Abstract
Promoter-proximal pausing of RNAPII coincides with the formation of the cap structure at the 5′ end of pre-mRNA, which is bound by the cap-binding protein complex (CBC). Although the positive transcription elongation factor b (P-TEFb) stimulates the release of RNAPII from pausing and promotes transcription elongation and alternative splicing by phosphorylating the RNAPII C-terminal domain at Ser2 (S2-P RNAPII), it is unknown whether CBC facilitates these events. In this study, we report that CBC interacts with P-TEFb and transcriptionally engaged RNAPII and is globally required for optimal levels of S2-P RNAPII. Quantitative nascent RNA immunoprecipitation and ChIP experiments reveal that depletion of CBC attenuates HIV-1 Tat transactivation and impedes transcription elongation of investigated CBC-dependent endogenous genes by decreasing the levels of P-TEFb and S2-P RNAPII, leading to accumulation of RNAPII in the body of these genes. Finally, CBC is essential for the promotion of alternative splicing through facilitating P-TEFb, S2-P RNAPII, and splicing factor 2/alternative splicing factor occupancy at a splicing minigene. These findings disclose a vital role of CBC in connecting pre-mRNA capping to transcription elongation and alternative splicing via P-TEFb.
Keywords: Cdk (Cyclin-dependent Kinase), Gene Regulation, RNA Polymerase II, RNA Splicing, Transcription Elongation Factors, CBC, P-TEFb, Alternative Splicing, Promoter-proximal Pausing, Transcription Elongation
Introduction
The mature mRNA is generated by the synthesis and cotranscriptional processing of pre-mRNA. These mutually coupled processes are executed by RNA polymerase II (RNAPII)2 and coordinated by the C-terminal domain (CTD) of its biggest subunit, Rpb1, which serves as a platform for binding pre-mRNA processing and chromatin-modifying factors (1). Differential recruitment of these complexes at distinct phases of the RNAPII transcription cycle is dictated by the phosphorylation pattern of the RNAPII CTD, whose multiple heptapeptide Y1S2P3T4S5P6S7 repeats undergo phosphorylation at all serine residues (2, 3).
After assembling with the transcription preinitiation complex at the promoter, RNAPII enters into transcription initiation and promoter clearance, which coincide with phosphorylation of the RNAPII CTD at Ser5 (S5-P RNAPII) and Ser7 (S7-P RNAPII) by Cdk7 of the transcription factor II H (4). The S5-P RNAPII mark is required for binding capping enzymes (CE) to modify the 5′ end of pre-mRNA by the addition of a 7-methyl G(5′)ppp(5′)N cap as soon as the transcript is 25–50 nucleotides long (5). This cap structure is cotranscriptionally and cooperatively bound by the cap-binding protein complex (CBC), which consists of the cap-binding protein (CBP) 20 and CBP80 (6, 7). Notably, CBC plays a critical role in processes accompanying transcription, including pre-mRNA splicing and 3′ end processing, and post-transcriptional events such as mRNA nuclear export and nonsense-mediated decay (8–12).
Importantly, pre-mRNA capping coincides with the promoter-proximal pausing of RNAPII that occurs soon after promoter clearance at most eukaryotic genes and is enabled by cooperative actions of the negative elongation factor and the DRB-sensitivity inducing factor (13). The release of RNAPII from this pause can be stimulated by the kinase activity of the positive transcription elongation factor b (P-TEFb), which is composed of the catalytic subunit Cdk9 and one of the three cyclin (Cyc) regulatory subunits CycT1, CycT2a, and Cyc2Tb (14). Upon its recruitment to paused RNAPII, P-TEFb phosphorylates the RNAPII CTD at Ser2 (S2-P RNAPII) and the Spt5 subunit of the DRB-sensitivity inducing factor, resulting in productive RNAPII elongation. Of note, a prototypical example of transcription regulation through promoter-proximal pausing is transcription of HIV-1 (14). Here, the release of RNAPII into elongation is directed by the viral transactivator (Tat), which recruits P-TEFb to stalled RNAPII through cooperative binding with the transactivation response element present at the 5′ end of HIV-1 transcript (15). In addition to stimulating transcription elongation, P-TEFb promotes transcription-coupled processes (16, 17). For instance, P-TEFb in yeast and Drosophila is required for cotranscriptional 3′ end processing, and human P-TEFb stimulates alternative splicing of pre-mRNA (18–20).
Although different phases of the RNAPII transcription cycle are being elucidated in great detail, mechanisms enabling efficient transcription elongation remain to be fully understood. In addition to the recruitment of P-TEFb to paused RNAPII by transcriptional activators and the double bromodomain-containing protein Brd4 (13, 21, 22), alternative modes of tethering P-TEFb for stimulating RNAPII elongation may exist. Given that CBC binds the pre-mRNA cap structure concomitant with RNAPII pausing, we hypothesized that CBC might mark the completion of transcript capping and in turn play a role in mediating productive transcription elongation. Furthermore, that both CBC and P-TEFb affect cotranscriptional pre-mRNA splicing and 3′ end processing led us to postulate that these two complexes could function in cooperation. In this study, we demonstrate a novel role of CBC in promoting transcription elongation by interacting with P-TEFb and facilitating its occupancy at target genes. We further disclose that CBC is required for modulating P-TEFb-dependent alternative splicing in human cells. Collectively, our findings reveal how CBC orchestrates the coupling of pre-mRNA capping to transcription elongation and alternative splicing.
