Abstract
An increasing progress on the role of Hedgehog (Hh) signaling for carcinogenesis has been achieved since the link of Hh pathway to human cancer was firstly established. In particular, the critical role of Hh signaling in the development of Basal cell carcinoma (BCC) has been convincingly demonstrated by genetic mutation analyses, mouse models of BCCs, and successful clinical trials of BCCs using Hh signaling inhibitors. In addition, the Hh pathway activity is also reported to be involved in the pathogenesis of Squamous Cell Carcinoma (SCC), melanoma and Merkel Cell Carcinoma. These findings have significant new paradigm on Hh signaling transduction, its mechanisms in skin cancer and even therapeutic approaches for BCC. In this review, we will summarize the major advances in the understanding of Hh signaling transduction, the roles of Hh signaling in skin cancer development, and the current implications of “mechanism-based” therapeutic strategies.
Keywords: Hedgehog, Smoothened, PTCH1, skin cancer, signal transduction, therapy
1. Introduction
Major advances in understanding the Hedgehog (Hh) pathway have been achieved since the Hh gene was identified in 1980 through genetic analyses of Drosophila segmentation by the Nobel laureates Eric Wieschaus and Christiane Nüsslein-Volhard[1]. As an essential signaling pathway in embryonic development, Hh pathway is critical for maintaining tissue polarity both for invertebrate and vertebrate embryos. Since inactivation of this pathway was linked to the hereditary developmental disorder holoprosencephaly in 1996[2,3], several human syndromes have been linked to genetic alterations in Hh pathway genes[4]. The most significant achievement is the link between the Hh pathway signaling activation and human cancer[5–8].
During the past fifteen years, studies revealed activation of Hh pathway in basal cell carcinoma, medulloblastoma, leukemia, gastrointestinal, lung, ovarian, breast, liver, pancreatic and prostate cancer[8–13]. The initial link between Hh signaling and human cancers was made from the discovery that loss-of-function mutations of human PTCH1 on human chromosome 9q22 are associated with a rare and hereditary form of BCC-basal cell nevus syndrome(BCNS), also called Gorlin syndrome[14,15]. Gorlin syndrome is a rare autosomal genetic disease with two distinct sets of phenotypes: predisposition to develop cancer such as BCC and medulloblastoma, and developmental defects such as bifid ribs and ectopic calcification. The tumor suppressor role of PTCH1 was demonstrated in knockout mice, where Ptch1+/− mice develop tumors in addition to other features observed in patients with Gorlin syndrome, such as spina bifida occulta[16–18].
More insights on the role of Hh pathway in human cancer was revealed from studies of basal cell carcinoma and medulloblastoma[8,11,19–21]. Recent evidence indicate thatthat activation of the Hh pathway plays an important role in other kinds of human skin cancer, such as SCC[22,23], melanoma[24–26] and Merkel cell carcinoma[27]. Inhibitors and modulators of Hh pathway are believed to be promising drugs in the clinical application of human skin cancer treatment[20,28–30]. Delineation of this pathway may improve our understanding of the role of Hh pathway in skin cancer and may lead to rational medical therapy for skin cancer and possibly other tumors. We will first review recent advances in our understanding of Hh signaling before we discuss the role of this pathway in human cancer.
2. The Hedgehog signal transduction in vertebrates
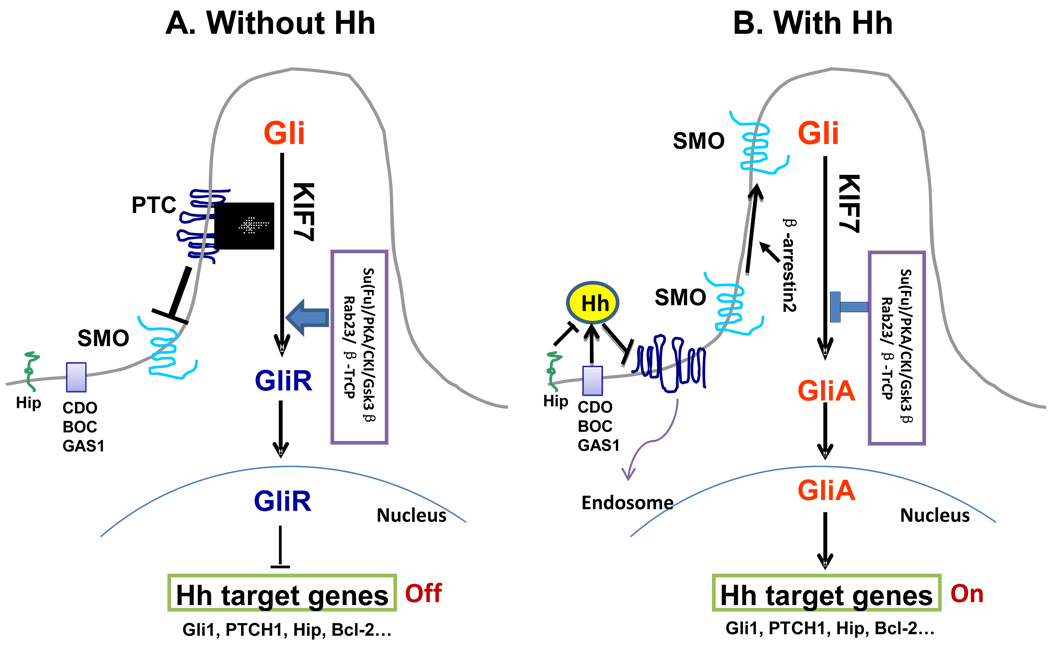
The Hh pathway was found to be highly conserved from fruit fly to human[31]. However, some significant differences between vertebrates and invertebrates in regard to Hh signaling components and their interplays have been discovered. Figure 1 shows a simplified diagram of the Hh signaling pathway in vertebrates.
Figure 1. A simplified model for Hh signaling in mammalian cells.
SMO is the key signal transducer of the Hh pathway. A, In the absence of Hh ligands, the PTCH receptor at the base of the primary cilium suppresses the function of SMO by preventing its entry into the cilium. Without SMO activation, Gli proteins are processed with Su(Fu), GSK3β, PKA, and CKI into a repressor form (Gli-R), which disable the Hh signaling pathway. B, In the presence of Hh, it binds to PTCH. The binding can be repressed by HIP and supported by Cdo, Gas1 and Boc. Upon binding, the Hh/PTCH complex becomes internalized in endosomes and later degraded. Without inhibition from PTCH, SMO becomes activated and facilitated Gli activation n (GliA), which stimulates Hh target gene expression.
Three hedgehog paralogous genes in vertebrates (one Hh in Drosophila) are Sonic Hh (Shh), Indian Hh (Ihh), and Desert Hh (Dhh) respectively. The Hh protein is synthesized as a precursor with 45 kDa in molecular weight, which undergoes auto-processing to generate the amino-terminal fragment (HhN). The HhN fragment becomes active after two post-translational modifications, covalent binding of a cholesterol moiety to their carboxyl terminus and palmitoylation at the N-terminus by hedgehog acyltransferase Skinny [32–36]. These lipid modifications contribute critically to the proper movement and full signaling potency of Hh proteins [37, 38]. After maturity, the Hh proteins are released from the producing cell. The secretion is dependent on dispatched (Disp)[39, 40], a 12-transmembrane transporter-like protein with structural homology to Hh receptor PTCH. In addition to Disp, there are several other molecules involved in this process, including metalloproteases[41], the heparan sulfate proteoglycans Dally-like (Dlp) and Dally[42, 43] or their regulators[44], as well as enzymes such as Sulfateless and Tout velu[45–47]. Then the Hh proteins can function either at short range to nearby cells or at long range to distant cells.
The signaling cascade of this pathway is initiated by binding of a Hh ligand to its membrane receptor PTCH[48]. In fact, there are two PTCH homolog genes in vertebrates, namely PTCH-1 and PTCH-2 (12-transmembrane proteins). PTCH-1 is critical for embryonic development, and is a human tumor suppressor gene. Smoothened (SMO), a 7-transmembrane protein on the cell surface, is a positive regulator of the signaling.
Data from a study in Drosophila showed direct evidence for SMO-coupling to G protein Gαi in the regulation of Hh pathway activation[49]. The precise mechanism by which SMO transduces the signaling cascade remains unclear. It is known, however, that a conformational change in SMO[50] and SMO protein translocation to the primary cilium are the two critical events for Hh signaling in vertebrates. In the absence of Hh, PTCH inhibits SMO[51], which inactivates target gene expression. In the presence of Hh, the repression of SMO by PTCH1 is however lifted, triggering activation of downstream Shh effectors, the glioma-associated (Gli) family of transcription factors. The Gli transcription factors (Gli1, Gli2, Gli3 in vertebrates and Ci in Drosophila) control the expression of Shh target genes including PTCH1 and Gli1, which provide negative and positive feedback for Hh signaling, respectively[52–56]. Several molecules are engaged in the reception of Hh ligands with PTCH. The Hh-interacting protein (HIP) can compete with PTCH to bind Hh, resulting in the negative regulation of Hh signaling[57]. On the other hand, CDO and BOC (its invertebrate homologue Ihog), GAS1, and Glypican-3 serve as co-receptors of Hh[58–65].
