Abstract
Insulin-degrading enzyme (IDE) is a Zn2+ metalloprotease with a characteristic inverted catalytic motif. IDE is ubiquitously expressed and degrades peptide substrates including insulin, endorphin, and the amyloid-β peptide. Although IDE is mainly expressed in the cytosol, it can also be found on the cell surface and in secreted form in extracellular fluids. As IDE lacks a characteristic signal sequence that targets the protein to the classical secretory pathway, release of the enzyme involves non-conventional mechanisms. However, functional domains of IDE involved in its secretion remain elusive. By bioinformatical, biochemical, and cell biological methods, we identified a novel amino acid motif (853EKPPHY858) close to the C terminus of IDE and characterized its function in the non-conventional secretion of the protein. Because of its close homology to an amino acid sequence found in bacterial proteins belonging to the SlyX family, we propose to call it the SlyX motif. Mutagenesis revealed that deletion of this motif strongly decreased the release of IDE, whereas deletion of a potential microbody-targeting signal at the extreme C terminus had little effect on secretion. The combined data indicate that the non-conventional secretion of IDE is regulated by the newly identified SlyX motif.
Keywords: Alzheimer Disease, Exocytosis, Metalloprotease, Protein Domains, Protein Secretion, Insulin-degrading Enzyme, MVB Exosome Pathways
Introduction
IDE4 or insulysin is a highly conserved Zn2+ peptidase and member of the M16 metallopeptidase family, characterized by an inverted sequence at the catalytic site (His-X-X-Glu-His). The enzyme was first isolated from beef liver and demonstrated to degrade insulin (1). Later, IDE was found to degrade additional peptide substrates including glucagon, transforming growth factor, and some others (for review, see Ref. 2). Interestingly, IDE substrates share little or no homology in primary amino acid sequence (3, 4). Accordingly, the secondary structure of peptide substrates appears to be a major determinant for the cleavage by IDE. Due to its involvement in the degradation of the amyloid-β peptide (Aβ), IDE has also gained considerable interest in Alzheimer disease-related research.
Like Aβ, most other substrates of IDE are extracellular or luminal peptides. However, IDE lacks a classical signal sequence that targets the protein to the conventional secretory pathway. Although abundantly expressed in the cytosol, fractions of IDE are also found in extracellular fluids and conditioned media of cultured cells (5–11). Thus, IDE could be grouped with other proteins including fibroblast growth factor-2 and interleukin-1β that follow non-conventional pathways for secretion (12). Recent evidence indicates that IDE could be released from cultured cells in association with exosomes, where it might come into contact with its extracellular substrates (13, 14).
However, signaling sequences within IDE that determine its secretion are unknown. Here, we identified a novel motif (EKPPHY) within IDE, which is also found in bacterial SlyX proteins. These proteins are usually less than 80 amino acids long, with the PPHY/W motif located at C terminus. Up to now, the function of SlyX proteins is unknown. Mutation of this motif strongly affected the secretion of IDE. Thus, this SlyX motif determines the non-conventional secretion of IDE.
MATERIALS AND METHODS
Reagents
All reagents for molecular biological and biochemical experiments were purchased from Sigma or Calbiochem/Merck if not stated otherwise. Restriction enzymes and T4 ligase were obtained from Fermentas and used as recommended. Kits from Promega were used for DNA purification and elution from agarose gel. Cell culture media and transfection reagent were ordered from Invitrogen.
Antibodies
The following antibodies were used: rabbit polyclonal antibody for IDE (Abcam catalog number ab25970); mouse monoclonal antibody for β-actin (Sigma, catalog number a1978); mouse monoclonal antibody for c-Myc, clone 9E10 (Abcam, catalog number ab32); mouse monoclonal antibody for GFP (Roche Applied Science, catalog number 11 814 460 001).
In Silico Analysis
The amino acid sequence of mouse IDE (NP_112419) was analyzed by free online tools according to the guidelines found on the corresponding web sites. The products used were PPSearch from the European Bioinformatics Institute-European Molecular Biology Laboratory (EMBL-EBI), SMART from the EMBL, and Motif Scan from MyHits.