EXPERIMENTAL PROCEDURES
Cell Culture
The HeLa-based HL3T1 and HH8 cell line expressing FLAG-tagged HEXIM1 (F.HEXIM1) were described (23, 24). Cells were grown at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum, 100 mm l-glutamine, and 50 μg each of penicillin and streptomycin per ml.
Plasmid DNAs and siRNAs
F.Tat was expressed from the pcDNA3.1 plasmid. pSVED-A Tot minigene cassette was described (25). γ-Methylphosphate-capping enzyme (MePCE) siRNA was purchased from Sigma-Genosys and had the sequence: 5′-rGrArArCUrArCUrArCrCrGrArAUrCrCrArATT-3′. Brd4 siRNA was described previously (19). CBP20 (sc-38249), CBP80 (sc-43669), and SF2/ASF (sc-38319) siRNAs were obtained from Santa Cruz Biotechnology. The control siRNA was purchased from Sigma.
HL3T1 or HeLa cells were transfected by plasmid DNAs and siRNAs using FuGENE6 reagent (Roche Applied Science) and Lipofectamine 2000 reagent (Invitrogen), respectively. For CAT reporter gene and alternative splicing assay, HL3T1 or HeLa cells, respectively, were seeded into 6-well plates and treated with 100 pmol of the respective siRNAs. For chromatin immunoprecipitation (ChIP) and quantitative nascent RNA immunoprecipitation (qNARIP) experiments, cells were seeded into 150-mm-diameter Petri dishes and treated with 1.4 nmol of the respective siRNAs. After 48 h, cells were transfected with the plasmid DNA and subjected to downstream procedures after additional 24 h.
Immunoreagents and Chemicals
The CBP20 (sc-48793), CBP80 (sc-48803), CycT1 (sc-10750), Cdk9 (sc-484), and Cdk4 (sc-601) antibodies, normal rabbit (sc-2027) and mouse IgG (sc-2025) were obtained from Santa Cruz Biotechnology. The GAPDH (ab4300), RNAPII CTD (ab5408), S2-P RNAPII CTD (ab5095), and S5-P RNAPII CTD (ab5131) antibodies were obtained from Abcam. The FLAG M2 (F3165) antibody was purchased from Sigma-Aldrich. The CBP80 antibody used in the ChIP assay was a kind gift from Dr. Elisa Izaurralde.
Immunoprecipitation Assay and Western Blotting
Immunoprecipitation assay and Western blotting were performed according to standard protocols. Whole cell extracts (WCEs) were prepared using buffer D (20 mm HEPES-KOH, pH 7.9, 15% glycerol, 0.2 mm EDTA, 0.2% Nonidet P-40, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride) containing 0.1 m KCl, and immune complexes were washed extensively with buffer D containing 0.3 m KCl. For antibodies, see the list of immunoreagents above. 0.8 μg of each antibody was used per individual immunoprecipitation experiment. Three independent experiments for each immunoprecipitation yielded similar results.
CAT Assay
A chloramphenicol acetyl transferase (CAT) enzymatic assay was performed using standard protocol. -Fold transactivation represents the ratio between the F.Tat-activated transcription and the activity of the reporter gene in cells transfected with the empty pcDNA3.1 plasmid. Results were obtained from at least three independent experiments.
RT-qPCR Assay
Total RNA from HeLa or HL3T1 cells was isolated using TRIzol Reagent (Invitrogen). Reverse transcription was performed with the Moloney murine leukemia virus reverse transcriptase (Invitrogen) using random hexamer primers. For quantitative PCR (qPCR) primers were designed using Integrated DNA Technologies PrimerQuest program (for sequences, see supplemental Table S1). qPCR was performed with Stratagene Mx3005P real-time PCR system and SYBR Green master mix (Applied Biosystems). For the quantification of mRNA levels from endogenous genes, absolute values were normalized to those of GAPDH mRNA. For the quantification of mRNA levels from transfected plasmids, absolute values were normalized to the levels of plasmid DNA. Results were obtained from at least three independent experiments.
ChIP Assay
Cross-linking was achieved by incubating HeLa or HL3T1 cells in 1% formaldehyde in medium for 10 min at room temperature. Cross-linking reactions were stopped by the addition of glycine to a final concentration of 0.125 m. Cells were washed with cold PBS, scraped, and pelleted in a conical tube. The crude nuclear extracts were obtained by incubating cells for 10 min on ice in 5 mm PIPES, pH 8.0, 85 mm KCl, and 0.5% Nonidet P-40. The pellets were washed in 50 mm Tris-HCl, pH 8.1, 10 mm EDTA. Sonication and immunoprecipitation were performed using a ChIP Assay kit (Upstate) according to the manufacturer's instructions. 0.5–1 × 107 cells were used for one immunoprecipitation. Antibodies used are described above. As a negative control, the normal rabbit and mouse IgG were used. Following reverse cross-linking, DNA was extracted with phenol-chloroform-isoamylalcohol, precipitated with ethanol, and dissolved in 30 μl of Tris-EDTA buffer. Each sample was diluted three times, and 1 μl of DNA was used with appropriate primer sets to amplify specific DNA fragments with qPCR (for sequences, see supplemental Table S2). Samples were analyzed by the Stratagene Mx3005P real-time PCR system and SYBR Green master mix. Absolute levels were normalized to input DNA. Results were obtained from at least three independent experiments.