It is not entirely clear how PTCH regulates SMO activity. It is believed that the primary cilium, a cell organelle present on most mammalian cells, plays an important role in the Hh signaling [66–71]. The current model for this regulatory mechanism involves trafficking of the PTCH and SMO in and out of the cilium as a key event in regulating SMO activity, not a physical interaction. In this model, PTCH1 is localized at the base of the primary cilium where it inhibits the activation of SMO. One molecule of PTCH can inhibit several molecules of SMO, suggesting that the inhibition of SMO by PTCH is in a catalytic way[51]. No solid evidence has been provided to support direct suppression of SMO by PTCH. The currently most favorite model is that PTC limits SMO signaling by transporting small endogenous molecules specifically targeted to SMO. Candidates of these small molecules include PI4P, lipoproteins, and pro-vitamin D3[20,72–75]. However, how these molecules regulate SMO signaling is unknown, and further studies are needed to further identify such small molecules
Upon binding of a Hh ligand, the receptor/ligand complex is translocated out of the primary cilium and internalize in endosomal vesicles, which triggers mobilization of SMO into cilium[20]. The level of SMO protein localized to the cilium is correlated with the Hh pathway activation. The exact mechanism of SMO ciliary translocation in response to Hh signaling remains unknown. Some evidence indicate that that β-arrestin 2 can regulate ciliary localization of SMO[76]. Recent studies suggest that ciliary mobilization and localization of SMO is not sufficient for SMO activation, and that its complete activation requires a second separate event [77,78]. The observation that cyclopamine, a SMO inhibitor, did not prevent the ciliary localization of SMO can be partly explained by the two-step activation ratiocination. Since not all events of the Hh signaling occur in cilium[79, 80], the role of cilium for Hh signaling downstream of SMO remains unclear.
It is not clear how SMO activation triggers a cascade of downstream events. Gli transcription factors can directly bind to a consensus binding site (5’-TGGGTGGTC-3’) in the promoter of target gene[82–85]. Regulation of Gli activity involves cytoplasmic-nuclear shuttling, protein phosphorylation, ubiquitination, acetylation, and protein degradation of Gli molecules. Protein kinase A can retain Gli1 protein in the cytoplasm via a PKA site in the nuclear localization signal domain[86], whereas activated Ras signaling induces Gli nuclear localization[26,87]. Protein kinase A (PKA), glycogen synthase kinase 3β (GSK3β), and casein kinase 1 (CK1) can sequentially phosphorylate the C-terminus phosphorylation sites of Gli3, targeting Gli3 to the proteasome for limited degradation. β-TrCP, cul3/BTB and numb/Itch mediate Gli ubiquitination[88–95]. β-TrCP E3 ligase also can process Gli3 into a transcriptional repressor[89]. Additionally, the protein kinases Cdc2l1, DYRK2, and MAP3K10, were identified in genome-wide screens as affecting Gli activity [96, 97].
Several molecules have been identified to be genetically downstream of SMO signaling in Drosophila, including COS2 and Fused. How their vertebrate homologues function in Hh signaling is yet established. Recent in vivo studies support that a COS2 homologue KIF7 functions in the Hh pathway, but no direct interaction between SMO and KIF7 is detected[98,99]. The phenotype of vertebrate Fused knockout mice is not similar to that observed in Shh null mice[100–102], and no changes of Hh signaling are observed in Fused null mice, suggesting that Fused is not critical for Hh signaling during early embryonic development of vertebrates. Suppressor of fused (Su(Fu)) is a negative regulator of mammalian Gli transcriptional factors[103, 104]. Su(Fu) is originally identified genetically in Drosophila, but itself is not required for pathway activity. In contrast, vertebrate Su(Fu) has an important role in the regulation of Gli activity. Su(Fu) null mouse mutants fail to repress the pathway, resulting in activation of Hh signaling [105, 106]. Su(Fu) can associate with all three Gli molecules [107,108] and may control nuclear translocation[103, 104] and degradation of these transcription factors[109]. Other evidence indicate that Su(Fu) may interact with the SCL/ TAL1 interrupting locus (SIL), which interferes with Su(Fu)'s repression on GLI1 [110]. Su(Fu) can also recruit SAP18 to occupy the Gli binding sites in the nucleus[111, 112], or recruit GSK3 beta for Gli3 processing[113]or interacts with Rab23 to inhibit Gli transcriptional activity (our unpublished data).
Several novel cytoplasmic regulators of Hh signaling in mammalian cells have been identified, for instance, Rab23[114] and tectonic[115] both are negative regulators downstream of SMO. Our data suggest that Rab23 may inhibit Gli1 activity in a Su(Fu)-dependent manner (our unpublished data), but the exact mechanism of action remains to be elucidated. Unlike many other Rab family members, Rab23could be localized in the nucleus and cytoplasm[116, our unpublished data], suggesting that Rab23 may have other uncharacterized functions apart from membrane trafficking.
Interestingly, PTCH, HIP, GAS1, and Gli1 are components as well as transcriptional targets of Hh pathway, suggesting that feedback regulatory loops are part of the mechanisms to maintain the level of Hh signaling and modulate the response of Hh signaling[117–120]. PTCH and HIP provide negative feedback regulation, whereas Gli1 forms a positive regulatory loop. On the other hand, GAS1 is down-regulated by the Hh pathway but it is a positive regulator for Hh signaling.
3. Skin cancers linked to aberrant Hedgehog pathway activity
Skin cancer is the most frequent cancer worldwide, and its incidence increases every year, leading to a huge health problem and financial burden in many countries. It is necessary, therefore, to get further understanding of molecular mechanism underlying skin carcinogenesis. In the past 15 years, convincing evidence indicate BCC etiology is highly dependent on the aberrant activation of hedgehog signaling pathway[19–21], which also plays an important role in SCC and MM[22–26] Other important alterations in SCC and MM include p53 pathway and the p16/Rb pathway.
3.1 Basal cell carcinoma
The first link between the Hh pathway and BCC came from the discovery of loss-of-function mutations of PTCH1 gene in Gorlin syndrome[14,15]. The individual patients with Gorlin syndrome are strongly predisposed to the development of basal cell carcinomas (BCCs) in the skin at early age as well as medulloblastomas. In addition, the incidence of meningioma, ovarian and heart fibroma, fetal rhabdomyoma (RM), and possibly rhabdomyosarcoma (RMS) is also higher in Gorlin Syndrome. Nevertheless, the majority of BCCs arise sporadically on sun exposed areas of the skin. Inactivating mutations of PTCH1 have been found in in most exons of the PTCH1 gene [121,122] in sporadic BCC. In the tumor, while one copy of PTCH1 is mutated, the other copy is lost through chromosome loss in the region where the PTCH gene resides. As a result of loss of PTCH1 function, SMO signaling is without control, leading to constitutive activation of the pathway. About 70% of BCCs contain genetic alterations in the PTCH1 gene.. Therefore, it is generally believed that BCC development require activation of Hh signaling pathway.
The mutations in other components of the Hh signaling pathway are also detected in BCCs, and may contribute to the formation of tumor. Shh has mitogenic effects in several tissues, including pre-somitic mesoderm, retina, and cerebellum[123]. The overexpression of Shh in mouse skin will develop many features of BCNS, including dysformation of the skeleton and skin, with multiple BCC-like epidermal proliferations after the first few days of skin development [124], indicating that Shh overproduction is sufficient to induce BCC in mice and can mimic the loss of PTCH function seen in human BCCs[124, 125]. The gain-of-function mutations of SMO could contribute to 6–21% of sporadic BCC development[126–128], and abnormalities of Hh pathway transcription factors GLI1 [129]and GLI2 [130, 131]can lead to constitutive activation of the pathway. Mutations in Su(Fu), a negative regulator of Hh pathway, have also been identified[132]. Recently, a truncating Su(Fu) germline mutation was found as a probable cause in the members of a family with Gorlin syndrome features, including MB and palmar/plantar pits, but not BCC. Notably, no PTCH1 mutations was detected in the affected family members[133].
Although genetic data are sufficient to support the Hh pathway as a ‘gatekeeper’ in BCC development [134], establishing animal models using tissue-specific activation of Hh signaling is still critical for further identifying and understanding Hh signaling in the carcinogenesis of BCC. Currently, mouse models for BCC are well established and provide a powerful tool for us to further our understanding of molecular Hh pathway-mediated development of BCC.
Of note, wild-type mice never develop BCCs, even after treatment with carcinogen, UV or ionizing radiation. However, Ptch1+/− mice are susceptible to BCC development following UV irradiation or ionizing radiation[18]. The frequency of BCC development under the conditional ablation of PTCH1 is from around 50% with 1 or 2 tumors per mouse to 100% penetrance by the age of 16 weeks[135, 136]. However, the Ptch1+/− mice rarely develop full-grown BCCs if kept under normal conditions. BCCs will occur after mice are exposed to ultraviolet (UV), or ionizing radiation, which is similar to BCC development in NBCCS patients, suggesting UV exposure is very important for BCC to develop. Due to the embryonic lethality of Ptch1−/−, tissue-specific knockout of PTCH1 has been generated[137]. By combining conditional gene knockout and the inducible activity of the keratin 6a promoter, Krt6a-cre:Ptch1neo/neo mice develop BCC following stimulation with retinoic acid[134]. The use of other skin-specific Cre-strains to drive PTCH1 ablation also produced BCC lesions [138]. These mouse experimental models reflect well BCC development observed in human with PTCH1 loss-of-function mutations, suggesting these animal models are very helpful in further studying molecular pathogenesis of BCC.