Cell Culture
COS7 cells were grown on 10-cm dishes (Greiner) or T75 flasks (Corning) in DMEM supplemented with 1% penicillin and streptomycin (v/v) and 10% fetal bovine serum (v/v). For transfection, cells were grown to 85% confluence and incubated with a mixture of the respective plasmids and Lipofectamine 2000 (Invitrogen) according to the supplier's protocol.
Cloning of IDE Variants
Specific primers with BamHI, EcoRI, and XhoI restriction sites, respectively, were designed for cloning of truncated IDE variants and used to amplify the corresponding DNA by PCR. For cloning of the IDE variant with deletion of the SlyX domain, internal primers were designed lacking this domain. The sequences of all primers used in the study are listed in supplemental Table S1. Amplified DNA fragments were digested with BamHI, EcoRI, and XhoI, respectively and cloned into pcDNA4/Myc-His vector using T4 ligase. Chemically competent strain of Escherichia coli DH5α was transformed with plasmid DNAs and used to inoculate 2 ml of LB medium supplemented with antibiotics and grown overnight at 37 °C with shaking at 220 rpm. Plasmid DNA was prepared using the kit from Fermentas and analyzed by DNA sequencing using sequencing primers listed in supplemental Table S1.
Protein Extraction and Western Immunoblotting
Briefly, cells were washed with PBS, resuspended in 800 μl of radioimmune precipitation buffer (150 mm NaCl, 10 mm Tris, pH 7.5, 1% IGEPAL, 5 mm EDTA), homogenized using a syringe with a 23-gauge needle, and centrifuged at 13,200 rpm for 15 min at 4 °C. The supernatant was boiled with loading buffer and separated by SDS-PAGE. Proteins were electrotransferred onto nitrocellulose membranes at 300 V for 2 h and detected with the indicated primary antibodies and HRP-conjugated secondary antibodies by enhanced chemiluminescence reagent (Amersham Biosciences). Signals were quantified with an ECL imager (Bio-Rad) and Quantity One software.
Immunocytochemistry
COS7 cells were seeded on coverslips in a 12-well plate (30,000 cells/well) and transfected with the respective constructs 24 h later. After an additional 48 h, the medium was removed, and cells were gently washed with warm PBS following fixation with 4% paraformaldehyde/PBS (w/v) for 20 min. After three washing steps with PBS, cells were permeabilized for 5 min with 0.25% Triton X-100 in PBS. Cells were blocked with 10% normal goat serum (v/v) or 10% BSA (w/v) in 0.125% Triton X-100 in PBS. The primary and secondary antibodies were incubated on cells for 1 h each. Intermediate washing steps were performed three times for 5 min with 0.125% Triton X-100. Coverslips were mounted on microscope slides using IMMU-Mount and dried overnight at room temperature.
Statistical Analysis
ECL signals were quantitated using an ECL imager (ChemiDoc XRS, Bio-Rad) and the Quantity One Software (Bio-Rad). Statistical analysis of three independent experiments (n = 3) was done by one-way analysis of variance using GraphPad PRISM 5. **, p < 0.01; ***, p < 0.001.