qNARIP Assay
Prior to sonication, cells were lysed for 10 min on ice in 700 μl of lysis buffer (50 mm HEPES, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) with protease and RNase inhibitors. Samples were sonicated 3 × 10 s at 10% amplitude using Bandelin homogenizer HD 3100. These settings yielded about 100-nucleotide-long fragments of nascent RNA. 5% of each sonicated sample was used for reverse cross-linking and isolation of input DNA and input RNA. Following centrifugation, chromatin solutions were precleared with 35 μl of protein A/G-agarose-PLUS beads (Santa Cruz Biotechnology) and then incubated with the appropriate antibody at 4 °C overnight. 1–2 107 cells were used for one immunoprecipitation. To immunoprecipitate nascent mRNA, the RNAPII CTD antibody (ab5408) was used, and the normal mouse IgG (sc-2025) served as a negative control. To collect immune complexes, 30 μl of the beads were added to each tube. Incubation and washes of the beads were performed as described in the ChIP Assay kit. Immune complexes were eluted twice by the addition of 75 μl of 1% SDS, 0.1 m NaHCO3 with RNase inhibitor. The complexes were reverse cross-linked in 200 mm NaCl, 10 mm EDTA, and 20 μg of proteinase K with RNase inhibitor at 65 °C for 2 h. RNA was isolated using TRIzol Reagent and treated with Turbo DNA-free DNase (Ambion). Reverse transcription was performed with the Superscript III reverse transcriptase (Invitrogen) using random hexamer primers. Samples were analyzed by the Stratagene Mx3005P real-time PCR system and SYBR Green master mix. qPCR was performed with primer pairs discriminating initiated and elongated nascent transcripts. For primer sequences, see supplemental Tables S3, S4, and S5. The levels of nascent mRNA were normalized to the levels of DNA in input samples amplified with the same primer pairs. The amplification of negative control samples yielded 10–15-fold lower values than the amplification of RNAPII-immunoprecipitated transcripts, reasserting the specificity of immunoprecipitation with the RNAPII antibody. qPCR of input DNA confirmed that expression of F.Tat was equally efficient in HL3T1 cells treated with the control or CBP20 siRNA. Results were obtained from at least three independent experiments.
RESULTS
CBC Interacts with P-TEFb and Is Required for Efficient Phosphorylation of the RNAPII CTD at Ser2
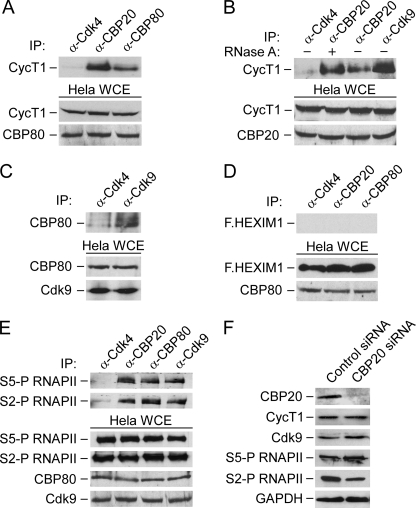
Functional similarities between CBC and P-TEFb in pre-mRNA processing could stem from their physical association. We therefore performed coimmunoprecipitation experiments between endogenous proteins in WCE of HeLa cells (Fig. 1). Indeed, CycT1 was readily detected in immunoprecipitations of CBP20 or CBP80 (Fig. 1A). This interaction was not abrogated upon treatment of WCE with RNase A, demonstrating that CBC and P-TEFb interact independently of RNA (Fig. 1B). Similarly, Cdk9 interacted with CBP80 in reciprocal coimmunoprecipitation experiments (Fig. 1C). The observed interactions were specific because an antibody against Cdk4 failed to immunoprecipitate the components of P-TEFb and CBC efficiently, even though it displayed immunoprecipitation efficacy similar to an antibody against Cdk9 (Fig. 1 and supplemental Fig. S1A). In addition and in contrast to P-TEFb, none of the CBC subunits bound HEXIM1, an inhibitor of P-TEFb within 7SK snRNP (22) (Fig. 1D and supplemental Fig. S1B). Although the stoichiometry of the interaction between CBC and P-TEFb is unclear at present, these findings indicate that CBC associates with the active pool of P-TEFb in cells.
FIGURE 1.
CBC and P-TEFb interact with each other as well as with S5-P and S2-P RNAPII, and depletion of CBC reduces total levels of S2-P RNAPII. A and B, WCEs of HeLa cells incubated (+) or not (−) with RNase A were subjected to immunoprecipitation (IP) with the indicated antibodies. Levels of CycT1 in immunoprecipitates (top) and those of CycT1 and CBP20 or CBP80 in WCEs (bottom) were detected by Western blotting. C, WCEs of HeLa cells were subjected to IP with the indicated antibodies. Levels of CBP80 in immunoprecipitates (top) and those of CBP80 and Cdk9 in WCEs (bottom) were detected by Western blotting. D, WCEs of HeLa cells expressing F.HEXIM1 were subjected to IP with the indicated antibodies. Levels of F.HEXIM1 in immunoprecipitates (top) and those of F.HEXIM1 and CBP80 in WCEs (bottom) were detected by Western blotting. E, WCEs of HeLa cells were subjected to IP with the indicated antibodies. Levels of S5-P RNAPII and S2-P RNAPII in immunoprecipitates (top) and those of S5-P RNAPII, S2-P RNAPII, CBP80, and Cdk9 in WCEs (bottom) were detected by Western blotting. F, levels of the indicated proteins in WCEs of HeLa cells treated with the control or CBP20 siRNA were detected by Western blotting.