In addition to the PTCH1 knockout mouse model, transgenic mice expressing SMO using Krt5 or Krt14 promoter also develop BCC-like tumors that look very similar to benign human basaloid follicular hamartomas [126,139]. However, these transgenic mice eventually lose the expression of SMO by an unknown mechanism. Using conditional knock-in technology, skin-specific SMOM2YFP (Krt14-creER: R26-SmoM2YFP or Krt14-cre:R26-SMOM2YFP) knock-in mice develop multiple microscopic BCCs at a very early age, providing an easy genetic assay for Hh signaling downstream of SMO[140]. Transgenic mice expressing Shh also develop multiple BCC-like epidermal proliferations [124]. Several transgenic mice have been developed using downstream transcriptional factors Gli1 and Gli2[129, 130]. The inducible expression of Gli2 in the skin results in BCCs after a few weeks. However, Su(Fu)+/− mice develop skin lesions resembling skin hyperplasia but not BCC-like tumors[105], even compound Ptch1+/−Su(Fu)+/− mice lack signs of obvious BCC lesions.
The cellular origin of BCC has been debated for a long time. Animal models can help us to dig it out. BCC are so named because the BCC cells histologically resemble to basal keratinocytes of the hair follicle(HF), sebaceous glands, and interfollicular epidermis (IFE), suggesting that BCCs may arise from the outer root sheet, or bulge of the HF, and the original cell is possibly a multipotent stem/progenitor cell. Several lines of evidence support this idea [124,141,142]. However, two recent studies gave different answers. Youssef et al. [143] localized the murine cell of origin of cutaneous Hh-driven tumors to be in the IFE, not in the hair follicle, by using cell-specific Cre to activate expression of a ROSA26-driven transgenic mutant SMO. Using cell fate tracking of X-ray induced BCCs in Ptch1+/_ mice, Wang et al. found that both cell fate mapping and enhancement of BCC carcinogenesis by deletion of p53 demonstrate conclusively the cell-of-origin of the BCCs from the hair follicle bulge stem cell (K15-expressing cells)[144]. The major difference between the two mouse models is that one used transgenic mutant SMO, and the other used Ptch1+/− mice. The underlying molecular mechanism remains unclear. Interestingly, conditional loss of p53 enhanced BCC carcinogenesis not only from the follicular bulge but also from the interfollicular epidermis in Ptch1+/− mice[144], likely due to enhanced expression of SMO, further suggesting that p53 may alter BCC carcinogenesis in part by affecting expression of signaling components[145].
As mentioned before, accumulating evidence indicates that an important organelle in Hh signal transduction in mammalian systems is primary cilia, projecting cylindrically from the membrane of most vertebrate cell types and containing multiple concentrated components of the Hh pathway, including Shh, PTCH1, SMO [51], Su(Fu) and Gli transcription factors[66–71]. Not surprisingly, questions come out whether primary cilia are required for Hh signaling-driven development of BCCs.
The function of primary cilium is regulated by protein complexes involved in intra-flagellar transport (IFT), which functions in retrograde and anterograde movement of cargo within the primary cilia[146]. Mice with mutation in IFT protein are shown to result in similar phenotypes with Hh or Gli2 loss of function [147–,150]. Gli3 processing is the most significantly affected event in IFT mutants[68,150,151]. In the mouse skin, disruption of laminin-511, an extracellular protein that plays an important role in promoting primary cilia formation and function, resulted in disturbance of mouse hair development [149,151]. It has been shown that a SMO mutant lacking a ciliary translocation signal can not mediate Hh signaling[67]. However, the translocation of SMO to cilium is not sufficient to activate Hh signaling[77,152]. Using tissue-specific gene knockout, Wong and colleagues have recently revealed that genetically disrupting cilia formation by conditional deletion of Kif3A or Ift88, both required for ciliogenesis, decreased BCC-like tumor formation in Rosa26-SMOM2/K14-cre mice, but increased tumor formation in Gli2 transgenic mice, suggesting a dual role of cilia in Hh signaling-mediated carcinogenesis in mice[153]. Namely, the primary cilia function can either promote or suppress BCC tumorigenesis. It is not difficult to understand this seemingly paradoxical effect since primary cilia which contain both positive and negative regulators for Hh signaling and are required for formation of both active and repressor forms of Gli [66–71]. However, from a therapeutic point of view, it should be cautious to consider their possible clinical implication as therapeutic targets in the treatment of BCC, and a precisely further analysis of molecular functions will be needed.
3.2 Squamous cell carcinoma (SCC)
Squamous cell carcinoma arises from squamous epithelium, frequently found in the lungs and skin, and occurring also in the anus, cervix, larynx, nose, and bladder. The majority of SCC from skin localizes in the head and neck, where the five-year survival rate after diagnosis for this type of cancer remains approximately 50%[154]. Although understanding of the molecular mechanism underlying SCC carcinogenesis was largely achieved in the past decade. It is currently believed that linkage of SCC to aberrant activation of Hh pathway is not as strong as that of BCC. Nevertheless, studies on several kinds of SCC show a role of Hh pathway inSCC.
In oral squamous cell carcinoma, PTCH was found two missense mutations which affected the conserved residue in the transmembrane domains of the gene product and in the intracellular loop at the C-terminal residue implicated in regulating the smoothened molecule, leading to ineffective Shh reception in SCC cells with PTCH mutations[155]. Moreover, transducing the wild-type PTCH1 gene back to human SCC cell lines, A431 and KA, that express only mutant patched mRNA, caused the cell proliferation drastically reduced. Furthermore, cyclopamine efficiently suppressed the growth of A431 and KA cells, indicating that Hh signaling is involved in the tumorigenesis of oral SCCs[156]. Ping provides further evidence that the PTCH gene is mutated in a subset of SCC from individuals with a history of multiple BCC[157]. Danaee et al. also found a high prevalence of a LOH at 9q22.3 in both BCC (75.5%) and SCC (60.8%)[158]. Wakabayashi showed some evidence of in an animal model. A carboxy-terminal polymorphism, between C57BL/6 and FVB/N strains of mice, in the Ptch1 gene leads to FVB/N mice highly susceptible to SCC. PTCH(FVB) overexpression in K5/Hras B6FVB F mice can promote SCC formation, but not required for tumor maintenance, suggesting a role of PTCH at an early stage of tumor development[159].
Most of studies on the relation of Hh pathway activation and SCC have been completed by immunohistochemistry staining and/or in situ hybridization. Overexpression of Shh had been observed in five cell lines among 14 human oral squamous cell carcinoma cell lines[160] and human lung squamous carcinoma (LK-2 and EBC-1) cell lines[161], and human squamous carcinoma tissues of lung[116,161,162], uterine cervix[163], esophagus[164–166] and stomach[167]. In addition to Shh, Hh target genes and major components, for instance, Ihh, PTCH, SMO, Gli-1, Gli-2 and Gli-3, were also highly expressed in the tumor [163,164,167]. These cells are also sensitive to cyclopamine, a specific Hh signaling inhibitor.
Recently, Schneider investigated the expression pattern of Hh pathway in squamous cell carcinoma of the skin, and head and neck[168]. Compared with healthy control tissues, they found significant overexpression of major components of the Hh pathway. Importantly, they observed that high expression of Shh correlates significantly with poor overall survival in patients with head and neck cancer, suggesting that activity of Hh pathway may serve as a prognostic factor in patients with head and neck SCC cancer[168]. This hypothesis is further supported by the fact that Gli1 nuclear expression is a strong and independent predictor of early relapse and poor prognosis in esophageal squamous cell carcinoma after chemoradiotherapy [169,170]. Additionally, SCC tumorgenesis is known to involve p53 pathway and WNT/catenin signaling, both of which have been shown to interact with the Hh pathway[145,171].
Taken all data together, evidence of Hh pathway in SCC carcinogenesis is clear but animal models for this mechanism have not been established yet.
3.3 Melanoma and Merkel cell carcinoma
Melanoma is one of the most aggressive cancers, accounting for approximately 4% of human skin cancers and yet 80% of deaths from cutaneous neoplasms[172]. Activating mutations in the oncogenes B-RAF and N-RAS are present in 70% and 15% of melanomas respectively[173–175]. However, Hh pathway activity in melanoma tumorigenesis was not revealed until recently. First, no genetic alterations in Hh pathway genes have been found in melanomas [176]. Second, no genetic mouse models for Hh signaling-mediated development of melanoma have been established. Nevertheless, the K5-Gli2 transgenic mice [130] can form hyperpigmented BCC-like tumors, and K5-SMO-M2 transgenic mice [139] show focal or global skin pigmentation, which support that Hh pathway activity is required for proliferation of normal human melanocytes[26].
Recently, several studies (11) suggested that the Hh pathway may play a role in melanoma progression. It was [25] [26] discovered that cyclopamine treatment delayed tumor growth of B16F0 melanoma cells in immunodeficient mice. Another study found that Gli1 expression was correlated with tumor progression and metastasis of human melanomas [177]. In a transgenic mouse model of melanoma induced by oncogenic NRAS in which Gli1 expression was elevated, melanoma tumor volume was drastically suppressed by cyclopamine treatment[26]. The most important result so far is from the laboratory of Alain Mauviel. Alexaki et al. demonstrated that the high expression of Gli2was associated with an invasive and metastatic phenotype in vitro and in vivo[178]. Melanoma cells with high GLI2 expression metastasized to bone more readily than cells with low GLI2 expression. Moreover, decreasing GLI2 expression can block bone metastasis of melanoma[178]. These and other data (the K5-Gli2 transgenic mice [130] present hyperpigmention), indicate that activation of GLI2 plays an important role in the cell invasion and metastasis of melanoma. GLI2 is show to be a direct target of TGF-b signaling[179], and it is well known that over-expression of TGF-b increases matrix protein secretion and promotes invasion of melanoma cells[180]. Taken together, it suggests that TGF-b regulates invasion and migration through Gli2, a Hh pathway component. Very recently, Johnson RW et al showed further evidence that Gli2 is required for TGF-β to stimulate PTHrP expression[181], however, the requirement is independent of canonical Hh signaling, suggesting that the molecular mechanism underlying which the Hh pathway components mediate melanoma invasion and metastasis still needs further investigations.
