Abstract
Although adeno-associated viral (AAV) vectors have been successfully used in hepatic gene transfer for treatment of hemophilia and other diseases in animals, adaptive immune responses blocked long-term transgene expression in patients on administration of single-stranded AAV serotype-2 vector. More efficient vectors have been developed using alternate capsids and self-complimentary (sc) genomes. This study investigated their effects on the innate immune profile on hepatic gene transfer to mice. A mild and transient up-regulation of myeloid differentiation primary response gene (88), TLR9, TNF-α, monocyte chemotactic protein-1, IFN-γ inducible protein-10, and IFN-α/β expression in the liver was found after single-stranded AAV vector administration, regardless of the capsid sequence. In contrast, scAAV vectors induced higher increases of these transcripts, upregulated additional proinflammatory genes, and increased circulating IL-6. Neutrophil, macrophage, and natural killer cell liver infiltrates were substantially higher on injection of scAAV. Some but not all of these responses were Kupffer cell dependent. Independent of the capsid or expression cassette, scAAV vectors induced dose-dependent innate responses by signaling through TLR9. Increased innate responses to scAAV correlated with stronger adaptive immune responses against capsid (but not against the transgene product). However, these could be blunted by transient inhibition of TLR9.
Introduction
Adeno-associated viral (AAV) vectors are widely used for stable in vivo gene transfer to terminally differentiated or quiescent cells such as muscle fibers, hepatocytes, neurons, retinal cells, and others. These vectors, derived from a nonpathogenic, replication-defective parvovirus with a small single-stranded (ss) DNA genome, have recently been successfully used in clinical gene transfer for inherited blindness and also show promise for other diseases.1,2
Eight years ago, Zaiss et al3 found that ssAAV serotype-2 vectors caused only a weak and highly transient innate immune response in the liver, suggesting that inflammatory reactions to AAV are negligible. Numerous animal studies have shown stable correction of genetic diseases by hepatic AAV gene transfer that may, in part, be because of the low innate immune profile of the vector, avoiding inflammatory signals.4 In humans, hepatic gene transfer with AAV2 has been hampered by pre-existing adaptive immunity after natural infection in the form of neutralizing antibodies and capsid-specific CD8+ T cells.5 Numerous changes to capsid and vector genomes have been developed in recent years in attempts to improve gene transfer efficacy and possibly evade immunity. For example, AAV8 shows substantially higher transduction efficiency in mouse liver and reduced activation of capsid-specific T cells, and it facilitates tolerance induction to transgenes.6,7 Furthermore, prevalence for neutralizing antibodies in humans is markedly lower to AAV8 than to AAV2.8 In another set of investigations, replacing surface-exposed tyrosine residues to phenylalanine has been shown to improve gene transfer for several serotypes. The resulting reduction in capsid phosphorylation in turn reduces accumulation in the cytoplasm (in favor of trafficking to the nucleus) and ubiquitination of capsid.9 AAV2 gene transfer to hepatocytes was most improved by a combination of 3 Tyr-Phe changes in amino acid residues 444, 500, and 730.10
Modifications of the recombinant AAV genome also can improve transduction rates. Being ss, the ssAAV genome has to be converted to a double-stranded form in the nucleus of an infected cell for transgene expression to occur. To overcome this rate-limiting step, self-complementary (sc)AAV vectors were developed by elimination of the terminal resolution site in one of the inverted terminal repeats (ITRs).11 For such a genome to be packaged into capsid, the size of the expression cassette has to be further reduced to not exceed the packaging limit. Two groups reported optimized scAAV vectors for treatment of the X-linked bleeding disorder hemophilia B (coagulation factor IX deficiency) by liver gene transfer.12,13
The hepatic microenvironment is more tolerogenic than that of many other tissues.14 For example, we were able to tolerize hemophilia B mice to human factor IX (hF.IX) by hepatic ssAAV2 gene transfer. This protocol was successful in several strains, with the exception of C3H.15 Nonetheless, AAV8 and AAV2(Y444/500/730F) vectors were able to tolerize this strain to hF.IX on gene transfer to the liver, prompting us to speculate that innate responses to these capsids may differ.7,10 Thus, we compared the innate immune profile of several ssAAV vectors in the murine liver, but we found an identical mild and highly transient response regardless of capsid sequence. However, changing the genome to scAAV substantially increased innate immunity in a TLR9-dependent manner.
Methods
Animal studies
C57BL/6, BALB/c, and C3H/OuJ mice (male, 6-8 weeks old) were purchased from The Jackson Laboratory. Hemophilia B C3H/HeJ mice (F9 gene deletion) were as published previously.16 AAV viral vectors (1 × 1011 vector genomes [vg] per mouse) or PBS was delivered to the portal circulation (via splenic capsule; C3H/OuJ, BALB/c, and hemophilia B mice) or by tail vein (intravenously, C57BL/6) injection.17 TLR9−/− C57BL/6J mice were obtained from Dr B. Beutler (The Scripps Research Institute).18 For blocking normal TLR9 function in wild-type C57BL/6J mice, 100 μg of TLR9 inhibitory oligodeoxynucleotide (ODN)2088 (TLR-9i) or TLR9 passive ODN control (InvivoGen) were coadministered intravenously with AAV vectors.19 For inactivation of Kupffer cells, 10 mg/kg gadolinium chloride (GdCl3) was injected intravenously 48 and 24 hours before vector administration as described previously.3 Liver samples were prepared for RNA isolation by freezing in RNAlater solution (Ambion) or for immunohistochemistry by either cryopreservation in optimal cutting temperature tissue freezing medium or by paraformaldehyde fixation followed by paraffin embedding.
AAV vectors
AAV vector constructs and serotypes were as follows: AAV2, AAV8, and AAV2-Y444/500/730F triple mutant (AAV-TM). Vector genomes were flanked by AAV2 ITRs. Self-complementary vectors (scAAV) lacked the terminal resolution sequence in 1 ITR as published by others.20 Single-stranded vectors (ssAAV) expressed hF.IX from the apolipoprotein E enhancer/human α1-anti–trypsin promoter. For scAAV vectors, hF.IX was expressed from the transthyretin promoter as published by others.12 In scAAV and ssAAV vectors, green fluorescent protein (GFP) expression was regulated by the cytomegalovirus enhancer/chicken β-actin promoter. Vectors were produced by triple transfection of human embryonic kidney (HEK)–293 cells using endotoxin-free plasmid preparations and purified by iodixanol gradient centrifugation (from Benzonase [Merck]–digested cell lysates) as published previously.21 Vector titers were determined by quantitative dot blot hybridization and confirmed by Western blot. Plasmid contamination during AAV vector preparation was not detectable in 1 × 1011 AAV particles in any of the viral vectors used. The SYBR-Green RT-PCR procedure using a primer pair for the ampicillin resistance gene (5′-TTACCAATGCTTAATCAGTG-3′, 3′-ATGAGTATTCAACATTTCCG-5′)] had a limit of detection of 250 plasmid molecules/1011 vg.