To characterize the relation between CBC and P-TEFb with the transcription apparatus, we asked whether they bind transcriptionally engaged RNAPII. Coimmunoprecipitation experiments between endogenous proteins revealed that both subunits of CBC as well as Cdk9 bind S5-P and S2-P RNAPII (Fig. 1E). These results are consistent with a previous report that identified proteins associated with the phosphorylated form of RNAPII using a comprehensive proteomic analysis (26). Because P-TEFb can regulate transcription elongation and pre-mRNA processing through S2-P RNAPII (17), we next investigated whether CBC affects this phosphorylation event. To this end, we measured the total levels of endogenous S5-P and S2-P RNAPII by Western blotting of WCE of HeLa cells treated with the control or CBP20 siRNA. Significant reduction of CBP20 amounts in cells treated with CBP20 siRNA did not change the expression of CycT1 or Cdk9 (Fig. 1F). On the contrary, we consistently observed a decrease in the levels of S2-P RNAPII in CBC-depleted cells, whereas those of S5-P RNAPII and GAPDH remained largely unaffected (Fig. 1F). Overall, these results demonstrate that endogenous CBC and P-TEFb interact in cells and that CBC is needed for optimal phosphorylation of the RNAPII CTD at Ser2. Furthermore, that CBC and P-TEFb associate with S5-P and S2-P RNAPII suggests their involvement in RNAPII transcription throughout the gene.
CBC Is Required for Transcription from the Tat-activated HIV-1 Promoter
The results above prompted us to consider a possible necessity of CBC for enabling the transcriptional effects of P-TEFb. First, we examined whether CBC is required for transcription from the HIV-1 promoter (Fig. 2). For this purpose, we used a HeLa-based HL3T1 cell line that contains an integrated HIV-1 long terminal repeat (LTR) controlling expression of the CAT reporter gene (23). To stimulate this transcription, we transiently expressed F.Tat in these cells. Strikingly, the treatment of HL3T1 cells with CBP20 siRNA inhibited Tat transactivation of the HIV-1 LTR 8.5-fold relative to the transactivation in cells treated with the control siRNA (Fig. 2B). Similar results were obtained in HL3T1 cells treated with CBP80 siRNA (supplemental Fig. S2B). Of note, CBP20 or CBP80 siRNA was effective because relative levels of targeted mRNA decreased 5-fold or 4-fold, respectively (Fig. 2A and supplemental Fig. S2A). Importantly, these treatments did not decrease the amounts of F.Tat (supplemental Fig. S2C). Next, we asked whether this inhibitory effect of CBC depletion on Tat-activated HIV-1 transcription could also be detected at the mRNA level. Indeed, the quantity of CAT mRNA decreased 3.3-fold in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 2C). Taken together, these findings suggest that CBC is required for HIV-1 Tat transactivation.
FIGURE 2.
Depletion of CBC abrogates Tat transactivation of the HIV-1 LTR CAT reporter gene and provokes accumulation of nascent transcripts within the body of the gene. A, relative quantity of CBP20 mRNA was determined by RT-qPCR using total RNA isolated from HL3T1 cells that expressed F.Tat and were treated with the control (white bars) or CBP20 siRNA (black bars). Absolute mRNA levels were normalized to GAPDH mRNA levels. B, CAT assay of WCEs of HL3T1 cells containing an integrated HIV-1 LTR CAT reporter gene is shown. Cells were treated with the control (white bars) or CBP20 siRNA (black bars) and expressed F.Tat as indicated below the CAT data. C, relative quantity of CAT mRNA was determined by RT-qPCR using total RNA samples isolated from HL3T1 cells that expressed F.Tat and were treated with the control (white bars) or CBP20 siRNA (black bars). Absolute CAT mRNA levels were normalized to the levels of transfected F.Tat gene. D, qNARIP was used to analyze HIV-1 LTR CAT nascent transcripts in HL3T1 cells expressing F.Tat. Cells were treated with the control (white bars) or CBP20 siRNA (black bars). Nascent mRNA was immunoprecipitated with the RNAPII antibody. Results are presented as percent of input DNA. Two different primer pairs were used for the amplification of transcripts at the 5′ region (5′r) or downstream region (Dr) as indicated below the schematic of the HIV-1 LTR CAT reporter gene. Arrow and pA depict transcription start site and polyadenylation signal, respectively. Primer pairs used for the amplification of DNA in ChIP-qPCR analysis (Fig. 3) at the promoter region (Pr) or coding region (Cr) are indicated above the gene schematic. Numbers below the schematic represent nucleotide positions of the gene in respect to the transcription start site. E, qNARIP was used to analyze HIV-1 nascent transcripts in HeLa expressing F.Tat and HIV-1 genome from pNL4–3 plasmid. Cells were treated with the control (white bars) or CBP20 siRNA (black bars). Nascent mRNA was immunoprecipitated with the RNAPII antibody. Results are presented as percent of input DNA. Four different primer pairs were used for the amplification of transcripts at the 5′ region (5′r) or downstream regions (Dr1, Dr2, and Dr3) as indicated below the schematic of the HIV-1 genome. Arrow and pA depict transcription start site and polyadenylation signal, respectively. Numbers below the schematic represent nucleotide positions of the gene in respect to the transcription start site.