In contrast to BCC, Merkel cell carcinoma (MCC) is a rare but aggressive skin tumor. While BCC is the most common type of skin cancer, with slow growth and almost without metastasis, MCC is extremely rare and more aggressive and thus has a higher mortality rate. Although Hh signaling pathway is important for human development and carcinogenesis in various malignancies, the relationship between MCC and Hh pathway activity was recently uncovered. The expression of Shh, Ihh, PTCH, SMO, Gli-1, Gli-2, and Gli-3 was detected immunohistochemically on tissue microarray with samples from 25 MCC patients and found frequently and intensely over-expressed[182]. It has also been found that high levels of PTCH and Ihh were significantly associated with an increase in patients overall and recurrence-free survival respectively, suggesting that the Hh pathway is strongly activated in MCC and thus may play a role in its carcinogenesis.
4. Models of Hh pathway activity in cancer and implications for therapy
Three basic models have been proposed on aberrant Hh signaling in human cancer[10,13]. Type I cancers are ligand-independent. The growth of cancers is driven by pathway-activating mutations in components of the Hh pathway, leading to over-proliferation, such as basal cell carcinomas. (b) Type II cancers have ligand-dependent autocrine Hh signaling. The cancer cells can secrete Hh ligand, and the ligand binds to the receptor on the same cells, leading to cell-autonomous pathway activation. (c) Type III cancers are ligand-dependent paracrine. The cancer cells can secrete Hh, and the ligand binds to the receptor on the stromal cells, leading to pathway activation in the stroma cells. Then the stroma cells feed other growth signals (not Hh siganling) back to stimulate tumor cell growth. Type IIIb cancers are ligand-dependent ‘reverse paracrine’. The cancer cells receive Hh ligand secreted from stromal cells, leading to pathway activation in the tumor.
Animal models indicate that development of BCC is driven constitutive Hh signaling activation, not by the presence of Hh ligands. However, autocrine and paracrine Hh signaling cannot be excluded in BCC[183,184]. Recent data indicatet that the surrounding PTCH1+/− stroma may affect BCC development in the NBCCS patients [185]. Therefore, it is very important to understand the signaling mechanisms by which Hh signaling mediates carcinogenesis.
5. Inhibitors and modulators of Hh pathway and their application in skin cancers
Hh signaling plays a significant role in skin cancers, especially in BCC, suggesting that certain types of skin cancer may be cured by targeted inhibition of the Hh pathway. At present, more than 50 compounds have been identified to have inhibitory effects on Hh signaling. Currently, there are 3 major targeting sites for Hh signaling inhibitors identified so far: Hh molecules (Shh neutralizing antibodies, small molecule Robotnikinin), SMO protein (cyclopamine and its derivatives IPI-926, Cyc-T, and synthetic compounds GDC-0449, Cur61414, XL-139, and LDE-225), and Gli transcriptional factor (HPI-1, HPI-2, GANT-56, and GANT-61)[12,20]. According their nature and derivation, all of these inhibitors could be divided into three groups: natural products (cyclopamine and its derivatives, and other natural products); synthetic small molecules; and Hh signaling modulators[120]. Table 1 lists major current Hh signaling inhibitors.
Table 1.
Hedgehog signaling inhibitors and modulators
| Compounds name | Hh target | 50% maximal inhibition (IC50) |
In vitro/in vivo studies | References |
|---|---|---|---|---|
| Robotnikinin | Shh | >10µM | In vitro | [204] |
| Cyclopamine | SMO | 300 nM | In vivo & in vitro | [205] |
| KAAD-cyclopamine | SMO | 20 nM | In vitro cultured cells | [206] |
| Jervine | SMO | 500 nM | In vitro and cultured embryos | [207] |
| Cyc-T | SMO | 20 nM | In vitro & in vivo studies | [191] |
| Cur-61414 | SMO | 200 nM | Phase I clinical trial halted | [208] |
| Sant-1,2,3,4 | SMO | 20–200 nM | In vitro studies | [209] |
| Compound 5 | SMO | <100 nM | In vitro studies | [210] |
| Compound Z | SMO | <1 nM | In vitro studies | [211] |
| IPI-926 | SMO | <20 nM | Phase I clinical trial | [212] |
| GDC-0449 | SMO | <20 nM | Phase I/II/III clinical trials | [197] |
| BMS-833923 (XL139) | SMO | <20 nM | Phase II clinical trial | NCI clinical trial database |
| LDE-225 | SMO | <20 nM | Phase II clinical trial | NCI clinical trial database |
| PF-04449913 | SMO | Phase I clinical trial | NCI clinical trial database | |
| Hhantag691/ Hhantag | SMO | In vitro studies | [213] | |
| Statin | SMO | In vitro studies | [214] | |
| Vitamin D3 | SMO | 100µM | In vitro | [75, 214] |
| Itraconazole | SMO? | <1.5 µM | In vitro and xenograft | [203] |
| Recombinant Hip | Hip | In vitro studies | [215] | |
| HPI-1, 2,3,4 | Gli | <10µ mol/L | In vitro | [199] |
| Gant-58,61 | Gli | 5µM | In vitro & in vivo studies | [216] |
5.1 Natural products (cyclopamine, its derivatives, and others)
Cyclopamine was the first SMO protein antagonist [186] discovered for treatment and chemoprevention of BCCs. Cyclopamine is a naturally occurring sterol alkaloid, derived from the Veratrum californicum (corn lily) plant. Identification of specific, small molecule antagonists of SMO has revealed exciting new prospects for targeted therapy of human cancers associated with Hh signaling.
In several mouse models, the in vivo effect of cyclopamine on tumor shrinkage has been demonstrated. Chronic oral delivery of cyclopamine blocks the growth of UV-induced BCCs in Ptch1+/− mice by with a 90% reduction in preexisting BCCs and a 50% reduction in new tumors [135], and also prevents development of additional microscopic BCCs, implying a cancer prevention potential of cyclopamine. Topical application of cyclopamine also significant reduce the size of human BCCs[187]. Clinical improvement was observed with 2 days’ topical use of cyclopamine in 4 patients with nevoid BCC syndrome.
However, the susceptibility to acid degradation and poor water solubility of cyclopamine hinders its utility as an administrable drug. Efforts to improve those parameters with additional modifications on cyclopamine have resulted in several derivatives with increasing acid stability and aqueous solubility, such as KAAD-cyclopamine, IPI-609, IPI-926, and Cyc-T[188–191]. IPI-926 is now at use in a Phase II clinical trial.
5.2 Synthetic Hh signaling antagonists
Because of the limitations of nature products for Hh pathway inhibition, a major focus of study has been to identify synthetic Hh antagonists with higher potency than cyclopamine, with most compounds targeting SMO. CUR61414 is a synthetic aminoproline that specifically inhibits SMO. CUR61414 decreased Gli1 levels, suppressed proliferation, and induced apoptosis of basaloid nests in murine embryonic skin. CUR61414 also caused regression of UV-induced tumors in Ptch1+/− knockout mice[192–195]. However, a Phase I clinical trial in 2005 for patients with BCC was halted because of its low efficacy, possibly due to poor penetration of the target tissue.
GDC-0449 is an orally active small molecule inhibitor of SMO[196] that has been studied in metastatic and locally advanced BCC. Van Hoff et al treated 33 BCC patients with oral GDC-0449[197]. Eighteen patients had a 50% objective response, suggesting that GDC-0449 is the most promising molecule in the treatment of locally advanced or metastatic basal-cell carcinoma[196]. Rapid tumor shrinkage was also observed in the clinical trial of GDC-0449 in a medulloblastoma patient, however, drug resistance happened very quickly after drug treatment due to a SMO mutation (disabling the binding of GDC-0449 to SMO)[198], implying a need for novel alternative strategies for treatment of cancers associated with Hh signaling. There are also several small molecules targeting at Shh or Gli[199]. Due to the wide spread existence of non-conical regulation of Gli transcription factors and the potential resistance to SMO inhibitors, antagonists of the Gli-factors, downstream of the Hh pathway, constitute a valuable resource for developing chemotherapeutic strategies against Hh pathway-related cancers.
5.3 Hh signaling modulators
Recent studies indicated that vitamin D3, whose secretion can be facilitated by PTCH1, can inhibit SMO signaling through direct binding to SMO. In transfection assays with mouse mesenchymal cells, addition of vitamin D3 inhibited GLI protein activity[75,200]. This finding raises a possibility to treat BCCs with nutritional supplements. Tazarotene, a retinoid with retinoic acid receptor (RAR) beta/gamma, was shown to be effective in a mouse model of BCCs, and the effect could be sustained even after drug withdrawal[201]. Several natural products, for instance genistein, EGCG and resveratrol, have been discovered to affect Hh signaling in a mouse model of prostate cancer[202]. A commonly used antifungal agent itraconazole, is shown to suppress Hh pathway activity and the growth of medulloblastoma in a mouse allograft model, acting on SMO by a mechanism distinct from that of cyclopamine and other known SMO antagonists[203]. Although the detailed molecular mechanisms of action for these signaling modulators remain elusive, elucidating the mechanisms of these modulators will open a new field for the treatment of Hh pathway-associated cancers.