Gene expression profiling
Messenger RNA was isolated from 30 mg of liver samples (stored in RNAlater) using the RNeasy kit (QIAGEN), and first stranded synthesis (cDNA) was performed using the SABiosciences kit. Quantitative RT-PCR was performed in duplicate using 500 ng of cDNA template, SYBR-Green PCR master mix, and primers in precoated 96-well plates (SABiosciences) in a total volume of 25 μL. Gene expression of IL-1α/β, IL-6, TNF-α, IL-12α interferon (IFN)-α/β, regulated on activation normal T cell expressed and secreted (RANTES), Kupffer cells (KCs), monocyte chemotactic protein-1 (MCP-1), macrophage inflammatory protein-1, IFN-γ inducible protein-10 (IP-10), TLR1 to 9, and Myeloid differentiation primary response gene (88) (MyD88) was compared with that of glyceraldehyde-3-phosphate dehydrogenase. Fold-change was determined using the 2ΔΔCt quantification method. ΔCt is the Ct value of the selected gene compared with Ct of glyceraldehyde-3-phosphate dehydrogenase, and ΔΔCT is the ΔCt of PBS-injected mice compared with ΔCt of vector-injected mice. The Ct value is the PCR cycle at which the log of normalized fluorescence crosses an established threshold. An icycler/MyiQ fluorescent detection system with iQ5 operating software Version 2.0 (Bio-Rad Laboratories) was used to generate and analyze data. Based on the range of ΔCt values, it was determined that differences > 2.5-fold were above the variation of the assay.
Cytokine levels in liver and plasma
ELISA kits and protocols (eBiosciences) were used to determine IL-6, TNF-α, and MCP-1 levels from liver protein extracts or plasma samples. Liver homogenates were prepared by adding 50 mg of liver to 750 μL of lysis buffer (150mM NaCl, 0.1% NP-40, 50mM Tris-HCl, and PicD/PicW [1:1000]protease inhibitor cocktail) for 30 minutes at room temperature (RT) followed by 5-second sonication bursts at 40% amplitude (5 times). Total protein concentration was analyzed using the BCA Protein Assay kit (Thermo Fisher Scientific). Samples containing equal amounts of whole extracted protein (150 μg) were loaded in duplicate per micro-well. Plasma samples were diluted 1:2 before measurement by ELISA.
Neutrophil staining
Sections (8 μm) from paraformaldehyde-fixed and paraffin-embedded liver tissue were mounted on poly-l-lysine slides and stained using a Naphthol AS-D Chloroacetate (Specific Esterase) kit (Sigma-Aldrich). In brief, slides were deparaffinized by heating to 55°C for 5 minutes and then immediately placed in coplin jars with xylene. Slides were rehydrated by transferring slides to 100%, 95%, and 75% ethanol and then rinsed in distilled H2O (dH2O). Rehydrated tissue sections were transferred to neutrophil staining buffer (prewarmed to 37°C in coplin jars) and incubated for 10 minutes at 37°C. Slides were rinsed in dH2O and counterstained with hematoxylin and coverslipped with Aqua-Mount (Lerner Lab) nonorganic mounting media. Neutrophil-specific chloracetate-esterase staining buffer was prepared according to the manufacturer's instructions (Sigma-Aldrich); Fast Cornith V salt was dissolved in 37°C prewarmed 6.3 Trizmal solution and immediately added to naphthol AS-D chloroacetate solution and thoroughly mixed.
Macrophage staining
Sections (8 μm) from liver tissue blocks cyropreserved in optimal cutting temperature were mounted on poly-l-lysine slides. Sections were fixed for 10 minutes at RT in acetone and then air-dried for 15 minutes. A HRP immunohistochemistry staining protocol for Mac-1 was used with hematoxylin counter stain and mounted with toluene-based Permount media (Thermo Fisher Scientific). In brief, sections were blocked (5% goat serum in PBS) for 30 minutes at RT, followed by PBS rinse, 15 minutes at RT of 1 to 2 drops of Avidin block (Vector Laboratories), PBS rinse, and 15 minutes of 1 to 2 drops of Biotin Block (Vector Laboratories). Mac-1 biotinylated primary antibody (eBiosciences) was added at 1:100 in PBS with 1% goat serum for 30 minutes at RT, followed by PBS rinse and then peroxidase block for 10 minutes at RT with 0.5% H2O2 in PBS. After PBS rinse, streptavidin-HRP (BD Biosciences) was added at 1:100 in PBS with 1% goat serum for 30 minutes at RT in the dark. After PBS rinse 3,3-diaminobenzidine buffer kit (Vector Laboratories; [2 mL of dH2O, 2 drops of pH buffer, 4 drops of 3,3-diaminobenzidine, and 2 drops of H2O2) was added at RT for 1 minute.
Natural killer cell staining
Liver sections were prepared and fixed as described under Macrophage staining. A horseradish peroxidase immunohistochemistry staining protocol for CD335 (Nkp 46) was used with hematoxylin counterstain. Sections were blocked (5% goat serum in PBS) for 30 minutes at RT, followed by PBS rinse. Rat IgG α-CD335 (unconjugated) primary antibody (eBiosciences) was added at 1:100 in PBS with 1% rat serum for 30 minutes at RT, followed by PBS rinse, 15 minutes at RT of 1 to 2 drops of Avidin block (Vector Laboratories), PBS rinse, and 15 minutes of 1 to 2 drops of Biotin block (Vector Laboratories). Biotinylated secondary α-rat IgG (Abcam) at 1:100 in PBS with 1% rat serum was added for 30 minutes at RT, followed by PBS-rinse, and then peroxidase block for 10 minutes at RT with 0.5% H2O2 in PBS. After PBS rinse, streptavidin-horseradish peroxidase (BD Biosciences) was added as described under Macrophage staining.
Data analysis of stained liver sections
For neutrophil observations, 40× images were captured using an Eclipse 80i fluorescence microscope (Nikon) and Retiga 2000R digital camera (QImaging). For macrophages and natural killer (NK) cells, 20× images were captured. Neutrophils, NK cells, or macrophages were counted manually from captured images. Data were collected from 20 images captured from each of the 3 sections prepared, for a total of 60 images per mouse liver. Data were calculated per square centimeter.