RESULTS
In Silico Analysis of Mouse IDE
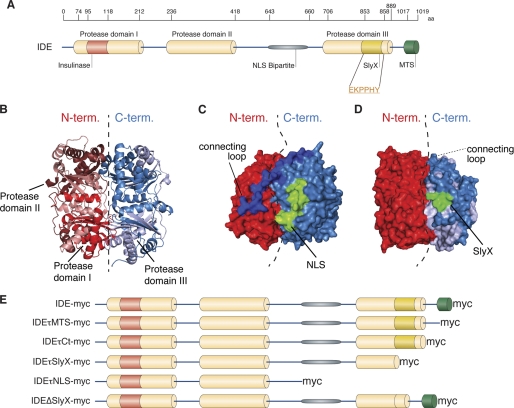
To identify functional domains in the primary amino acid sequence of IDE, we first performed an in silico analysis using online bioinformatical tools. Our analysis revealed that IDE contains three protease domains, one active with the catalytic core (aa 74–212) and two inactive ones (aa 236–418 and 706–889) (Fig. 1A), which are typical for M16 peptidases. Protease domains I and II are localized in the N-terminal half, whereas protease domain III is in the C-terminal half of the protein (Fig. 1, A and B). A nuclear localization sequence (NLS) was also identified between the second and third protease domains (positions 643–660 aa) (Fig. 1, A and C) (15). Three-dimensional modeling revealed that the NLS is present on the surface of IDE (Fig. 1C). Consistent with a functional NLS, a fraction of IDE has been detected previously in the nucleus (16). In addition, a microbody-targeting signal (MTS) is localized at the C terminus of the protein (17).
FIGURE 1.
In silico analysis of mouse IDE revealed new functional domains. A, diagram of IDE summarizing the results of in silico analysis. The relative size scale is shown above the diagram to illustrate the positions of featured domains: inactive protease domains II (aa 236–418) and III (aa 706–889); NLS (aa 643–660) between protease domains II and III; SlyX domain (EKPPHY, aa 853–858) at the end of protease domain III; and a previously described MTS domain at the C-terminal end of the protein (aa 1017–1019). B, protease domains I and II are forming the N-terminal half (N-term.) of the protein responsible for catalytic activity, and the protease domain III is localized in the C-terminal half (C-term.) and involved in substrate binding. C and D, three-dimensional mapping of NLS and SlyX domains demonstrated that these domains are located at the protein surface. E, schematic representation of IDE constructs with truncations and deletion cloned to assess the role of newly identified domains.
Interestingly, we also identified a novel sequence motif EKPPHY located at the end of the inactive protease domain III (aa 853–858) (Fig. 1A). This motif shares 100% identity with the motif within the C terminus of the bacterial SlyX protein (InterPro Database number, IPR007236; Pfam Database number, PF04102), a member of the SlyX superfamily with a conservative motif PPHY/W of unknown function (18). Notably, this motif is also localized at the surface of IDE in a three-dimensional model (Fig. 1D).
To investigate a potential functional relevance of the identified SlyX sequence, we first generated truncated variants of IDE (Fig. 1E). In addition, a specific deletion mutant of the SlyX motif was also generated. To distinguish the recombinant IDE variants from the endogenous protein, mutant IDE proteins were tagged with C-terminal Myc epitope. As control, a full-length IDE construct carrying a C-terminal Myc epitope was also generated.
Expression and Subcellular Localization of IDE Variants
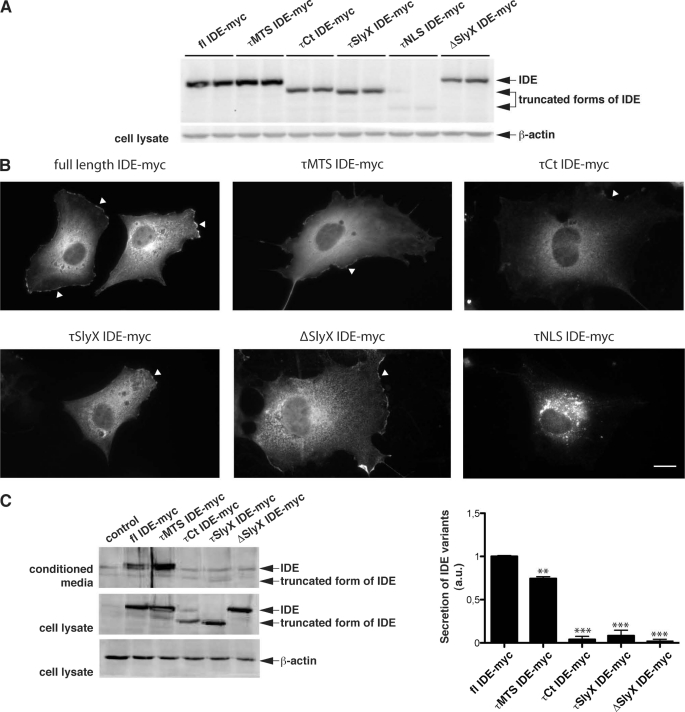