6. Conclusions
In summary, recent advances have linked Hh signaling activation to a variety of human cancer, including skin cancers BCC, SCC, melanomas and MCC. The successful clinical trials of BCCs using Hh signaling inhibitor GDC-0449 further indicate the clinical implications of targeted inhibition of the Hh pathway. While a curative breakthrough for BCCs may occur in the foreseeable future, further basic understanding of the molecular mechanisms by which Hh signaling mediated cancer development is required for additional development and clinical application of hedgehog inhibitors. In particular, animal models for hedgehog signaling-mediated carcinogenesis for melanoma, SCC and MCC have not been established yet. It is anticipated that the next five year will be a fruitful period of time in the study of Hh signaling in skin cancers and the clinical implications.
Acknowledgments
Grants: from NSFC30572099, NSFC 30872264, NCET-05-0914 and NCI CA94160
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: No potential conflict of interests to be disclosed
References
- 1.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996 Nov;14(3):357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 3.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996 Nov;14(3):353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin. Genet. 67(2005):193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 5.Epstein EH. Basal cell carcinomas: Attack of the hedgehog. Nat Rev Cancer. 2008;8(10):743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J. Hedgehog signaling pathway: Development of antagonists or cancer therapy. Curr Oncol Rep. 2008;10(2):107–113. doi: 10.1007/s11912-008-0018-7. [DOI] [PubMed] [Google Scholar]
- 7.Xie J. Molecular biology of basal and squamous cell carcinomas. Adv Exp Med Biol. 2008;624:241–251. doi: 10.1007/978-0-387-77574-6_19. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation andmaintenance. Nat. Rev. Cancer. 3(2003):903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 10.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 11.Teglund S, Toftgård R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010 Apr;1805(2):181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010 Jan 28;29(4):469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 13.Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009 Jun;30(6):303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Mutations of the human homolog of drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr, Scott MP. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 16.Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of gorlin syndrome. Nat Med. 1998;4(5):619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich LV, Milenkovic L, Higgins KM, et al. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 18.Aszterbaum M, Beech J, Epstein EH., Jr Ultraviolet radiation mutagenesis of hedgehog pathway genes in basal cell carcinomas. J Investig Dermatol Symp Proc. 1999;4(1):41–45. doi: 10.1038/sj.jidsp.5640179. [DOI] [PubMed] [Google Scholar]
- 19.Anna Saran. Basal cell carcinoma and the carcinogenic role of aberrant Hedgehog signaling. Future Oncol. 2010;6(6):1003–1014. doi: 10.2217/fon.10.49. [DOI] [PubMed] [Google Scholar]
- 20.Heretsch P, Tzagkaroulaki L, Giannis A. Modulators of the hedgehog signaling pathway. Bioorg Med Chem. 2010 Sep 15;18(18):6613–6624. doi: 10.1016/j.bmc.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 21.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009 Sep;9(7):873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 22.Zhu W, You Z, Li T, Yu C, Tao G, Hu M, Chen X. Correlation of Hedgehog Signal Activation with Chemoradiotherapy Sensitivity and Survival in Esophageal Squamous Cell Carcinomas. Jpn J Clin Oncol. 2010 Dec;1 doi: 10.1093/jjco/hyq217. [DOI] [PubMed] [Google Scholar]
- 23.Schneider S, Thurnher D, Kloimstein P, Leitner V, Petzelbauer P, Pammer J, Brunner M, Erovic BM. Expression of the Sonic hedgehog pathway in squamous cell carcinoma of the skin and the mucosa of the head and neck. Head Neck. 2011 Feb;33(2):244–250. doi: 10.1002/hed.21437. [DOI] [PubMed] [Google Scholar]
- 24.Alexaki VI, Javelaud D, Van Kempen LC, Mohammad KS, Dennler S, Luciani F, Hoek KS, Juàrez P, Goydos JS, Fournier PJ, Sibon C, Bertolotto C, Verrecchia F, Saule S, Delmas V, Ballotti R, Larue L, Saiag P, Guise TA, Mauviel A. GLI2-mediated melanoma invasion and metastasis. J Natl Cancer Inst. 2010 Aug 4;102(15):1148–1159. doi: 10.1093/jnci/djq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng L, Cuneo KC, Cooper MK, Wang H, Sekhar K, Fu A, Hallahan DE. Hedgehog signaling in the murine melanoma microenvironment. Angiogenesis. 2007;10(4):259–267. doi: 10.1007/s10456-007-9078-9. [DOI] [PubMed] [Google Scholar]
- 26.Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz I Altaba A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007 Apr 3;104(14):5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunner M, Thurnher D, Pammer J, Heiduschka G, Petzelbauer P, Schmid C, Schneider S, Erovic BM. Expression of hedgehog signaling molecules in Merkel cell carcinoma. Head Neck. 2010 Mar;32(3):333–340. doi: 10.1002/hed.21191. [DOI] [PubMed] [Google Scholar]
- 28.Kolterud A, Toftgard R. Strategies for Hedgehog inhibition and its potential role in cancer treatment. Drug Discovery Today: Therapeutic Strategies. 2007;4(4):229–235. [Google Scholar]
- 29.So PL, Tang JY, Epstein EH. Novel investigational drugs for basal cell carcinoma. Expert Opin Investig Drugs. 2010 Sep;19(9):1099–1112. doi: 10.1517/13543784.2010.504714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caro I, Low JA. The role of the hedgehog signaling pathway in the development of basal cell carcinoma and opportunities for treatment. Clin Cancer Res. 2010 Jul 1;16(13):3335–3339. doi: 10.1158/1078-0432.CCR-09-2570. [DOI] [PubMed] [Google Scholar]
- 31.Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7(11):841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 32.Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266(5190):1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 33.Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374(6520):363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 34.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274(5285):255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Zhang H, Litingtung Y, Chiang C. Cholesterol modification restricts the spread of Shh gradient in the limb bud. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6548–6553. doi: 10.1073/pnas.0600124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for n-palmitoylation of sonic hedgehog. J Biol Chem. 2008;283(32):22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, McMahon AP. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- 38.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and longrange signaling in vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111(1):63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 40.Caspary T, García-García MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, Alcorn HL, Anderson KV. Mouse dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr Biol. 2002;12(18):1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- 41.Dierker T, Dreier R, Petersen A, Bordych C, Grobe K. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells. J Biol Chem. 2009;284(12):8013–8022. doi: 10.1074/jbc.M806838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckett K, Franch-Marro X, Vincent JP. Glypican-mediated endocytosis of hedgehog has opposite effects in flies and mice. Trends Cell Biol. 2008;18(8):360–363. doi: 10.1016/j.tcb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of hedgehog pathway components by RNAi in drosophila cultured cells. Science. 2003;299(5615):2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 44.Baena-Lopez LA, Rodriguez I, Baonza A. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci U S A. 2008;105(28):9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyoda H, Kinoshita-Toyoda A, Fox B, Selleck SB. Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J Biol Chem. 2000;275(29):21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- 46.Bellaiche Y, The I, Perrimon N. Tout-velu is a drosophila homologue of the putative tumour suppressor EXT-1 and is needed for hh diffusion. Nature. 1998;394(6688):85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 47.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of ihh signaling during endochondral ossification. Dev Cell. 2004;6(6):801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 49.Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ. G protein galphai functions immediately downstream of smoothened in hedgehog signalling. Nature. 2008;456(7224):967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450(7167):252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 51.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of smoothened. Nature. 2002;418(6900):892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 54.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 56.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 57.Chuang PT, McMahon AP. Vertebrate hedgehog signalling modulated by induction of a hedgehog-binding protein. Nature. 1999;397(6720):617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 58.Martinelli DC, Fan CM. Gas1 extends the range of hedgehog action by facilitating its signaling. Genes Dev. 2007;21(10):1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen BL, Tenzen T, McMahon AP. The hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21(10):1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seppala M, Depew MJ, Martinelli DC, et al. Gas1 is a modifier for holoprosencephaly and genetically interacts with sonic hedgehog. J Clin Invest. 2007;117(6):1575–1584. doi: 10.1172/JCI32032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10(5):647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444(7117):369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- 63.Zhang W, Kang JS, Cole F, Yi MJ, Krauss RS. Cdo functions at multiple points in the sonic hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev Cell. 2006;10(5):657–665. doi: 10.1016/j.devcel.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Yao S, Lum L, Beachy P. The ihog cell-surface proteins bind hedgehog and mediate pathway activation. Cell. 2006;125(2):343–357. doi: 10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 65.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits hedgehog signaling during development by competing with patched for hedgehog binding. Dev Cell. 2008;14(5):700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 66.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 68.Huangfu D, Anderson KV. Cilia and hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102(32):11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoover AN, Wynkoop A, Zeng H, Jia J, Niswander LA, Liu A. C2cd3 is required for cilia formation and hedgehog signaling in mouse. Development. 2008;135(24):4049–4058. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiprilov EN, Awan A, Desprat R, et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J. Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl Acad. Sci. USA. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.YYavari A, Nagaraj R, Owusu-Ansah E, Folick A, Ngo K, Hillman T, Call G, Rohatgi R, Scott MP, Banerjee U. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev Cell. 2010;19(1):54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khaliullina H, Panáková D, Eugster C, Riedel F, Carvalho M, Eaton S. Patched regulates smoothened trafficking using lipoprotein-derived lipids. Development. 2009;136(24):4111–4121. doi: 10.1242/dev.041392. [DOI] [PubMed] [Google Scholar]