In vitro TLR9 and inflammasome activation
An HEK-293 reporter cell line stably transfected with TLR9 was obtained from InvivoGen. TLR9 activation in this HEK-293-TLR9 cell line results in expression and secretion of IL-8 into the culture media. Cells were grown to 75% confluence in a 48-well tissue culture tray and infected with AAV viral vectors at multiplicity of infection (MOI) of 105 vg/cell for 6 hours in 200 μL of DMEM with 2% FBS at 37°C in 5% CO2. After infection, media were collected for measurement of IL-8 levels by an ELISA kit (Thermo Fisher Scientific). As a positive control, cells were transfected with 10 μg/mL TLR9 stimulatory ODN2216 (InvivoGen) using Lipofectamine 2000 (Invitrogen). To assay for inflammasome activation, phorbol 12-myristate 13-acetate–differentiated human THP-1 (macrophages) cells were infected with scAAV2-GFP or ssAAV2-GFP vectors (at an MOI of 105 vg) for 6 hours followed by Western blot for IL-1β activation as published previously.22 Adenovirus infection was used as positive control at an MOI of 102.
Systemic hF.IX and antibody levels against AAV2 capsid
Antibody titers against AAV capsid and hF.IX as well as hF.IX levels in plasma samples were measured by IgG subclass-specific ELISA as published previously.15,16,23 For anti-capsid assays, ELISA plates were coated with 4 × 107 intact AAV2 viral particles/micro-titer well.
ELISpot analyses
Frequencies of AAV2 capsid-specific CD8+ T cells in BALB/c mice and of hF.IX-specific CD4+ and CD8+ T cells in hemophilia B C3H/HeJ were determined by ELISpot using published protocols and epitopes (peptide antigens).16,24 Measurements were done at the peak of the response, which is 1 week for capsid and 4 weeks for hF.IX. Further details are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Statistical analyses
Differences between 2 experimental groups were analyzed with 2-tailed Student t test. Statistical analysis of gene expression increases between vector groups was calculated by 2-way ANOVA with “vector delivered” and “genes up-regulated” as the independent variables. For comparison of levels of cellular infiltrates between all experimental groups, 1-way ANOVA with variance was applied using Prism Version 5.0 software (GraphPad Software).
Results
Gene expression comparison of innate immune response to AAV vectors
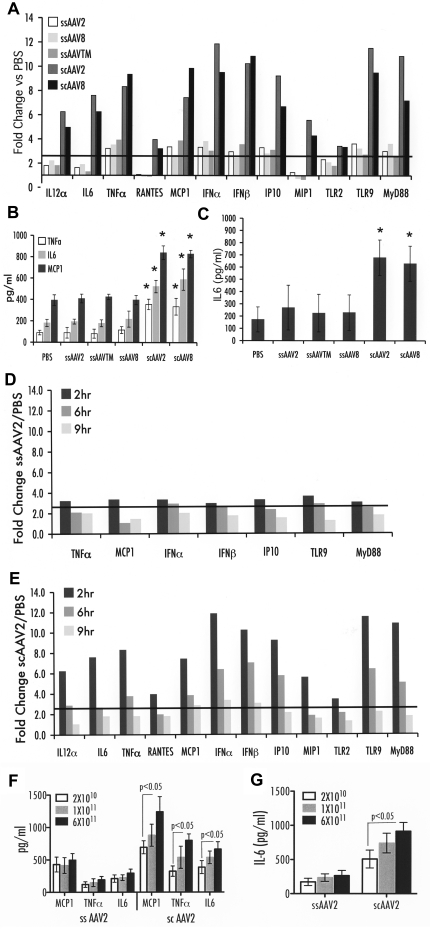
Prior work showed that ssAAV2 vectors mildly up-regulated expression of proinflammatory genes during the first 2 hours after hepatic vector administration.3 Using an RT-PCR array for a well-established set of transcripts indicative for innate immune responses, we examined the effect of the AAV capsid and genome 2, 6, and 9 hours after administration of 1 × 1011 vg/mouse to the portal circulation of C3H/OuJ mice. Regardless of capsid sequence (AAV2, Y444/500/730F, AAV8), all ssAAV-F.IX vectors induced only a mild response at 2 hours after delivery. Relative to PBS injection, a 3- to 4-fold increase in TLR9, MyD88, TNF-α, IFN-α/β, IP-10, and MCP-1 (CCL2) expression was evident, compared with a 4- to 12-fold increase after delivery of scAAV2-F.IX vector (Figure 1A). In addition, the scAAV-F.IX vector provoked increases (from 3- to 8-fold) in IL-6, IL-12α, RANTES (CCL5), MIP-1 (CCL3), and TLR2 expression that were not observed on injection of ssAAV vectors. Several other innate response genes showed no differences in expression compared with PBS-injected mice, which included IL-1α/1β, KCs, TLR1, and TLR3 to 8. All responses were transient and declined by 6 hours and further by 9 hours (Figure 1D-E). However, several transcripts remained overexpressed at 6 hours after scAAV injection. Figure 1B shows that scAAV vector delivery caused a significant 2- to 3-fold increase in levels of IL-6, MCP-1, and TNF-α cytokines in hepatic protein extracts compared with ssAAV vectors (2-hour time point). Although systemic IL-6 levels of mice injected with ssAAV vectors were similar to those of PBS-injected mice, animals that received scAAV vector showed significantly increased levels of IL-6 in plasma by 2 hours (Figure 1C). No increase in systemic TNF-α was observed (data not shown). In addition, an scAAV8-GFP vector was tested to assess effects of the transgene; this vector yielded very similar increases in innate immunity as the scAAV2-F.IX vector (Figure 1A-C). To further investigate the time frame of the innate response, we also examined MCP-1, TNF-α, and IL-6 protein levels in the liver at 1 hour after vector administration, at which time scAAV already showed a heightened response (supplemental Figure 1). Systemic IL-6 levels were not yet increased at 1 hour (supplemental Figure 1). Together, these data indicate that the responses to ssAAV vectors are mild and very transient regardless of the composition of the capsid, whereas the scAAV vectors augment, broaden, and extend innate response. To generate additional support for this point, a dose-response study was performed. Changes in cytokine levels remained minimal over a wide range of vector doses for ssAAV at 2 hours (up to 6 × 1011 vg/mouse; Figure 1F-G). In contrast, there was a dose-dependent response to the administration of scAAV vector, with the lowest dose (2 × 1010 vg/mouse) tested giving a stronger response than the highest dose of ssAAV vector (30-fold difference in vector dose; Figure 1F-G).
Figure 1.