CBC Facilitates the Gene Occupancy of P-TEFb and S2-P RNAPII for Tat-stimulated HIV-1 Transcription Elongation
To test directly whether depletion of CBC antagonized transcription elongation from the HIV-1 LTR, we developed a qNARIP assay (for details, see “Experimental Procedures,” supplemental Fig. S3, and supplemental Control Experiments) and used it for analyzing the nascent CAT transcripts in HL3T1 cells expressing F.Tat. Briefly, we cross-linked the cells, sonicated nuclear extracts, immunoprecipitated the pre-mRNA with an antibody directed against RNAPII, and measured relative levels of nascent transcripts at their 5′ region (5′r; initiated transcripts) and downstream region (Dr; elongated transcripts) by using two different primer pairs (Fig. 2D). Although the levels of initiated CAT transcripts did not change considerably, those of elongated transcripts increased 1.6-fold in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 2D). In addition, we wanted to confirm that this accumulation of RNAPII-bound elongated transcripts also occurs at several regions within the body of a transcription unit. Here, we employed a qNARIP assay for determining nascent HIV-1 transcripts in HeLa cells expressing F.Tat and the whole HIV-1 genome encoded by the pNL4–3 plasmid (Fig. 2E). Indeed, although the levels of nascent transcripts at the 5′ region of the genome remained largely unaltered, the levels of elongated transcripts at pol, vpr, and env genes increased 2.4-, 1.9-, and 2.6-fold in cells treated with CBP20 siRNA compared with the control siRNA, respectively (Fig. 2E). Accumulation of RNAPII-associated pre-mRNA in the body of the genes suggests that depletion of CBC restrains efficient transcription elongation. In addition, that qNARIP assay detects considerably more initiated than elongated transcripts indicates the occurrence of RNAPII promoter-proximal pausing at both transcription units (Fig. 2, D and E, compare 5′r and Dr).
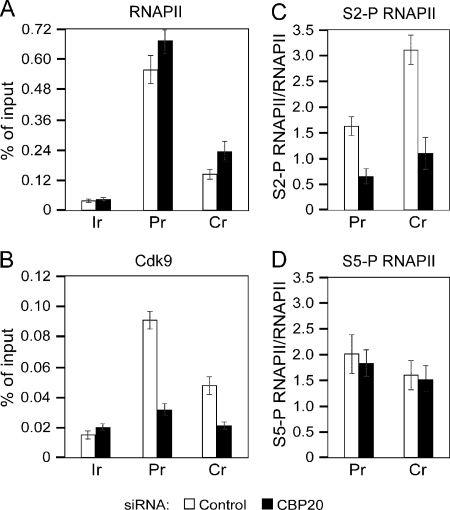
To address the mechanism by which the depletion of CBC antagonizes P-TEFb-dependent transition into effective transcription elongation, we performed quantitative chromatin immunoprecipitation (ChIP-qPCR) analysis at the promoter and coding region of the HIV-1 LTR CAT reporter gene in HL3T1 cells expressing F.Tat. In agreement with the qNARIP analysis of this gene, we observed accumulation of total RNAPII at the coding region of the reporter gene in cells treated with CBP20 siRNA, suggesting deficient RNAPII elongation (Fig. 3A). This interpretation is in agreement with the effect of depleting a Ser/Arg-rich (SR) splicing factor SC35, which led to a deficient occupancy of Cdk9 and S2-P RNAPII at SC35-dependent genes, resulting in the accumulation of RNAPII in the body of these genes (27). Critically, the occupancy of Cdk9 at the promoter and coding region decreased 2.7-fold and 2.3-fold under the same experimental conditions, respectively (Fig. 3B). Concomitantly, the presence of S2-P RNAPII decreased 3-fold when normalized to the levels of total RNAPII at both gene regions upon depleting CBC (Fig. 3C), whereas the occupancy of S5-P RNAPII remained largely unchanged (Fig. 3D). Levels of S2-P and S5-P RNAPII presented as percent of input DNA are shown in supplemental Fig. S4, A and B. CBC was efficiently depleted in cells treated with CBP20 siRNA compared with the control siRNA because the occupancy of CBP80 at the gene regions decreased 3.2-fold and 2.9-fold, respectively (supplemental Fig. S4C). These findings demonstrate that CBC promotes the presence of P-TEFb at the HIV-1 promoter and coding region of the reporter gene for effective phosphorylation of the RNAPII CTD at Ser2 and efficient transcription elongation along the gene. Moreover, the continuous presence of P-TEFb and CBC with elongating RNAPII (Fig. 3 and supplemental Fig. S4) underscores further their active role in transcription elongation and/or cotranscriptional processes throughout the coding region of genes.
FIGURE 3.
Depletion of CBC leads to accumulation of RNAPII within the body of HIV-1 LTR CAT reporter gene and reduces the levels of Cdk9 and S2-P RNAPII at the promoter and coding region of the gene. ChIP-qPCR analysis was performed at the promoter (Pr) and coding region (Cr) of the HIV-1 LTR CAT gene and at the intergenic region (Ir) of the α-actin gene in HL3T1 cells expressing F.Tat. Cells were treated with the control (white bars) or CBP20 siRNA (black bars), and chromatin was immunoprecipitated with antibodies against the proteins indicated above the graphs. Results are presented as percent of input DNA or relative to the levels of total RNAPII as indicated.
