our analyses demonstrated consistently that the sensitivity of E. coli O157 detection by using photopolymerization was found to be 100–1000 CFU/mL in all validation experiments measuring assay sensitivity. This level of sensitivity was similar to a previous report of E. coli O157 detection with fluorescent methods (Call et al., 2001). In the present study, detection at cell concentrations <100 CFU/mL was variable (data not shown), due potentially to an effect of PCR inhibitors or to inefficient cell recovery during the assay, as reported previously (Call et al., 2001). Thus, the results from the present study demonstrated that the sensitivity of E. coli O157 detection by using photopolymerization with DNA microarrays was similar to fluorescence. Previously, similar detection sensitivities for photopolymerization and fluorescence were reported for influenza A virus identification with a low-density microarray (Kuck and Taylor, 2008). Additionally, our study demonstrated that photopolymerization resulted in the rapid genotyping of E. coli O157 in only 2–4 h, a significant improvement when compared to our results with fluorescence detection methods that required several days for virulent strain identification (Quiñones et al., 2007). Future work is aimed at further development of DNA microarrays with photopolymerization by adding pathogen-specific probes for identifying virulent strains of E. coli O157 and non-O157 from distinct geographical locations and sources. The pathogen-specific probes on the expanded DNA microarrays will target several non-O157 serovars implicated in severe human disease (Brooks et al., 2005; Bettelheim, 2007; Karmali, 2009; Buchanan and Appel, 2010). Additional probes will be included to target virulence determinants that code for adhesins, proteases, cytotoxins, and effectors and that are considered to be necessary for E. coli strains to be pathogenic (Bettelheim, 2007; Karmali, 2009; Buchanan and Appel, 2010).
Conclusions
Photopolymerization is a simple, rapid, and quantitative DNA microarray-based detection method for assessing the genetic composition of E. coli O157 strains by examining genes encoding Shiga toxin and selected virulence determinants that have been associated with pathogenic E. coli. This approach facilitates the evaluation of the potential virulence of E. coli O157 strains and provides relevant information to the food industry for determining the prevalence and risks of pathogenic E. coli in food production environments.
Acknowledgments
This work was supported by the U.S. Department of Agriculture, Agricultural Research Service CRIS project number 5325-42000-045, and by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant number U01A1070276. The authors would like to thank Edith Pierre-Jerome for excellent technical assistance and to Robert E. Mandrell and Michael Cooley (USDA/ARS/WRRC, Albany, CA) for providing the bacterial strains used in this study.
Disclosure Statement
B.Q. and M.S.S. declare no competing financial interests. E.D.D. and A.W.T. are both paid employees of InDevR, Inc., a for-profit entity.
References
- Bettelheim KA. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit Rev Microbiol. 2007;33:67–87. doi: 10.1080/10408410601172172. [DOI] [PubMed] [Google Scholar]
- Bettelheim KA. Beutin L. Rapid laboratory identification and characterization of verocytotoxigenic (Shiga toxin producing) Escherichia coli (VTEC/STEC) J Appl Microbiol. 2003;95:205–217. doi: 10.1046/j.1365-2672.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- Bilge SS. Vary JC., Jr. Dowell SF. Tarr PI. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot K. Huletsky A. Peytavi R. Turcotte S. Veillette V. Boissinot M. Picard FJ. Martel EA. Bergeron MG. Rapid exonuclease digestion of PCR-amplified targets for improved microarray hybridization. Clin Chem. 2007;53:2020–2023. doi: 10.1373/clinchem.2007.091157. [DOI] [PubMed] [Google Scholar]
- Brooks JT. Sowers EG. Wells JG. Greene KD. Griffin PM. Hoekstra RM. Strockbine NA. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis. 2005;192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- Buchanan RL. Appel B. Combining analysis tools and mathematical modeling to enhance and harmonize food safety and food defense regulatory requirements. Int J Food Microbiol. 2010;139(Suppl 1):S48–S56. doi: 10.1016/j.ijfoodmicro.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Call DR. Challenges and opportunities for pathogen detection using DNA microarrays. Crit Rev Microbiol. 2005;31:91–99. doi: 10.1080/10408410590921736. [DOI] [PubMed] [Google Scholar]
- Call DR. Brockman FJ. Chandler DP. Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int J Food Microbiol. 2001;67:71–80. doi: 10.1016/s0168-1605(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Cooley M. Carychao D. Crawford-Miksza L. Jay MT. Myers C. Rose C. Keys C. Farrar J. Mandrell RE. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One. 2007;2:e1159. doi: 10.1371/journal.pone.0001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson ED. Taylor AW. Smagala JA. Rowlen KL. Molecular detection of Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis. Mol Biotechnol. 2009;42:117–127. doi: 10.1007/s12033-009-9143-2. [DOI] [PubMed] [Google Scholar]