Innate immune response after liver gene transfer with ssAAV2, -TM, or -8 vectors or with scAAV2 or -8 vectors to C3H/OuJ mice (n = 4 per experimental group). All vectors contained expression cassettes for hF.IX, except for scAAV8-GFP vector. (A) Fold-changes in hepatic mRNA levels (2 hours after vector administration at 1 × 1011 vg) compared with PBS-injected mice are shown. A change > 2.5-fold (horizontal line) was considered greater than the variability of the assay. (B) Cytokine levels of TNF-α, IL-6, and MCP-1 in liver protein extract 2 hours after vector administration at 1 × 1011 vg. (C) Systemic IL-6 levels 2 hours after vector administration at 1 × 1011 vg (in picograms per milliliter of plasma). Data are average per experimental group ± SD. (D-E) Fold-change in hepatic mRNA levels compared with PBS-injected mice as a function of time after vector administration shown for ssAAV2-hF.IX (D) and scAAV2-hF.IX (E) vectors was statistically different between vector groups at 2 hours, P < .001, and at 6 hours, P < .01, calculated by 2-way ANOVA of variance using “vectors delivered” and “genes up-regulated” as independent variables. Cytokine levels of TNF-α, IL-6, and MCP-1 in liver protein extract (F) or systemic IL-6 levels 2 hours after vector administration of either 2 × 1010, 1 × 1011, or 6 × 1011 vg (G); statistics were done using 2-tailed Student t test. *P < .05 in comparison with ssAAV2-transduced liver as calculated by 2-tailed Student t test.
Innate responses to scAAV are partially KC-dependent
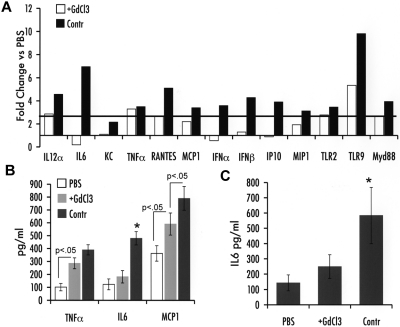
The low proinflammatory cytokine response to ssAAV was found previously to depend on KCs.3 Therefore, we inactivated these resident hepatic antigen-presenting cells by repeated GdCl3 administration before injection of an scAAV2-GFP vector given at a dose of 1 × 1011 vg/mouse. In control animals (with intact KCs), a response pattern similar to that outlined here for scAAV vectors was observed (Figure 2A-C). Inactivation of KCs significantly blunted most but not all these responses to scAAV. In particular, expression of transcripts for TNF-α and TLR2 was not significantly changed after GdCl3 treatment. TLR9, MyD88, IL-12α, MCP-1, and MIP-1 showed a moderate reduction in gene expression. In contrast, IFN-α/β, IP-10, and IL-6 mRNA induction was completely ablated by KC inactivation (Figure 2A). Consistent with the RT-PCR data, GdCl3 treatment prevented a rise in hepatic and systemic IL-6 levels, whereas TNF-α and MCP-1 induction in the liver was intermediate between PBS- and scAAV-treated control mice (Figure 2B-C).
Figure 2.
Impact of KC depletion on innate immune response 2 hours after liver gene transfer with scAAV2 vector in C3H/OuJ mice (n = 4 per experimental group). (A) Fold-changes in hepatic mRNA levels (compared with PBS-injected mice) are shown for mice that did or did not receive GdCl3 treatment before scAAV2-GFP vector administration. A change > 2.5-fold (horizontal line) was considered greater than the variability of the assay. (B) Cytokine levels of TNF-α, IL-6, and MCP-1 in liver protein extract. (C) Systemic IL-6 levels. *P < .05 in comparison with ssAAV2-transduced liver as calculated by 2-tailed Student t test.
Innate immune responses to scAAV are TLR9-dependent
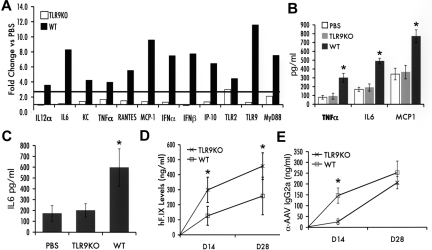
Given that a change in the configuration of the DNA genome caused an increase in innate immune responses and that recent data from Yang and colleagues implicated TLR9,25 an innate endosomal pathogen pattern recognition receptor that senses DNA, in immunity to ssAAV, we tested for innate immune response to scAAV in livers of TLR9−/− mice. Because these mice are on a C57BL/6 background, wild-type (WT) C57BL/6 mice were tested in parallel. All mice received liver-directed gene transfer with 1011 vg of scAAV2-F.IX vector. In WT mice, innate immunity was comparable to that in C3H/OuJ mice receiving scAAV vectors (Figure 3A-C). In contrast, TLR9−/− mice, with the exception of a marginal increase in transcripts for TLR2, lacked induction of other transcripts that were measured as markers for innate immune responses in the liver (Figure 3A). IL-6, MCP-1, and TNF-α levels in the liver (as well as circulating IL-6 levels) were comparable to those of PBS-injected WT mice (Figure 3B-C). Gene transfer resulted in circulating hF.IX levels that were 2- to 3-fold lower in WT compared with TLR9−/− mice (Figure 3D). Antibody formation to AAV2 capsid in TLR9−/− mice was delayed compared with that in WT mice (Figure 3E).
Figure 3.
Innate immune response after liver gene transfer with scAAV2-hF.IX vector in TLR9−/− (TLR9KO) or WT C57BL/6J mice (n = 5 per group). (A) Fold-changes in hepatic mRNA levels (2 hours after vector administration) compared with PBS-injected mice of same strain are shown. A change > 2.5-fold (horizontal line) was considered greater than the variability of the assay. (B) Cytokine levels of TNF-α, IL-6, and MCP-1 in liver protein extract 2 hours after vector administration. (C) Systemic IL-6 levels 2 hours after vector administration. Systemic hF.IX levels (D) and antibody titers (IgG2a; E) against AAV capsid as a function of time after vector administration (average ± SD). PBS, WT control mice injected with PBS. *P < .05 as calculated by 2-tailed Student t test (comparison of WT and TLR9−/− mice).
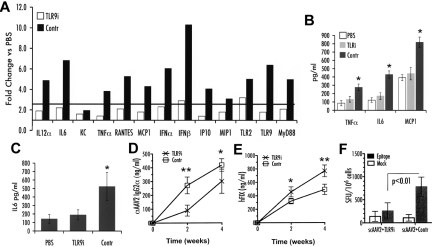
TLR9−/− mice have MyD88 mRNA levels that are ∼ 5-fold lower than those in WT mice, which could theoretically dampen signaling from TLRs other than TLR9. To further address the TLR9 dependence of innate immune responses to scAAV and to rule out other TLRs that signal through MyD88, an inhibitory ODN was used to silence TLR9 signaling in WT C57BL/6J mice at the time of hepatic gene transfer. As shown in Figure 4A-C, coadministration of the TLR9 inhibitory ODN with scAAV2-F.IX vector effectively and broadly blunted the innate immune response. In addition, use of TLR9i increased systemic F.IX expression (Figure 4E).