CBC Promotes the Gene Occupancy of P-TEFb and S2-P RNAPII for Transcription Elongation of Endogenous Genes Exhibiting RNAPII Promoter-proximal Pausing
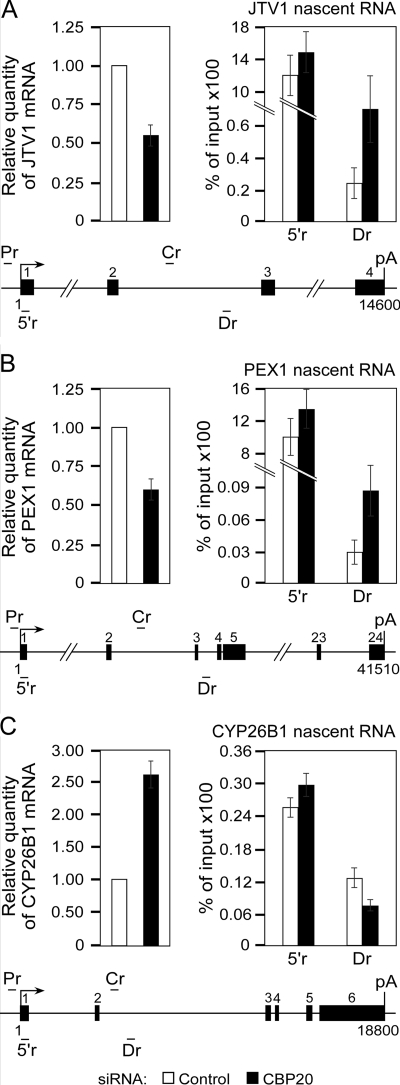
Next, we investigated whether depletion of CBC also affects transcription elongation of endogenous genes (Fig. 4). To this end, we analyzed aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (JTV1) and peroxisomal biogenesis factor 1 (PEX1) genes that were previously demonstrated to be down-regulated 2.4-fold and 3.2-fold, respectively, in CBC-depleted cells (28). We also examined the cytochrome P450, family 26, subfamily B, polypeptide 1 (CYP26B1) gene that was up-regulated 3.2-fold under the same experimental conditions (28). To corroborate these results, we first performed RT-qPCR using total RNA from HeLa cells treated with the control or CBP20 siRNA. Indeed, whereas 3.5-fold reduction of CBP20 mRNA levels (supplemental Fig. S5) yielded a 2-fold and 1.8-fold decrease in mRNA levels of JTV1 and PEX1 genes, respectively, CYP26B1 mRNA levels increased 2.6-fold (Fig. 4, A–C, left). Next, we performed qNARIP analysis of nascent JTV1, PEX1, and CYP26B1 transcripts in HeLa cells expressing the control or CBP20 siRNA. Similar to the qNARIP analyses of the HIV-1 LTR-driven transcription units (Fig. 2, D and E), the levels of initiated transcripts at all examined genes remained largely unchanged, whereas those of elongated JTV1 and PEX1 transcripts increased 3-fold in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 4, A–C, right). In contrast, the amounts of elongated CYP26B1 transcripts decreased 0.56-fold under the same experimental conditions (Fig. 4C, right). The observed accumulation of RNAPII-associated pre-mRNA in the body of the genes down-regulated in CBC-depleted cells again suggests inefficient transcription elongation. Furthermore and in contrast to the situation at the CYP26B1 gene, substantially higher levels of initiated than elongated JTV1 and PEX1 transcripts indicate that JTV1 and PEX1 genes exhibit promoter-proximal pausing of transcriptionally engaged RNAPII (Fig. 4, A–C, right, compare 5′r and Dr).
FIGURE 4.
Depletion of CBC results in accumulation of RNAPII-associated nascent RNA within the body of CBC-down-regulated endogenous genes. Left, relative quantities of JTV1, PEX1, and CYP26B1 mRNAs were determined by RT-qPCR using total RNA samples isolated from HeLa cells that were treated with the control (white bars) or CBP20 siRNA (black bars). Right, qNARIP analysis of JTV1, PEX1, or CYP26B1 nascent transcripts in HeLa cells. Cells were treated with the control (white bars) or CBP20 siRNA (black bars). Nascent mRNA was immunoprecipitated with the RNAPII antibody. Results are presented as percent of input DNA. Two different primer pairs were used for the amplification of transcripts at the 5′ region (5′r) or downstream region (Dr) as indicated below the schematic of each gene. Arrow and pA depict transcription start site and polyadenylation signal, respectively. Exons are numbered and represented with black rectangles. Primer pairs used for the amplification of DNA in ChIP-qPCR analysis (Fig. 5) at the promoter region (Pr) or coding region (Cr) are indicated above each gene schematic. Numbers below each schematic represent nucleotide positions of the gene in respect to the transcription start site.