- 74.Callejo A, Culi J, Guerrero I. Patched, the receptor of hedgehog, is a lipoprotein receptor. Proc Natl Acad Sci USA. 2008;105(3):912–917. doi: 10.1073/pnas.0705603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bijlsma MF, Spek CA, Zivkovic D, van de Water S, Rezaee F, Peppelenbosch MP. Repression of smoothened by patched-dependent (pro-)vitamin d3 secretion. PLoS Biol. 2006;4(8):e232. doi: 10.1371/journal.pbio.0040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz RJ. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320(5884):1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl. Acad. Sci. USA. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of gli protein function by Su(Fu) in hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23(16):1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia J, Kolterud A, Zeng H, Hoover A, Teglund S, Toftgård R, Liu A. Suppressor of fused inhibits mammalian hedgehog signaling in the absence of cilia. Dev Biol. 2009;330(2):452–460. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The gli gene is a member of the kruppel family of zinc finger proteins. Nature. 1988;332(6162):371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 83.Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, Seuanez HN, O'Brien SJ, Vogelstein B. The gli-kruppel family of human genes. Mol Cell Biol. 1988;8(8):3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kinzler KW, Vogelstein B. The Gli gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10(2):634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124(7):1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 86.Sheng T, Chi S, Zhang X, Xie J. Regulation of gli1 localization by the camp/protein kinase a signaling axis through a site near the nuclear localization signal. J Biol Chem. 2006;281(1):9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- 87.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007 May 11;282(19):14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 88.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26(9):3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang B, Li Y. Evidence for the direct involvement of {beta}trcp in gli3 protein processing. Proc Natl Acad Sci USA. 2006;103(1):33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. Numb is a suppressor of hedgehog signalling and targets gli1 for itch-dependent ubiquitination. Nat Cell Biol. 2006;8(12):1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 91.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, Pelloni M, De Simone G, Pedone EM, Gallinari P, Giorgi A, Steinkühler C, Vitagliano L, Pedone C, Schinin ME, Screpanti I, Gulino A. Histone deacetylase and cullin3-ren(kctd11) ubiquitin ligase interplay regulates hedgehog signalling through gli acetylation. Nat Cell Biol. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 92.Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev. Biol. 2009;326:177–189. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J Biol. Chem. 2007;282:10846–10852. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- 94.Tempe D, Casas M, Karaz S, Blanchet-Tournier MF, Concorde t JP. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol. Cell. Biol. 2006;26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang J. Regulation of hh/gli signaling by dual ubiquitin pathways. Cell Cycle. 2006;5(21) doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- 96.Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, Ojala P, Taipale J. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 97.Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, Ye W, Davis DP, de Sauvage FJ. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci. Signal. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- 98.Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KK, Briscoe J, Hui CC. The kinesin protein kif7 is a critical regulator of gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2(76):ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 99.Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, Peterson AS. The mammalian cos2 homolog kif7 plays an essential role in modulating hh signal transduction during development. Curr Biol. 2009;19(15):1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 100.Wilson CW, Nguyen CT, Chen MH, Yang JH, Gacayan R, Huang J, Chen JN, Chuang PT. Fused has evolved divergent roles in vertebrate hedgehog signalling and motile ciliogenesis. Nature. 2009;459(7243):98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merchant M, Evangelista M, Luoh SM, Frantz GD, Chalasani S, Carano RA, van Hoy M, Ramirez J, Ogasawara AK, McFarland LM, Filvaroff EH, French DM, de Sauvage FJ. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25(16):7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen MH, Gao N, Kawakami T, Chuang PT. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25(16):7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt B, Toftgård R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 104.Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 105.Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 106.Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 107.Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 108.Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 2004;24:8627–8641. doi: 10.1128/MCB.24.19.8627-8641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yue S, Chen Y, Cheng SY. Hedgehog signaling promotes the degradation of tumor suppressor Su(Fu) through the ubiquitin-proteasome pathway. Oncogene. 2009;28(4):492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- 110.Kasai K, Inaguma S, Yoneyama A, Yoshikawa K, Ikeda H. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res. 2008;68:7723–7729. doi: 10.1158/0008-5472.CAN-07-6661. [DOI] [PubMed] [Google Scholar]
- 111.Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem. J. 2004;378:353–362. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cheng SY, Bishop JM. Suppressor of fused represses gli-mediated transcription by recruiting the sap18-msin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99(8):5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kise Y, Morinaka A, Teglund S, Miki H. Su(Fu) recruits gsk3beta for efficient processing of gli3. Biochem Biophys Res Commun. 2009;387(3):569–574. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 114.Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse sonic hedgehog signalling pathway. Nature. 2001;412(6843):194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 115.Reiter JF, Skarnes WC. Tectonic, a novel regulator of the hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20(1):22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang S, Yang L, An Y, Ma X, Zhang C, Xie G, Chen ZY, Xie J, Zhang H. Expression of hedgehog signaling molecules in lung cancer. Acta Histochem, Acta Histochem. 2010 Jul 23; doi: 10.1016/j.acthis.2010.06.003. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 117.Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007 May 15;21(10):1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007 May 15;21(10):1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cohen MM., Jr Hedgehog signaling update. Am J Med Genet A. 2010 Aug;152A(8):1875–1914. doi: 10.1002/ajmg.a.32909. [DOI] [PubMed] [Google Scholar]
- 120.Liu H, Gu D, Xie J. Clinical implications of hedgehog signaling pathway inhibitors. Chin J Cancer. 2011 Jan;30(1):13–26. doi: 10.5732/cjc.010.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Undén AB, Dean M, Brash DE, Bale AE, Toftgård R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 122.Kim M Y, Park HJ, Baek SC, Byun DG, Houh D. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J Dermatol Sci. 2002 May;29(1):1–9. doi: 10.1016/s0923-1811(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 123.Hahn H, Wojnowski L, Miller G, Zimmer A. The patched signaling pathway in tumorigenesis and development: lessons from animal models. J Mol Med. 1999;77:459–468. doi: 10.1007/s001099900018. [DOI] [PubMed] [Google Scholar]
- 124.Oro AE, Higgins KM, Hu Z, Bonifas JM, Epstein EH, Jr, Scott M. Basal cell carcinoma in mice overexpressing sonic hedgehog. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 125.Fan H, Oro AE, Scott MP, Khavari PA. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic Hedgehog. Nat Med. 1997;3:788–792. doi: 10.1038/nm0797-788. [DOI] [PubMed] [Google Scholar]