Figure 4.
Innate and adaptive immune response after liver gene transfer with scAAV2-hF.IX vector in WT C57BL/6J mice (n = 4 per group). Vector was coadministered with TLR9 inhibitory (TLR9i) or passive (contr) ODN. (A) Fold-changes in hepatic mRNA levels (2 hours after vector/ODN administration) compared with PBS-injected mice are shown. A change > 2.5-fold (horizontal line) was considered greater than the variability of the assay. (B) Cytokine levels of TNF-α, IL-6, and MCP-1 in liver protein extract 2 hours after vector/ODN or PBS administration. (C) Systemic IL-6 levels 2 hours after vector/ODN or PBS administration. (D) IgG2a antibody titers against AAV2 capsid 2 and 4 weeks after injection, measured by ELISA. (E) Systemic hFIX levels 2 and 4 weeks after delivery, measured by ELISA. (F) CD8+ T cell responses to AAV2 capsid 1 week after gene transfer, measured as IFN-γ spot-forming units (SFU) per 106 cells using ELISpot. *P < .05, **P < .01 calculated by 2-tailed Student t test for comparison of TLR9i or control ODN.
scAAV vectors show increased signaling through TLR9 but fail to activate the inflammasome
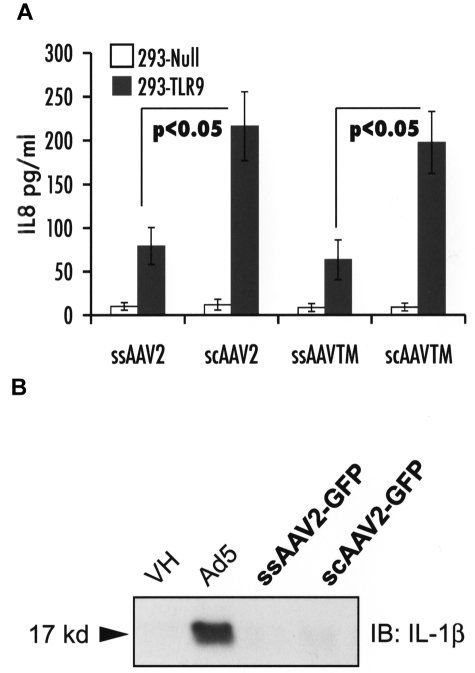
HEK-293-TLR9 reporter cells were used to evaluate TLR9 sensing in vitro on their transduction with ss or scAAV vectors. Levels of IL-8 secreted into conditioned media, which correlate with the extent of TLR9 signaling in these cells, were measured. Figure 5A shows that a scAAV2 vector causes a 3-fold increase in IL-8 compared with ssAAV2 vectors, regardless of the expression cassette (F.IX or GFP) or changes in surface-exposed tyrosine residues (WT or Y444/500/730F AAV2 capsid). Very minor induction of IL-8 was measured for AAV8 vectors, which do not efficiently transduce these cells in vitro (data not shown). In contrast to adenovirus, both ss- and scAAV2 vectors failed to activate the inflammasome in macrophage cells (Figure 5B).
Figure 5.
In vitro innate immune sensing of AAV vectors. (A) Strength of TLR9 signaling as determined by levels of IL-8 secretion by HEK-293-null and -293-TLR9 cells 6 hours after infection with ssAAV2-GFP or scAAV2-GFP or ssAAV-TM-hF.IX or scAAV-TM-hF.IX. (B) Activation of the inflammasome in THP-1 cells in response to adenovirus (positive control), ssAAV2-GFP, or scAAV2-GFP.
Neutrophil, macrophage, and NK cell infiltration in liver
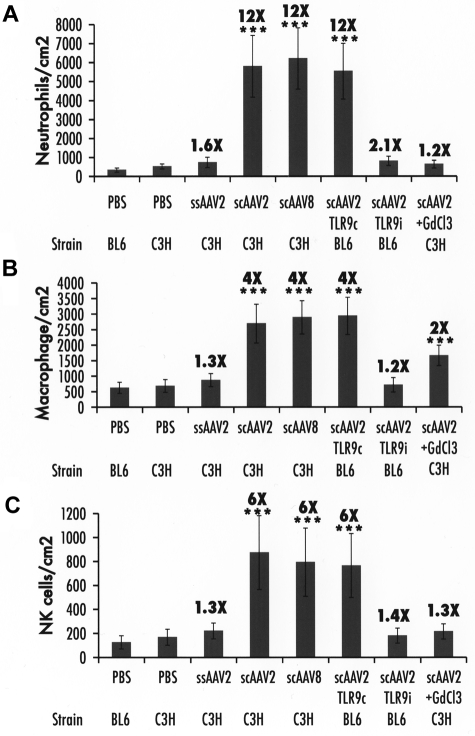
The increase of cytokine levels in the liver after scAAV administration could potentially increase hepatotoxic responses. This was analyzed by determining the number of neutrophils, macrophages, and NK cells in the liver. For ssAAV vectors, a < 2-fold increase in neutrophil and macrophage infiltration was seen in the liver after 2 hours, which reached statistical significance (P < .05) by Student t test comparison with PBS-injected mice but not when all experimental groups were compared with ANOVA (Figures 6–7). In contrast, neutrophils were elevated 12-fold, macrophages 6-fold, and NK cells 4-fold on scAAV vector administration, reaching high statistical significance (Figures 6–7). Coadministration of the TLR9 inhibitory ODN prevented infiltration by macrophages, NK cells, and almost completely that by neutrophils (Figures 6–7). KC inactivation had a similar effect on neutrophil and NK cell recruitment but caused only a moderate reduction of macrophage accumulation, which remained significantly elevated (Figures 6–7).
Figure 6.
Cellular infiltrates in liver sections 2 hours after vector administration (representative examples). (A-G) Neutrophils. (H-N) Macrophages. (O-U) NK cells. (A) C3H-OuJ control mice injected with PBS. Other panels shown are for hepatic gene transfer with ssAAV2-hF.IX in C3H-OuJ (B, I, P), scAAV2-hF.IX/TLR9i ODN in C57BL/6J (C, J, Q), scAAV2-GFP/GldCl3-treated C3H-OuJ (D, K, R), scAAV2-hF.IX in C3H-OuJ (E, L, S), scAAV8-GFP in C3H-OuJ (F, M, T), or (G) scAAV2-hF.IX/TLR9 control-ODN in C57BL/6J mice (G, N, U). Original magnification: ×40 (neutrophils) or ×20 (macrophages and NK cells).
Figure 7.