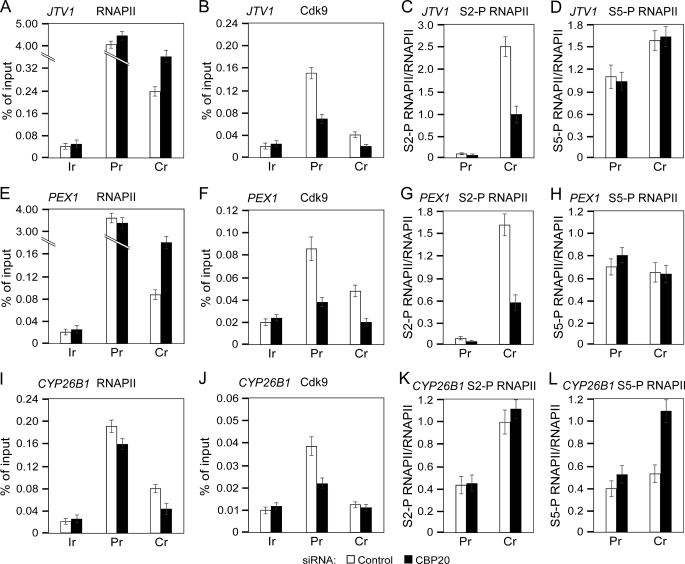
To provide additional evidence for altered transcription elongation at the endogenous genes, we analyzed the mechanism by which depletion of CBC impacts transcription of JTV1, PEX1, and CYP26B1 genes using ChIP-qPCR (Fig. 5). In agreement with the qNARIP results, both genes requiring CBC for their expression contained considerably higher occupancy of RNAPII at the promoter region compared with coding region in cells treated with either siRNA, indicating the occurrence of RNAPII promoter-proximal pausing at these genes (Fig. 5, A and E). Furthermore, both genes displayed the accumulation of RNAPII at their coding regions in response to the depletion of CBC, which together with RT-qPCR and qNARIP results underscore a decrease in RNAPII elongation along these genes (Fig. 5, A and E). Importantly, the occupancy of Cdk9 at the promoter and coding region of both CBC-dependent genes decreased ∼2-fold in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 5, B and F). Critically, upon depleting CBC the levels of S2-P RNAPII decreased 2-fold at the promoter region of both genes when normalized to the occupancy of total RNAPII. This decrease became even more apparent at the coding region, reaching 2.5-fold for JTV1 and 2.7-fold for PEX1 gene (Fig. 5, C and G). In contrast, the levels of S5-P RNAPII relative to the total RNAPII remained unchanged at both regions of the examined genes (Fig. 5, D and H). The ChIP-qPCR results at the up-regulated CYP26B1 gene were very different. They revealed a 0.55-fold decrease in the occupancy of total RNAPII at the coding region in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 5I), suggesting an increase in RNAPII elongation, although the levels of S2-P RNAPII relative to the total RNAPII did not change upon CBC depletion (Fig. 5K). Unexpectedly, the amount of S5-P RNAPII increased 2-fold relative to the total RNAPII at the coding region of the gene in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 5L). Finally, the occupancy of Cdk9 decreased 1.7-fold at the promoter region in CBC-depleted cells compared with the control cells, but could hardly be detected at the coding region of this gene in cells treated with either siRNA (Fig. 5J). Also, levels of S2-P and S5-P RNAPII presented as percent of input DNA for all genes examined are shown in supplemental Fig. S6. CBC was efficiently lowered in cells treated with CBP20 siRNA compared with the control siRNA because the occupancy of CBP80 at the genes decreased 2–3-fold (supplemental Fig. S6, C, F, and I). Collectively, these findings demonstrate that in addition to HIV-1, CBC facilitates the occupancy of P-TEFb at JTV1 and PEX1 genes for effective phosphorylation of the RNAPII CTD at Ser2. That these genes show accumulation of RNAPII and its associated pre-mRNA at their coding regions in response to depletion of CBC suggests a requirement for CBC in promoting transcription elongation of genes displaying RNAPII promoter-proximal pausing. On the contrary, a reversal of these hallmarks at the CYP26B1 gene implicates that CBC and P-TEFb are dispensable for transcription of genes that are up-regulated in CBC-depleted cells.
FIGURE 5.
Depletion of CBC leads to accumulation of RNAPII within the body of CBC-down-regulated endogenous genes and reduces the occupancy of Cdk9 and S2-P RNAPII at the promoter and coding region of these genes. ChIP-qPCR analysis was performed at the promoter (Pr) and coding region (Cr) of the indicated endogenous genes and at the intergenic region (Ir) of the α-actin gene in HeLa cells expressing the control (white bars) or CBP20 siRNA (black bars). Chromatin was immunoprecipitated with antibodies against the proteins indicated above the graphs. Results are presented as percent of input DNA or relative to the levels of total RNAPII as indicated.
Depletion of CBC Antagonizes the Promotion of Alternative Splicing by P-TEFb
Several reports have revealed that CBC as well as P-TEFb influence cotranscriptional processes (8–11, 18–20). Therefore, we next addressed the impact of CBC on P-TEFb-dependent alternative splicing (17) (Fig. 6). In these experiments, we investigated alternative splicing of pSVED-A Tot minigene containing a weak exon (EDA) with a suboptimal 3′ splice site (25). The alternative splicing of nascent transcript of the minigene yields two mRNAs that include or lack EDA (Fig. 6A). The inclusion of this exon into the processed transcript depends on the binding of SR splicing factor 2/alternative splicing factor (SF2/ASF) to its exonic splicing enhancer in the pre-mRNA and on the recruitment of SF2/ASF to the transcription apparatus via S2-P RNAPII (26, 29). Moreover and importantly for this work, derepression of P-TEFb upon depletion of 7SK MePCE, a protein critical for the assembly of core 7SK snRNP, led to increased levels of S2-P RNAPII and SF2/ASF at the minigene for promoting the EDA inclusion (19).
FIGURE 6.