- He Z. Zhou J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl Environ Microbiol. 2008;74:2957–2966. doi: 10.1128/AEM.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali MA. Host and pathogen determinants of Verocytotoxin-producing Escherichia coli-associated hemolytic uremic syndrome. Kidney Int Suppl. 2009;112:S4–S7. doi: 10.1038/ki.2008.608. [DOI] [PubMed] [Google Scholar]
- Kawasaki S. Fratamico PM. Horikoshi N. Okada Y. Takeshita K. Sameshima T. Kawamoto S. Multiplex real-time polymerase chain reaction assay for simultaneous detection and quantification of Salmonella species, Listeria monocytogenes, and Escherichia coli O157:H7 in ground pork samples. Foodborne Pathog Dis. 2010;7:549–554. doi: 10.1089/fpd.2009.0465. [DOI] [PubMed] [Google Scholar]
- Kimura R. Mandrell RE. Galland JC. Hyatt D. Riley LW. Restriction-site-specific PCR as a rapid test to detect enterohemorrhagic Escherichia coli O157:H7 strains in environmental samples. Appl Environ Microbiol. 2000;66:2513–2519. doi: 10.1128/aem.66.6.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck LR. Taylor AW. Photopolymerization as an innovative detection technique for low-density microarrays. Biotechniques. 2008;45:179–186. doi: 10.2144/000112889. [DOI] [PubMed] [Google Scholar]
- Letowski J. Brousseau R. Masson L. Designing better probes: effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J Microbiol Methods. 2004;57:269–278. doi: 10.1016/j.mimet.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Mandrell RE. Enteric humans pathogens associated with fresh produce: sources, transport, and ecology. In: Fan X, editor; Niemira B, editor; Doona CJ, editor; Feeherry F, editor; Gravani RB, editor. Microbial Safety of Fresh Produce. Ames, IA: IFT Press/Wiley-Blackwell Publishing; 2009. pp. 5–41. [Google Scholar]
- Nataro JP. Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ. Kang CH. Chae JC. Rhee SK. Metagenome microarray for screening of fosmid clones containing specific genes. FEMS Microbiol Lett. 2008;284:28–34. doi: 10.1111/j.1574-6968.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- Quiñones B. Massey S. Friedman M. Swimley MS. Teter K. Novel cell-based method to detect Shiga toxin 2 from Escherichia coli O157:H7 and inhibitors of toxin activity. Appl Environ Microbiol. 2009;75:1410–1416. doi: 10.1128/AEM.02230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones B. Parker CT. Janda JM., Jr. Miller WG. Mandrell RE. Detection and genotyping of Arcobacter and Campylobacter isolates from retail chicken samples by use of DNA oligonucleotide arrays. Appl Environ Microbiol. 2007;73:3645–3655. doi: 10.1128/AEM.02984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V. Brett K. Hornitzky MA. Dowton M. Bettelheim KA. Walker MJ. Djordjevic SP. Distribution of intimin subtypes among Escherichia coli isolates from ruminant and human sources. J Clin Microbiol. 2003;41:5022–5032. doi: 10.1128/JCM.41.11.5022-5032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel JM. Sparling PH. Crowe C. Griffin PM. Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PR. Hobbs M. Valvano MA. Skurnik M. Whitfield C. Coplin D. Kido N. Klena J. Maskell D. Raetz CRH. Rick PD. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- Rozen S. Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, editor; Krawetz SA, editor. Bioinformatics Methods and Protocols. Vol. 132. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Serna A. Boedeker EC. Pathogenesis and treatment of Shiga toxin-producing Escherichia coli infections. Curr Opin Gastroenterol. 2008;24:38–47. doi: 10.1097/MOG.0b013e3282f2dfb8. [DOI] [PubMed] [Google Scholar]
- Shimizu T. Yamasaki S. Tsukamoto T. Takeda Y. Analysis of the genes responsible for the O-antigen synthesis in enterohaemorrhagic Escherichia coli O157. Microb Pathog. 1999;26:235–247. doi: 10.1006/mpat.1998.0253. [DOI] [PubMed] [Google Scholar]
- Sikes HD. Hansen RR. Johnson LM. Jenison R. Birks JW. Rowlen KL. Bowman CN. Using polymeric materials to generate an amplified response to molecular recognition events. Nat Mater. 2008;7:52–56. doi: 10.1038/nmat2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo B. He Y. Tu SI. Shi X. A multiplex real-time polymerase chain reaction for simultaneous detection of Salmonella spp., Escherichia coli O157, and Listeria monocytogenes in meat products. Foodborne Pathog Dis. 2010;7:619–628. doi: 10.1089/fpd.2009.0430. [DOI] [PubMed] [Google Scholar]
- Uttamchandani M. Neo JL. Ong BN. Moochhala S. Applications of microarrays in pathogen detection and biodefence. Trends Biotechnol. 2009;27:53–61. doi: 10.1016/j.tibtech.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdnick D. Handran S. Pickett S. Key considerations for accurate microarray scanning and image analysis. In: Kamberova G, editor. DNA Image Analysis: Nuts and Bolts. Salem, MA: DNA Press; 2002. pp. 83–98. [Google Scholar]
- Vora GJ. Meador CE. Anderson GP. Taitt CR. Comparison of detection and signal amplification methods for DNA microarrays. Mol Cell Probes. 2008;22:294–300. doi: 10.1016/j.mcp.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang WL. Kohler B. Oswald E. Beutin L. Karch H. Morabito S. Caprioli A. Suerbaum S. Schmidt H. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 2002;40:4486–4492. doi: 10.1128/JCM.40.12.4486-4492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]