Summary of cellular infiltrates in the liver 2 hours after vector administration. (A) Neutrophils. (B) Macrophages. (C) NK cells. Data were collected from 20 images captured from each of the 3 sections prepared for a total of 60 images per mouse liver. Data are presented as average number of cells per square centimeter per mouse (n = 4 per group) ± SD. Fold-increase in comparison to PBS-injected mice of the same strain is indicated. ***P < .001 (ANOVA comparison with PBS-injected mice).
Effects on adaptive immune responses to vector and transgene product
To test for potential effects of increased innate immunity on adaptive responses, serotype-2 vectors were used for hepatic gene transfer in BALB/c mice, because the dominant CD8+ T cell epitope for capsid is known and the response is more robust than in C57BL/6 mice. Similarly, AAV2-hF.IX vectors were administered to hemophilia B (F9−/−) C3H/HeJ mice, which have stronger B and T cell responses to hF.IX compared with other strains. At equal dose, scAAV vectors gave higher CD8+ T cell and antibody responses against capsid compared with ssAAV vectors (supplemental Figures 2-3). However, the adaptive response to the FIX transgene product was not increased (supplemental Figure 4). Interestingly, inhibition of TLR9 substantially reduced CD8+ T cell responses and delayed antibody formation against viral particles (Figure 4D-F).
Discussion
Data presented here show that injection of AAV vectors, in which the genome was changed from ss to scDNA, causes a substantial increase in the innate immune response in the liver regardless of the strain of mouse or the transgene/promoter cassette. These responses were TLR9-dependent and increased adaptive responses against the vector (but not the transgene product), and scAAV vectors signaled more strongly through this endosomal receptor for DNA.
Role of TLR9 in innate responses to AAV vectors in the liver
A recent study showed TLR9-dependent induction of IFN-α/β expression in plasmacytoid dendritic cells (pDCs) pulsed with ssAAV in vitro and demonstrated TLR9-MyD88–dependent B and T cell responses to ssAAV vector and its transgene product on in vivo muscle gene transfer.25 TLR9 is located in endosomes and recognizes unmethylated cytosine guanine dinucleotide motifs that are typically present in viral or bacterial but not mammalian DNA.18,26 The resulting signaling, through the MyD88 adapter molecule, activates NF-κB, resulting in expression of proinflammatory cytokines, such as IL-6, IL-12α, and TNF-α, and induces expression of type I interferons (IFN-α and -β) via IFN-regulatory factors 3 and 7.27,28 The effects on adaptive responses were largely attributed to IFN production by pDCs, because no evidence was found for induction of proinflammatory cytokines on in vitro pulsing of conventional DCs, pDC, or macrophages with ssAAV.25
Our in vivo studies support that ssAAV vectors only induce low and very transient inflammatory responses. In contract, we clearly demonstrate TLR9-dependent induction of IL-6, TNF-α, and other proinflammatory cytokines and chemokines in the liver by scAAV vectors (in addition to interferon). KC function is required in vivo for several components of the innate cytokine response. It is thought that induction of proinflammatory cytokines after TLR9 signaling occurs via activation of the classic NF-κB pathway. This is consistent our recent report, demonstrating rapid activation of this pathway after infusion of AAV vector into the liver.23 However, more studies are required to dissect the relative contributions of IFN and of inflammation on adaptive responses to AAV.
IL-6, among other functions, regulates neutrophil trafficking and suppresses apoptosis in neutrophils. Induction of IL-6 expression was KC-dependent; and in the absence of KC function, neutrophil infiltration was mostly suppressed. In contrast, KC function was less critical for production of MCP-1, which attracts macrophages, and of TNF-α, a cytokine mostly expressed by macrophages. Thus, KC inactivation only partially blocked macrophage infiltration in the liver on scAAV gene transfer. Finally, we observed enhanced NK cell infiltration in response to scAAV, which was eliminated by blocking KC function. The increased levels of IL-12, RANTES, IP-10, and MIP-1 expression after scAAV vector administration are substantially reduced after KC inactivation, all of which have been involved in NK cell chemoattraction.29,30 In addition, it has been shown that NK cell activation, but not recruitment, and innate immune elimination of adenoviral vectors is dependent on type 1 IFNs and ultimately reduces transduction efficiency.31
Complement plays a role in innate and adaptive immune responses to AAV in that the capsid binds iC3b complement protein, leading to macrophage activation and contributing to antibody formation against vector particles.32 However, this mechanism is not essential for innate inflammatory responses to AAV in the liver.32 Induction of inflammatory gene expression and of IFN-α/β in the liver was entirely TLR9-dependent, indicating that sensing of the viral DNA in the endosome is critical for innate immunity to scAAV. These characteristics are in some ways similar to responses to adenovirus, a nonenveloped double-stranded DNA virus. TLR9 is the most critical TLR in innate immunity to adenovirus, and cellular immune responses to adenoviral transduced hepatocytes are blunted in TLR9−/− mice.33 TLR2, a cell surface receptor for molecular structure on bacteria, fungi, and viral particles, also contributes to innate immunity to adenovirus, whereas other TLRs do not seem to be involved.34,35 However, the response to adenovirus is much broader and includes activation of the inflammasome and sensing of the viral genome in the cytoplasm.22,36
IFN-α and -β are important for differentiation into fully functional effector T cells, for enhancement of antiviral B cell responses, and for activation of NK cells, and they can down-regulate promoter activity.37,38 It is likely that lack of an IFN-α/β response to scAAV in TLR9−/− mice contributed to the delay in anti-AAV formation and to increased levels of transgene expression. Further studies in IFN-α/βR−/− mice may help address this point. Yang and colleagues25 found increased secretion of IFN-α/β by pDCs pulsed with ssAAV in vitro. Although pDCs are considered the most potent producers of type I interferons, induction of IFN-α/β expression in the liver was dependent on KC function, possibly because of a requirement for sequestration of AAV particles in the liver by KCs, perhaps facilitating pDC activation.3,39
A general concern in using viral vectors produced by transfection of plasmid DNA is contamination of viral preparations with bacterial components such as endotoxin, which can contribute to innate immunity via activation of TLR4. However, several points argue against an endotoxin effect in our study. The heightened response was only seen for scAAV, whereas ssAAV produced in the same facility caused only very low innate immunity. Furthermore, endotoxin-free plasmid preparations were used during vector production, the response to scAAV was effectively blocked using a TLR9-specific inhibitor, and TLR4 expression remained unchanged.