- 126.Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, de Sauvage FJ. Activating Smoothened mutations in sporadic basal cell carcinoma. Nature. 1998 Jan 1;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 127.Lam CW, Xie J, To KF, Ng HK, Lee KC, Yuen NW, Lim PL, Chan LY, Tong SF, McCormick F. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18:833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 128.Reifenberger J, Wolter M, Weber RG, Megahed M, Ruzicka T, Lichter P, Reifenberger G. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- 129.Nilsson M, Undèn AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgård R. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI1. Proc. Natl Acad. Sci. USA. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, Dlugosz AA. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 131.Sheng H, Goich S, Wang A, Grachtchouk M, Lowe L, Mo R, Lin K, de Sauvage FJ, Sasaki H, Hui CC, Dlugosz AA. deletion of an NH(2)terminal fragment alters skin tumor phenotype. Cancer Res. 2002;62:5308–5316. [PubMed] [Google Scholar]
- 132.Reifenberger J, Wolter M, Knobbe CB, Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka T, Reifenberger G. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br. J. Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 133.Pastorino L, Ghiorzo P, Nasti S, Battistuzzi L, Cusano R, Marzocchi C, Garrè ML, Clementi M, Scarrà GB. Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am. J. Med. Genet. 149A(2009):1539–1543. doi: 10.1002/ajmg.a.32944. [DOI] [PubMed] [Google Scholar]
- 134.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 135.Athar M, Li C, Tang X, Chi S, Zhang X, Kim AL, Tyring SK, Kopelovich L, Hebert J, Epstein EH, Jr, Bickers DR, Xie J. Inhibition of smoothened signaling prevents ultraviolet b-induced basal cell carcinomas through regulation of fas expression and apoptosis. Cancer Res. 2004;64(20):7545–7552. doi: 10.1158/0008-5472.CAN-04-1393. [DOI] [PubMed] [Google Scholar]
- 136.Mancuso M, Pazzaglia S, Tanori M, Hahn H, Merola P, Rebessi S, Atkinson MJ, Di Majo V, Covelli V, Saran A. Basal cell carcinoma and its development: Insights from radiation-induced tumors in ptch1-deficient mice. Cancer Res. 2004;64(3):934–941. doi: 10.1158/0008-5472.can-03-2460. [DOI] [PubMed] [Google Scholar]
- 137.Ellis T, Smyth I, Riley E, Graham S, Elliot K, Narang M, Kay GF, Wicking C, Wainwright B. Patched 1 conditional null allele in mice. Genesis. 2003;36(3):158–161. doi: 10.1002/gene.10208. [DOI] [PubMed] [Google Scholar]
- 138.Siggins SL, Nguyen NY, McCormack MP, Vasudevan S, Villani R, Jane SM, Wainwright BJ, Curtis DJ. The Hedgehog receptor Patched1 regulates myeloid and lymphoid progenitors by distinct cell-extrinsic mechanisms. Blood. 2009;114:995–1004. doi: 10.1182/blood-2009-03-208330. [DOI] [PubMed] [Google Scholar]
- 139.Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A, de Sauvage F, Dlugosz AA. The magnitude of hedgehog signaling activity defines skin tumor phenotype. EMBO J. 2003;22(11):2741–2751. doi: 10.1093/emboj/cdg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the hedgehog pathway. Cancer Res. 2006;66(20):10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Donovan J. Review of the hair follicle origin hypothesis for basal cell carcinoma. Dermatol. Surg. 2009;35:1311–1323. doi: 10.1111/j.1524-4725.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 142.Fazaa B, Cribier B, Zaraa I, Zermani R, Zeglaoui F, Zouari B, Ben Jilani S, Maalej M, Kamoun MR. Low-dose X-ray depilatory treatment induces trichoblastic tumors of the scalp. Dermatology. 2007;215:301–307. doi: 10.1159/000107623. [DOI] [PubMed] [Google Scholar]
- 143.Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, Sotiropoulou PA, Blanpain C. Identification of the cell lineage at the origin of basal cell carcinoma. Nat. Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 144.Wang GraceYing, Wang Joy, Mancianti Maria-Laura, Epstein ErvinH., Jr Basal cell carcinomas arise from hair follicle stem cells in Ptch1+/− mice. Cancer Cell. 2011 January 18;19:1–11. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ho L, Alman B. Protecting the hedgerow: p53 and hedgehog pathway interactions. Cell Cycle. 2010 Feb 1;9(3):506–511. doi: 10.4161/cc.9.3.10552. [DOI] [PubMed] [Google Scholar]
- 146.Scholey JM, Anderson KV, Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125(3):439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 147.Lehman JM, Laag E, Michaud EJ, Yoder BK. An essential role for dermal primary cilia in hair follicle morphogenesis. J Invest. Dermatol. 2009;129:438–448. doi: 10.1038/jid.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mill P, Mo R, Fu H, Grachtchouk M, Kim PC, Dlugosz AA, Hui CC. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins [J] Nature. 2003;426(6962):3–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 150.Cortellino S, Wang C, Wang B, Bassi MR, Caretti E, Champeval D, Calmont A, Jarnik M, Burch J, Zaret KS, Larue L, Bellacosa A. Defective ciliogenesis, embryonic lethality and severe impairment of the sonic hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene ift122/wdr10, partially overlapping with the DNA repair gene med1/mbd4. Dev Biol. 2009;325(1):225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287(2):378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 152.Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One. 2009;4(4):e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Jr, Dlugosz AA, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathwaydependent tumorigenesis. Nat. Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 155.Michimukai E, Kitamura N, Zhang Y, Wang H, Hiraishi Y, Sumi K, Hayashido Y, Toratani S, Okamoto T. Mutations in the human homologue of the Drosophila segment polarity gene patched in oral squamous cell carcinoma cell lines. In Vitro Cell Dev Biol Anim. 2001 Jul–Aug;37(7):459–464. doi: 10.1290/1071-2690(2001)037<0459:mithho>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 156.Koike C, Mizutani T, Ito T, Shimizu Y, Yamamichi N, Kameda T, Michimukai E, Kitamura N, Okamoto T, Iba H. Introduction of wild-type patched gene suppresses the oncogenic potential of human squamous cell carcinoma cell lines including A431. Oncogene. 2002 Apr 18;21(17):2670–2678. doi: 10.1038/sj.onc.1205370. [DOI] [PubMed] [Google Scholar]
- 157.Ping XL, Ratner D, Zhang H, Wu XL, Zhang MJ, Chen FF, Silvers DN, Peacocke M, Tsou HC. PTCH mutations in squamous cell carcinoma of the skin. J Invest Dermatol. 2001 Apr;116(4):614–616. doi: 10.1046/j.1523-1747.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- 158.Danaee H, Karagas MR, Kelsey KT, Perry AE, Nelson HH. Allelic loss at Drosophila patched gene is highly prevalent in basal and squamous cell carcinomas of the skin. J Invest Dermatol. 2006;126:1152–1158. doi: 10.1038/sj.jid.5700209. [DOI] [PubMed] [Google Scholar]
- 159.Wakabayashi Y, Mao JH, Brown K, Girardi M, Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007 Feb 15;445(7129):761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- 160.Nishimaki H, Kasai K, Kozaki K, Takeo T, Ikeda H, Saga S, Nitta M, Itoh G. A role of activated Sonic hedgehog signaling for the cellular proliferation of oral squamous cell carcinoma cell line. Biochem Biophys Res Commun. 2004 Feb 6;314(2):313–320. doi: 10.1016/j.bbrc.2003.12.097. [DOI] [PubMed] [Google Scholar]
- 161.Fujita E, Khoroku Y, Urase K, Tsukahara T, Momoi MY, Kumagai H, Takemura T, Kuroki T, Momoi T. Involvement of Sonic hedgehog in the cell growth of LK-2 cells, human lung squamous carcinoma cells. Biochem Biophys Res Commun. 1997 Sep 18;238(2):658–664. doi: 10.1006/bbrc.1997.7262. [DOI] [PubMed] [Google Scholar]
- 162.Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, Jones D, Castro CY, Logrono R, Haque A, Zwischenberger J, Tyring SK, Zhang H, Xie J. Activation of the hedgehog pathway in a subset of lung cancers. Cancer Lett. 2006 Nov 28;244(1):53–60. doi: 10.1016/j.canlet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 163.Xuan YH, Jung HS, Choi YL, Shin YK, Kim HJ, Kim KH, Kim WJ, Lee YJ, Kim SH. Enhanced expression of hedgehog signaling molecules in squamous cell carcinoma of uterine cervix and its precursor lesions. Mod Pathol. 2006 Aug;19(8):1139–1147. doi: 10.1038/modpathol.3800600. [DOI] [PubMed] [Google Scholar]
- 164.Ma X, Sheng T, Zhang Y, Zhang X, He J, Huang S, Chen K, Sultz J, Adegboyega PA, Zhang H, Xie J. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer. 2006 Jan 1;118(1):139–148. doi: 10.1002/ijc.21295. [DOI] [PubMed] [Google Scholar]
- 165.Mori Y, Okumura T, Tsunoda S, Sakai Y, Shimada Y. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology. 2006;70(5):378–389. doi: 10.1159/000098111. [DOI] [PubMed] [Google Scholar]
- 166.Isohata N, Aoyagi K, Mabuchi T, Daiko H, Fukaya M, Ohta H, Ogawa K, Yoshida T, Sasaki H. Hedgehog and epithelial-mesenchymal transition signaling in normal and malignant epithelial cells of the esophagus. Int J Cancer. 2009 Sep 1;125(5):1212–1221. doi: 10.1002/ijc.24400. [DOI] [PubMed] [Google Scholar]
- 167.Ma XL, Sun HJ, Wang YS, Huang SH, Xie JW, Zhang HW. Study of Sonic hedgehog signaling pathway related molecules in gastric carcinoma. World J Gastroenterol. 2006 Jul 7;12(25):3965–3969. doi: 10.3748/wjg.v12.i25.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schneider S, Thurnher D, Kloimstein P, Leitner V, Petzelbauer P, Pammer J, Brunner M, Erovic BM. Expression of the Sonic hedgehog pathway in squamous cell carcinoma of the skin and the mucosa of the head and neck. Head Neck. 2011 Feb;33(2):244–250. doi: 10.1002/hed.21437. [DOI] [PubMed] [Google Scholar]
- 169.Zhu W, You Z, Li T, Yu C, Tao G, Hu M, Chen X. Correlation of Hedgehog Signal Activation with Chemoradiotherapy Sensitivity and Survival in Esophageal Squamous Cell Carcinomas. Jpn J Clin Oncol. 2010 De;1 doi: 10.1093/jjco/hyq217. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 170.Yoshikawa R, Nakano Y, Tao L, Koishi K, Matsumoto T, Sasako M, Tsujimura T, Hashimoto-Tamaoki T, Fujiwara Y. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br J Cancer. 2008 May 20;98(10):1670–1674. doi: 10.1038/sj.bjc.6604361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schneider FT, Schänzer A, Czupalla CJ, Thom S, Engels K, Schmidt MH, Plate KH, Liebner S. Sonic hedgehog acts as a negative regulator of {beta}-catenin signaling in the adult tongue epithelium. Am J Pathol. 2010 Jul;177(1):404–414. doi: 10.2353/ajpath.2010.091079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Houghton AN, Polsky D. Focus on melanoma. Cancer Cell. 2002;2(4):275–278. doi: 10.1016/s1535-6108(02)00161-7. [DOI] [PubMed] [Google Scholar]
- 173.Bastian BC. Molecular genetics of melanocytic neoplasia: practical applications for diagnosis. Pathology. 2004;36(5):458–461. doi: 10.1080/00303020412331282717. [DOI] [PubMed] [Google Scholar]