Different variants of IDE were expressed in COS7 cells and analyzed by Western immunoblotting and immunocytochemistry. All variants of IDE could be expressed in these cells (Fig. 2A). Full-length IDE and the τMTS IDE mutant were detected at sizes around 115 kDa, and τCt and τSlyX were detected at 100 kDa. The shortest mutant of IDE, τNLS, was detected at ∼75 kDa. Immunofluorescence microscopy demonstrated that the full-length IDE, as well as a τMTS mutant of IDE, were mainly localized in the cytoplasm. In addition, prominent association with the plasma membrane was also observed (Fig. 2B). Cells expressing shorter variants of IDE, τCt, τSlyX, and ΔSlyX, demonstrated a similar pattern and were indistinguishable from the full-length IDE and τMTS variants, i.e. the protein was found in the cytoplasm and at the membrane. The τNLS variant of IDE, however, was present in vesicular-like structures around the nucleus (Fig. 2B). The overall expression of this variant was significantly lower as compared with other forms of IDE, probably due to decreased stability. Therefore, this mutant was not analyzed further.
FIGURE 2.
Importance of the SlyX domain in the secretion of IDE. A, expression of truncated and deletion variants of IDE in COS7 cells. Western immunoblot detection with anti-Myc antibody demonstrated expression of full-length (fl) IDE (∼115 kDa), τMTS (∼115 kDa), τCt (∼100 kDa), τSlyX (∼100 kDa), and the ΔSlyX IDE variant (∼110 kDa) at comparable levels. Expression of the τNLS IDE variant (∼75 kDa) was very low, probably due to low stability. B, immunocytochemical analysis of COS7 cells transfected with various IDE constructs using anti-Myc antibodies. Full-length IDE and τMTS, τCt, and τSlyX IDE are present in the cytoplasm and at the plasma membrane (arrowheads). ΔSlyX variant of IDE demonstrated very similar distribution. τNLS IDE was detected in punctate structures with very little if any cytoplasmic or membrane localization. Scale bar, 20 μm. C, Western immunoblot analysis of IDE secretion. Cells were transfected with the indicated IDE variants and incubated for 48 h, IDE was then detected in the cell lysates and conditioned media by Western immunoblotting. Weak bands represent endogenous IDE. The secretion of the τCt, τSlyX, and ΔSlyX variants was strongly reduced as compared with full-length IDE, whereas deletion of the MTS (τMTS) had minor effect on secretion. Quantification of IDE secretion was done by ECL imaging; values represent means of three independent experiments ± S.E. (secretion level of full-length IDE was set as 1). **, p < 0.01; ***, (p < 0.001). a.u., arbitrary units.
Importance of the SlyX Motif in the Non-conventional Secretion of IDE
It has been shown previously that IDE can be secreted via a non-conventional pathway in association with exosomes (13, 14). Thus, we analyzed the secretion of the different IDE variants. Consistent with previous data, significant amounts of full-length IDE were readily detected in the conditioned media of transfected COS7 cells (Fig. 2C).
The IDE mutant with deleted MTS motif showed slightly decreased secretion (75 ± 2% as compared with full-length IDE; Fig. 2C). Notably, secretion of shorter variants was markedly decreased as compared with full-length IDE or τMTS IDE (4 ± 3% for τCt, 8 ± 6% for τSlyX, and 3 ± 2% for ΔSlyX; Fig. 2C). These data indicate that the C-terminal region and/or the SlyX motif play a role in non-conventional secretion of IDE. Evidence for a critical involvement of the SlyX domain in secretion came from the analysis of the ΔSlyX variant of IDE that only lacks the EKPPHY sequence. These combined results demonstrate the importance of the identified SlyX motif in the non-conventional secretion of IDE.