Self-complementary genomes heighten innate immunity to AAV vector
On receptor-mediated endocytosis, AAV vectors enter endosomes. Intracellular trafficking through early and late endosomes is influenced by cell type and MOI.40 Although AAV particles are more resistant to endosomal degradation than some other parvoviruses, capsid modifications occur in the acidic endosomal environment, including VP1/VP2 externalization and partial disassembly.40–42 Ultimately, the virus has to escape the endosome to traffic to the nucleus via the cytoplasm. During trafficking, some particles may be degraded, thereby releasing their DNA contents. Because of the packaging mechanism and space constrains within the capsid, it is thought that the scAAV genome is present in ss form in the particle but readily becomes double-stranded after release from the capsid.11 Our data do not differentiate between sensing of ss or double-stranded DNA by TLR9 in endosomes. It is known that TLR9 signaling is stimulated by both double-stranded and ss unmethylated DNA molecules,43–47 although sc (containing double 5′-end, sense-antisense sequences) TLR9 agonists induce more robust immune stimulation compared with single 5′-end cytosine guanine dinucleotide oligonucleotides.48 It is feasible that the capsids of scAAV vectors are less stable in the endosomes and that thus more genomes are released and made available for TLR9 sensing as remains to be studied. Regardless, our data demonstrate that changing the configuration of a single-stranded viral genome to a sc/double-stranded form increases the immunogenicity of the viral particle. During the innate response, TLR9 transcript levels in the liver was increased, which may be because of an up-regulation of expression by resident cells, infiltration by highly TLR9-expressing cells, or both.
Our data implicate that the genome configuration rather than characteristics of the capsid determines the strength of inflammatory responses to hepatic AAV gene transfer, although capsid sequences are known to substantially affect adaptive responses, in particular to the transgene product, involving mechanisms that depend on T help and costimulatory pathways.49 In addition, the level of gene transfer to professional antigen presenting cells such as DCs and the transduction pattern and distribution of the antigen in the tissue impact the response.7,50
Implications for gene therapy
The innate response to scAAV was self-limited and resolved within < 12 hours after vector administration. Part of the appeal of scAAV vectors is the potential to achieve therapeutic efficacy at lower vector doses, in which case increased innate immunity may be less of a concern. Certainly, differences in innate immunity in the liver did not correlate with transduction efficiency of hepatocytes. For example, the Y444/500/730F mutation increases gene transfer to hepatocytes with an AAV2 vector by > 10-fold, and AAV8 vectors direct 100- to 500-fold higher hF.IX expression levels in mouse liver, with a dose of 1011 vg transducing nearly all hepatocytes.7,10 Yet, the innate immune response to scAAV8 vectors was of similar magnitude to that of scAAV2, whereas ssAAV2 and ssAAV8 vectors were equally low in immunogenicity.
High vector doses of scAAV vectors, should these be required, may increase recruitment of inflammatory cells, a source of hepatotoxicity. Increased innate immunity to scAAV is linked to higher adaptive responses to the viral capsid. Hence, blockage of TLR9 substantially reduced the CD8+ T cell response to capsid. Interestingly, we found no evidence for increased responses to the transgene product. This is probably because of the short-lived nature of the innate response to AAV in the liver. During the first 12 hours, ie, the time at which inflammatory signals are present, the only antigen present at high level is the input capsid, whereas F.IX is only expressed subsequently after viral uncoating and transcriptional activation. However, affects on adaptive responses also may be more severe in other tissues that, in contrast to the liver, may be less able to control inflammatory signals.16 With regard to design therapeutic protocols, our data show that innate and adaptive responses to scAAV vectors can be effectively suppressed by blocking TLR9 signaling (albeit the effect on antibodies against capsid was largely transient). A more general implication of these results is that a change in the genome configuration (from ssDNA to sc/double-stranded) can alter the immunogenicity of a viral particle.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01 AI51390 (R.W.H.), P01 HL078810 (H.C.J.E., R.W.H.), and R01 HL087836 (B.L.) and by grants from the Canadian Institutes of Health Research and The Alberta Heritage Foundation for Medical Research (D.A.M.). D.M.M. was supported by F32 HL096281, and M.S. was supported by K99 HL098692.
Footnotes
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.T.M., M.S., D.A.M., B.L., and R.W.H. designed experiments; A.T.M., M.S., D.M.M., I.Z., R.C.R., B.M., and D.A.M. performed experiments and analyzed data; B.L. and R.W.H. directed the study and analyzed data; H.C.J.E. assisted with data analysis and interpretation; and A.T.M., H.C.J.E., and R.W.H. wrote the manuscript.
Conflict-of-interest disclosure. R.W.H. has been receiving royalty payments from Genzyme Corp for license of AAV-F.IX technology. The remaining authors declare no competing financial interests.
Correspondence: Roland W. Herzog, University of Florida, Cancer and Genetics Research Center, 2033 Mowry Rd, Room 203, Gainesville, FL 32610; e-mail: rherzog@ufl.edu.
References
- 1.Herzog RW, Cao O, Srivastava A. Two decades of clinical gene therapy–success is finally mounting. Discov Med. 2010;9(45):105–111. [PMC free article] [PubMed] [Google Scholar]
- 2.Mátrai J, Chuah MK, VandenDriessche T. Preclinical and clinical progress in hemophilia gene therapy. Curr Opin Hematol. 2010;17(5):387–392. doi: 10.1097/MOH.0b013e32833cd4bd. [DOI] [PubMed] [Google Scholar]
- 3.Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76(9):4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somanathan S, Breous E, Bell P, Wilson JM. AAV vectors avoid inflammatory signals necessary to render transduced hepatocyte targets for destructive T cells. Mol Ther. 2010;18(5):977–982. doi: 10.1038/mt.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mingozzi F, High KA. Immune responses to AAV in clinical trials. Curr Gene Ther. 2007;7(5):316–324. doi: 10.2174/156652307782151425. [DOI] [PubMed] [Google Scholar]
- 6.Vandenberghe LH, Wang L, Somanathan S, et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat Med. 2006;12(8):967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 7.Cooper M, Nayak S, Hoffman BE, Terhorst C, Cao O, Herzog RW. Improved induction of immune tolerance to factor IX by hepatic AAV-8 gene transfer. Hum Gene Ther. 2009;20(7):767–776. doi: 10.1089/hum.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutin S, Monteilhet V, Veron P, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21(6):704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 9.Zhong L, Li B, Mah CS, et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci U S A. 2008;105(22):7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markusic DM, Herzog RW, Aslanidi GV, et al. High-efficiency transduction and correction of murine hemophilia b using AAV2 Vectors devoid of multiple surface-exposed tyrosines. Mol Ther. 2010;18(12):2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16(10):1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, Sun J, Zhang T, et al. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol Ther. 2008;16(2):280–289. doi: 10.1038/sj.mt.6300355. [DOI] [PubMed] [Google Scholar]