- 174.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 175.Crowson AN, Magro C, Miller A, Mihm MC., Jr The molecular basis of melanomagenesis and the metastatic phenotype. Semin Oncol. 2007;34(6):476–490. doi: 10.1053/j.seminoncol.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 176.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 177.Das S, Harris LG, Metge BJ, Liu S, Riker AI, Samant RS, Shevde LA. The Hedgehog pathway transcription factor GLI1 promotes malignant behavior of cancer cells by up-regulating osteopontin. J. Biol. Chem. 2009;284:22888–22897. doi: 10.1074/jbc.M109.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Alexaki Vasileia-Ismini, Javelaud Delphine, Van Kempen LeonCL, Mohammad KhalidS, Dennler Sylviane, Luciani Flavie, Hoek KeithS, Juàrez Patricia, Goydos JamesS, Fournier PierrickJ, Sibon Claire, Bertolotto Corine, Verrecchia Franck, Saule Simon, Delmas Veronique, Ballotti Robert, Larue Lionel, Saiag Philippe, Guise TheresaA, Mauviel Alain. Gli2-Mediated Melanoma invasion and Metastasis. J Natl Cancer Inst. 2010;102:1148–1159. doi: 10.1093/jnci/djq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dennler S, André J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67(14):6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 180.Javelaud D, Alexaki VI, Mauviel A. Transforming growth factor-beta in cutaneous melanoma. Pigment Cell Melanoma Res. 2008;21(4):123–132. doi: 10.1111/j.1755-148X.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- 181.Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, Matrisian LM, Mundy GR, Sterling JA. TGF-{beta} Promotion of Gli2-Induced Expression of Parathyroid Hormone-Related Protein, an Important Osteolytic Factor in Bone Metastasis, Is Independent of Canonical Hedgehog Signaling. Cancer Res. 2011 Feb 1;71(3):822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Brunner M, Thurnher D, Pammer J, Heiduschka G, Petzelbauer P, Schmid C, Schneider S, Erovic BM. Expression of hedgehog signaling molecules in Merkel cell carcinoma. Head Neck. 2010 Mar;32(3):333–340. doi: 10.1002/hed.21191. [DOI] [PubMed] [Google Scholar]
- 183.Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 184.Tanese K, Fukuma M, Yamada T, Mori T, Yoshikawa T, Watanabe W, Ishiko A, Amagai M, Nishikawa T, Sakamoto M. G-protein-coupled receptor GPR49 is upregulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am. J. Pathol. 2008;173:835–843. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Valin A, Barnay-Verdier S, Robert T, Ripoche H, Brellier F, Chevallier-Lagente O, Avril MF, Magnaldo T. PTCH1 dermal fibroblasts isolated from healthy skin of Gorlin syndrome patients exhibit features of carcinoma associated fibroblasts. PLoS ONE. 2009;4:e4818. doi: 10.1371/journal.pone.0004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev. 2002;16(21):2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tabs S, Avci O. Induction of the differentiation and apoptosis of tumor cells in vivo with efficiency and selectivity. Eur J Dermatol. 2004;14:96–102. [PubMed] [Google Scholar]
- 188.Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, Koorstra JB, Habbe N, Karikari C, Mullendore M, Gabrielson KL, Sharma R, Matsui W, Maitra A. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol. Cancer Ther. 2008;7:2725–2735. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tremblay MR, Nevalainen M, Nair SJ, Porter JR, Castro AC, Behnke ML, Yu LC, Hagel M, White K, Faia K, Grenier L, Campbell MJ, Cushing J, Woodward CN, Hoyt J, Foley MA, Read MA, Sydor JR, Tong JK, Palombella VJ, McGovern K, Adams J. Semisynthetic cyclopamine analogues as potent and orally bioavailable hedgehog pathway antagonists. J. Med. Chem. 2008;51:6646–6649. doi: 10.1021/jm8008508. [DOI] [PubMed] [Google Scholar]
- 190.Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, Yu LC, Behnke ML, Nair SJ, Hagel M, White K, Conley J, Manna JD, Alvarez-Diez TM, Hoyt J, Woodward CN, Sydor JR, Pink M, MacDougall J, Campbell MJ, Cushing J, Ferguson J, Curtis MS, McGovern K, Read MA, Palombella VJ, Adams J, Castro AC. Discovery of a potent and orally active hedgehog pathway antagonist (ipi-926) J Med Chem. 2009;52(14):4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- 191.Xie J, Garrossian M. The Board of Regents of the University of Texas System, Assignee. US patent application number WO 2009099625. Cyclopamine tartrate salt and uses thereof. 2009
- 192.Daya-Grosjean L, Couve-Privat S. Sonic hedgehog signaling in basal cell carcinomas. Cancer Lett. 2005;225:181–192. doi: 10.1016/j.canlet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 193.Szeimies RM, Karrer S. Towards a more specific therapy: targeting nonmelanoma skin cancer cells. Br J Dermatol. 2006;154 Suppl. 1:16–21. doi: 10.1111/j.1365-2133.2006.07232.x. [DOI] [PubMed] [Google Scholar]
- 194.Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, Kon C, Gatchalian C, Porter JA, Rubin LL, Wang FY. Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc Natl Acad Sci USA. 2003;100:4616–4621. doi: 10.1073/pnas.0732813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Athar M, Tang X, Lee JL, Kopelovich L, Kim AL. Hedgehog signalling in skin development and cancer. Exp Dermatol. 2006;15:667–677. doi: 10.1111/j.1600-0625.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 196.Robarge KD, Brunton SA, Castanedo GM, Cui Y, Dina MS, Goldsmith R, Gould SE, Guichert O, Gunzner JL, Halladay J, Jia W, Khojasteh C, Koehler MF, Kotkow K, La H, Lalonde RL, Lau K, Lee L, Marshall D, Marsters JC, Jr, Murray LJ, Qian C, Rubin LL, Salphati L, Stanley MS, Stibbard JH, Sutherlin DP, Ubhayaker S, Wang S, Wong S, Xie M. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009 Oct 1;19(19):5576–5581. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 197.Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, Darbonne WC, Marsters JC, Jr, de Sauvage FJ, Low JA. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 198.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009 Sep 17;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, Solow-Cordero DE, Jiang J, Rowitch DH, Chen JK. Small-molecule inhibitors reveal multiple strategies for hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106(33):14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tang JY, So PL, Epstein EH., Jr Novel Hedgehog pathway targets against basal cell carcinoma. Toxicol Appl Pharmacol. 2007;224:257–264. doi: 10.1016/j.taap.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.So PL, Fujimoto MA, Epstein EH., Jr Pharmacologic retinoid signaling and physiologic retinoic acid receptor signaling inhibit basal cell carcinoma tumorigenesis. Mol Cancer Ther. 2008;7(5):1275–1284. doi: 10.1158/1535-7163.MCT-07-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Slusarz A, Shenouda NS, Sakla MS, Drenkhahn SK, Narula AS, MacDonald RS, Besch-Williford CL, Lubahn DB. Common botanical compounds inhibit the hedgehog signaling pathway in prostate cancer. Cancer Res. 2010;70(8):3382–3390. doi: 10.1158/0008-5472.CAN-09-3012. [DOI] [PubMed] [Google Scholar]
- 203.Kim J, Tang JY, Gong R, Kim J, Lee JJ, Clemons KV, Chong CR, Chang KS, Fereshteh M, Gardner D, Reya T, Liu JO, Epstein EH, Stevens DA, Beachy PA. Itraconazole, a commonly used antifungal that inhibits hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17(4):388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, Taveras KM, Hyman JM, Lee SW, Koehler AN, Chen JK, Fox JL, Mandinova A, Schreiber SL. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol. 2009 Mar;5(3):154–156. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sanchez P, Ruiz i Altaba A. In vivo inhibition of endogenous brain tumors through systemic interference of hedgehog signaling in mice. Mech Dev. 2005;122(2):223–230. doi: 10.1016/j.mod.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 206.Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, Zhang K, Tang W, Stelter AA, Wang Q, Zhang H, Xie J. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006 Jul;27(7):1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 207.Mistretta CM, Liu HX, Gaffield W, MacCallum DK. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for Shh signaling in taste papilla development and patterning: fungiform papillae double in number and form in novel locations in dorsal lingual epithelium. Dev Biol. 2003 Feb 1;254(1):1–18. doi: 10.1016/s0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 208.Frank-Kamenetsky M, Zhang XM, Bottega S, Guicherit O, Wichterle H, Dudek H, Bumcrot D, Wang FY, Jones S, Shulok J, Rubin LL, Porter JA. Small-molecule modulators of hedgehog signaling: Identification and characterization of smoothened agonists and antagonists. J Biol. 2002;1(2):10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of smoothened activity. Proc Natl Acad Sci USA. 2002;99(22):14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Borzillo GV, Lippa B. The hedgehog signaling pathway as a target for anticancer drug discovery. Curr Top Med Chem. 2005;5(2):147–157. doi: 10.2174/1568026053507732. [DOI] [PubMed] [Google Scholar]
- 211.Rubin LL, de Sauvage FJ. Targeting the hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5(12):1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 212.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, Denicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Rückert F, Grützmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, McGovern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, Curran T. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/ˇ)p53(ˇ/ˇ) mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 214.Iwasaki JK, Srivastava D, Moy RL, Lin HJ, Kouba DJ. The molecular genetics underlying basal cell carcinoma pathogenesis and links to targeted therapeutics. J Am Acad Dermatol. 2010 Aug 25; doi: 10.1016/j.jaad.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 215.Vogt A, Chuang PT, Hebert J, Hwang J, Lu Y, Kopelovich L, Athar M, Bickers DR, Epstein EH., Jr Immunoprevention of basal cell carcinomas with recombinant hedgehog-interacting protein. J Exp Med. 2004 Mar 15;199(6):753–761. doi: 10.1084/jem.20031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lauth M, Bergström A, Shimokawa T, Toftgård R. Inhibition of gli-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA. 2007;104(20):8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]