To confirm the role of the SlyX domain in secretion, we also generated fusion proteins of GFP with the SlyX domain of IDE. The presence of the SlyX motif strongly increased the secretion of GFP into the conditioned medium, further indicating that this domain could promote secretion of cytosolic proteins (supplemental Fig. 1).
DISCUSSION
In this work, we sought to identify functional domains in the primary amino acid sequence of mouse IDE and found several motifs including inactive protease domains II and III, an NLS, and a novel SlyX sequence (EKPPHY). Inactive protease domains are a typical feature of M16 proteases. These domains do not exert catalytic activity but are involved in the interaction and positioning of peptide substrates for subsequent cleavage by the active protease domain. In IDE, the inactive protease domains II and III initially bind peptide substrates, which are subsequently cleaved by the catalytically active protease domain I in the N-terminal half of the enzyme (20).
Little is known about the specific mechanisms that target IDE to different cell compartments. In addition to the MTS (17), a mitochondria-targeting signal, which can only be found in a particular splice variant of IDE, has been described (21). A previous study showed that the MTS motif is crucial for IDE stability, and its deletion (DEL.pts) led to the destabilization of the protein (17). In another study, however, larger deletions of N- and C-terminal domains could be efficiently expressed (22). Here, we also show that C-terminal deletion variants, including τMTS, τCT, and the τSlyX, were expressed at similar levels and showed similar localizations as compared with the full-length variant of IDE. Only the τNLS variant that lacks the C-terminal 376 amino acids showed strongly decreased expression and aberrant localization, suggesting that this variant has decreased stability, likely due to impaired folding.
Interestingly, although IDE has no classical signal sequence for secretion, it is found in extracellular fluids including blood and cerebrospinal fluid and also in conditioned media of cultured cells (14, 19, 23). However, the mechanism by which IDE is secreted was largely unclear. Only recently has it been shown that IDE is predominantly secreted via a non-conventional secretion pathway, which involves multivesicular bodies and exosomes (13, 14). Notably, C-terminal deletions of IDE decreased the secretion of IDE variants. Although the deletion of the MTS motif slightly decreased secretion, larger truncations led to almost complete inhibition of IDE release. More importantly, the specific deletion of the newly identified SlyX motif also led to blockage of release, indicating that this sequence plays a crucial role in non-conventional secretion of IDE. Because the τCt form of IDE that contains the SlyX motif also showed decreased secretion, interaction of the SlyX motif with more C-terminal domains might also be involved in the secretion process. Thus, it will be interesting to further evaluate the structural requirements of IDE for efficient secretion.
Also, to assess involvement of the SlyX domain in protein secretion, we fused the SlyX motif to GFP. Its addition drastically increased secretion of GFP into the medium indicating that the SlyX motif may be sufficient for protein secretion via non-conventional secretory pathway.
Interestingly, the same domain is present in other proteins, which are reported to be secreted via non-conventional pathways, such as heparan sulfate proteoglycans (12) and phospholipid scramblase 1 (24). Together, these results demonstrate the importance of a novel hexapeptide sequence EKPPHY in non-conventional pathways for secretion of IDE that we call the SlyX motif. A selective targeting of this motif might help to modulate release of the enzyme and thereby regulate extracellular degradation of peptide substrates, including insulin and Aβ.
Supplementary Material
This work was supported in part by the Deutsche Forschungsgemeinschaft (Grants SFB645 and KFO177), the Federal Ministry for Education and Research (BMBF) (Grant 01GI0708), and the BONFOR Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- IDE
- insulin-degrading enzyme
- Aβ
- amyloid-β peptide
- NLS
- nuclear localization sequence
- MTS
- microbody-targeting signal
- aa
- amino acids
- Ct
- C terminus.