- 13.Nathwani AC, Gray JT, McIntosh J, et al. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109(4):1414–1421. doi: 10.1182/blood-2006-03-010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LoDuca PA, Hoffman BE, Herzog RW. Hepatic gene transfer as a means of tolerance induction to transgene products. Curr Gene Ther. 2009;9(2):104–114. doi: 10.2174/156652309787909490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mingozzi F, Liu YL, Dobrzynski E, et al. Induction of immune tolerance to coagulation factor IX antigen by in vivo hepatic gene transfer. J Clin Invest. 2003;111(9):1347–1356. doi: 10.1172/JCI16887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao O, Hoffman BE, Moghimi B, et al. Impact of the underlying mutation and the route of vector administration on immune responses to factor IX in gene therapy for hemophilia B. Mol Ther. 2009;17(10):1733–1742. doi: 10.1038/mt.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martino AT, Nayak S, Hoffman BE, et al. Tolerance induction to cytoplasmic beta-galactosidase by hepatic AAV gene transfer: implications for antigen presentation and immunotoxicity. PLoS ONE. 2009;4(8):e6376. doi: 10.1371/journal.pone.0006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 19.Stunz LL, Lenert P, Peckham D, et al. Inhibitory oligonucleotides specifically block effects of stimulatory CpG oligonucleotides in B cells. Eur J Immunol. 2002;32(5):1212–1222. doi: 10.1002/1521-4141(200205)32:5<1212::AID-IMMU1212>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10(26):2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 21.Zolotukhin S, Byrne BJ, Mason E, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6(6):973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 22.Muruve DA, Petrilli V, Zaiss AK, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 23.Jayandharan GR, Aslanidi G, Martino AT, et al. Activation of the NF-kappaB pathway by adeno-associated virus (AAV) vectors and its implications in immune response and gene therapy. Proc Natl Acad Sci U S A. 2011;108(9):3743–3748. doi: 10.1073/pnas.1012753108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Sabatino DE, Mingozzi F, Hui DJ, et al. Identification of mouse AAV capsid-specific CD8+ T cell epitopes. Mol Ther. 2005;12(6):1023–1033. doi: 10.1016/j.ymthe.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Huang X, Yang Y. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J Clin Invest. 2009;119(8):2388–2398. doi: 10.1172/JCI37607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 27.Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uematsu S, Akira S. Toll-like receptors and type I interferons. J Biol Chem. 2007;282(21):15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 29.Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155(8):3877–3888. [PubMed] [Google Scholar]
- 30.Bernardini G, Sciume G, Bosisio D, Morrone S, Sozzani S, Santoni A. CCL3 and CXCL12 regulate trafficking of mouse bone marrow NK cell subsets. Blood. 2008;111(7):3626–3634. doi: 10.1182/blood-2007-08-106203. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Huang X, Yang Y. A critical role for type I IFN-dependent NK cell activation in innate immune elimination of adenoviral vectors in vivo. Mol Ther. 2008;16(7):1300–1307. doi: 10.1038/mt.2008.88. [DOI] [PubMed] [Google Scholar]
- 32.Zaiss AK, Cotter MJ, White LR, et al. Complement is an essential component of the immune response to adeno-associated virus vectors. J Virol. 2008;82(6):2727–2740. doi: 10.1128/JVI.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerullo V, Seiler MP, Mane V, et al. Toll-like receptor 9 triggers an innate immune response to helper-dependent adenoviral vectors. Mol Ther. 2007;15(2):378–385. doi: 10.1038/sj.mt.6300031. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki M, Cerullo V, Bertin TK, et al. MyD88-dependent silencing of transgene expression during the innate and adaptive immune response to helper-dependent adenovirus. Hum Gene Ther. 2010;21(3):325–336. doi: 10.1089/hum.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appledorn DM, Patial S, McBride A, et al. Adenovirus vector-induced innate inflammatory mediators, MAPK signaling, as well as adaptive immune responses are dependent upon both TLR2 and TLR9 in vivo. J Immunol. 2008;181(3):2134–2144. doi: 10.4049/jimmunol.181.3.2134. [DOI] [PubMed] [Google Scholar]
- 36.Nociari M, Ocheretina O, Murphy M, Falck-Pedersen E. Adenovirus induction of IRF3 occurs through a binary trigger targeting Jun N-terminal kinase and TBK1 kinase cascades and type I interferon autocrine signaling. J Virol. 2009;83(9):4081–4091. doi: 10.1128/JVI.02591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown BD, Sitia G, Annoni A, et al. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109(7):2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Huang X, Yang Y. Type I IFN signaling on both B and CD4 T cells is required for protective antibody response to adenovirus. J Immunol. 2007;178(6):3505–3510. doi: 10.4049/jimmunol.178.6.3505. [DOI] [PubMed] [Google Scholar]
- 39.Lieber A, He CY, Meuse L, et al. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J Virol. 1997;71(11):8798–8807. doi: 10.1128/jvi.71.11.8798-8807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding W, Zhang L, Yan Z, Engelhardt JF. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 2005;12(11):873–880. doi: 10.1038/sj.gt.3302527. [DOI] [PubMed] [Google Scholar]
- 41.Ros C, Baltzer C, Mani B, Kempf C. Parvovirus uncoating in vitro reveals a mechanism of DNA release without capsid disassembly and striking differences in encapsidated DNA stability. Virology. 2006;345(1):137–147. doi: 10.1016/j.virol.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 42.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol. 2006;80(22):11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer S, Kirschning CJ, Hacker H, et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A. 2001;98(16):9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141(2):137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- 45.Agrawal S, Kandimalla ER. Synthetic agonists of Toll-like receptors 7, 8 and 9. Biochem Soc Trans. 2007;35(Pt 6):1461–1467. doi: 10.1042/BST0351461. [DOI] [PubMed] [Google Scholar]
- 46.Yu D, Putta MR, Bhagat L, et al. Agonists of Toll-like receptor 9 containing synthetic dinucleotide motifs. J Med Chem. 2007;50(25):6411–6418. doi: 10.1021/jm070881l. [DOI] [PubMed] [Google Scholar]
- 47.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5(6):471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 48.Struthers M, Bett AJ, Wisniewski T, et al. Synthesis and immunological activities of novel agonists of toll-like receptor 9. Cell Immunol. 2010;263(1):105–113. doi: 10.1016/j.cellimm.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Mays LE, Vandenberghe LH, Xiao R, et al. Adeno-associated virus capsid structure drives CD4-dependent CD8+ T cell response to vector encoded proteins. J Immunol. 2009;182(10):6051–6060. doi: 10.4049/jimmunol.0803965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Song S. Distinct immune responses to transgene products from rAAV1 and rAAV8 vectors. Proc Natl Acad Sci U S A. 2009;106(40):17158–17162. doi: 10.1073/pnas.0909520106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.