REFERENCES
- 1. Mirsky I. A., Broh-Kahn R. H. (1949) Arch. Biochem. 20, 1–9 [PubMed] [Google Scholar]
- 2. Fernandez-Gamba A., Leal M. C., Morelli L., Castano E. M. (2009) Curr. Pharm. Des. 15, 3644–3655 [DOI] [PubMed] [Google Scholar]
- 3. Guo Q., Manolopoulou M., Bian Y., Schilling A. B., Tang W. J. (2010) J. Mol. Biol. 395, 430–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malito E., Ralat L. A., Manolopoulou M., Tsay J. L., Wadlington N. L., Tang W. J. (2008) Biochemistry 47, 12822–12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernstein H. G., Ansorge S., Riederer P., Reiser M., Frölich L., Bogerts B. (1999) Neurosci. Lett. 263, 161–164 [DOI] [PubMed] [Google Scholar]
- 6. Behl M., Zhang Y., Zheng W. (2009) Cerebrospinal Fluid Res. 6, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crossgrove J. S., Li G. J., Zheng W. (2005) Exp. Biol. Med. (Maywood) 230, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strozyk D., Launer L. J., Adlard P. A., Cherny R. A., Tsatsanis A., Volitakis I., Blennow K., Petrovitch H., White L. R., Bush A. I. (2009) Neurobiol. Aging 30, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edbauer D., Willem M., Lammich S., Steiner H., Haass C. (2002) J. Biol. Chem. 277, 13389–13393 [DOI] [PubMed] [Google Scholar]
- 10. Kurochkin I. V., Goto S. (1994) FEBS. Lett. 345, 33–37 [DOI] [PubMed] [Google Scholar]
- 11. Leal M. C., Dorfman V. B., Gamba A. F., Frangione B., Wisniewski T., Castaño E. M., Sigurdsson E. M., Morelli L. (2006) J. Neuropathol. Exp. Neurol. 65, 976–987 [DOI] [PubMed] [Google Scholar]
- 12. Nickel W., Rabouille C. (2009) Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 13. Bulloj A., Leal M. C., Xu H., Castaño E. M., Morelli L. (2010) J. Alzheimers Dis. 19, 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamboli I. Y., Barth E., Christian L., Siepmann M., Kumar S., Singh S., Tolksdorf K., Heneka M. T., Lütjohann D., Wunderlich P., Walter J. (2010) J. Biol. Chem. 285, 37405–37414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akiyama H., Shii K., Yokono K., Yonezawa K., Sato S., Watanabe K., Baba S. (1988) Biochem. Biophys. Res. Commun. 155, 914–922 [DOI] [PubMed] [Google Scholar]
- 17. Chesneau V., Perlman R. K., Li W., Keller G. A., Rosner M. R. (1997) Endocrinology 138, 3444–3451 [DOI] [PubMed] [Google Scholar]
- 18. Bothmann H., Pluckthun A. (2000) J. Biol. Chem. 275, 17100–17105 [DOI] [PubMed] [Google Scholar]
- 19. Zhao J., Li L., Leissring M. A. (2009) Mol. Neurodegener. 4, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen Y., Joachimiak A., Rosner M. R., Tang W. J. (2006) Nature 443, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farris W., Leissring M. A., Hemming M. L., Chang A. Y., Selkoe D. J. (2005) Biochemistry 44, 6513–6525 [DOI] [PubMed] [Google Scholar]
- 22. Li P., Kuo W. L., Yousef M., Rosner M. R., Tang W. J. (2006) Biochem. Biophys. Res. Commun. 343, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 23. Qiu W. Q., Walsh D. M., Ye Z., Vekrellis K., Zhang J., Podlisny M. B., Rosner M. R., Safavi A., Hersh L. B., Selkoe D. J. (1998) J. Biol. Chem. 273, 32730–32738 [DOI] [PubMed] [Google Scholar]
- 24. Merregaert J., Van Langen J., Hansen U., Ponsaerts P., El Ghalbzouri A., Steenackers E., Van Ostade X., Sercu S. (2010) J. Biol. Chem. 285, 37823–37837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.