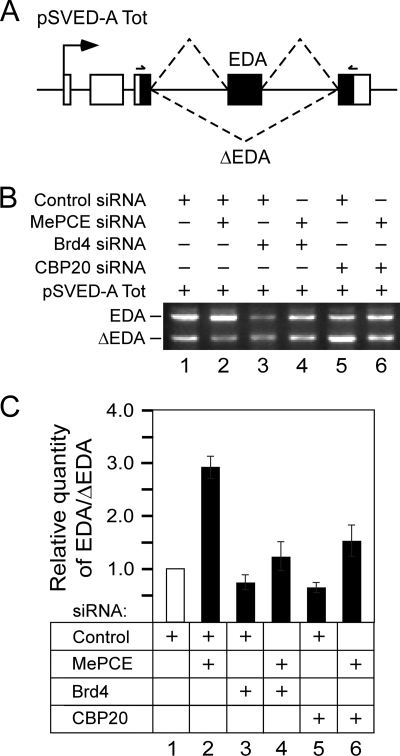
CBC is necessary for P-TEFb-dependent stimulation of alternative EDA exon inclusion. A, schematic of the pSVED-A Tot minigene. Arrow represents the transcription start site from the α-globin promoter. The third exon of the three α-globin exons (white rectangles) contains three fibronectin exons (black rectangles; exons 32–34) with the accompanying introns (solid lines). Dashed lines represent the two possible mature EDA and ΔEDA mRNAs that include or lack the second fibronectin exon named EDA, respectively. The psv5′j/psv3′j primer pair used for RT-PCR analysis is indicated. B, levels of EDA and ΔEDA mRNAs detected by RT-PCR using total RNA isolated from HeLa cells that expressed pSVED-A Tot cassette and were treated with the siRNAs as indicated above the agarose gel. C, ratio between relative quantities of EDA versus ΔEDA mRNA determined by RT-qPCR using total RNA isolated from HeLa cells that expressed pSVED-A Tot cassette and were treated with the siRNAs as indicated below the graph.
The levels of the two alternatively spliced mRNAs were followed by RT-PCR analysis of total RNA from HeLa cells expressing pSVED-A Tot mRNA using primers that annealed to exons adjacent to the EDA (Fig. 6, A and B) or by RT-qPCR analysis employing primer pairs that were specific for the two mRNAs (Fig. 6C). In agreement with the previous report (19), the depletion of MePCE enhanced the inclusion of EDA greatly, whereas the depletion of Brd4, an integral component of the transcriptionally active P-TEFb (22), had the opposite effect (Fig. 6B, lanes 1–3). In fact, in MePCE siRNA- and Brd4 siRNA-treated cells, the ratio between EDA-containing and EDA-less mRNA (EDA/ΔEDA) increased 3-fold and decreased 0.75-fold, respectively, compared with the control siRNA (Fig. 6C, bars 1–3). Significantly, akin to the depletion of Brd4, the knockdown of CBC also resulted in a 0.65-fold decrease in EDA/ΔEDA (Fig. 6B, lane 5, and Fig. 4C, bar 5). Most importantly, enhanced inclusion of EDA by derepressed P-TEFb in cells treated with MePCE siRNA was counteracted when the cells were treated with Brd4 or CBP20 siRNA in addition to MePCE siRNA (Fig. 6B, lanes 4 and 6). In this case, RT-qPCR revealed that EDA/ΔEDA increased only 1.25-fold or 1.5-fold, respectively, compared with the control siRNA (Fig. 6C, bars 4 and 6). The knockdown of all targeted mRNAs was efficient as evident from relative quantities of mRNA levels in cells treated with the control and CBP20, Brd4, or MePCE siRNAs (supplemental Fig. S7, A–C). Noteworthy, the relative quantity of pSVED-A Tot mRNA was not altered in cells treated with CBP20 siRNA (supplemental Fig. S8), indicating that the depletion of CBC does not affect the minigene transcription. Taken together, these results indicate that depletion of CBC counteracts P-TEFb-dependent inclusion of EDA into alternatively spliced mRNA.
CBC Facilitates Recruitment of P-TEFb for the Stimulation of Alternative Splicing via S2-P RNAPII and SF2/ASF
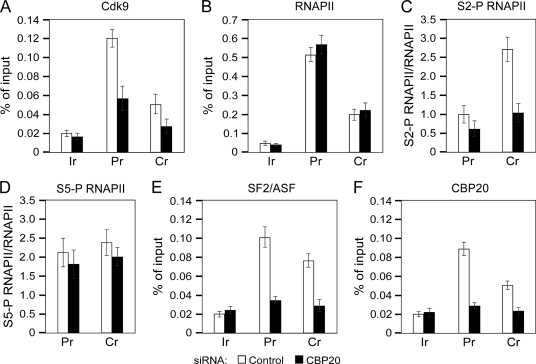
Finally, we sought to dissect a mechanism by which depletion of CBC antagonized P-TEFb-dependent alternative splicing. For this purpose, we performed ChIP-qPCR analysis at the promoter and coding region of the pSVED-A Tot minigene in HeLa cells treated with the control or CBP20 siRNA. Of note, the occupancy of Cdk9 at the promoter and coding region of the minigene decreased 2.1-fold and 1.7-fold, respectively, in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 7A). This drop coincided with a decrease in S2-P RNAPII levels, which were lowered 1.6-fold and 2.5-fold at both gene regions, respectively, when normalized to the levels of total RNAPII (Fig. 7C). In contrast, the relative levels of S5-P RNAPII remained largely unchanged (Fig. 7D). Levels of S2-P and S5-P RNAPII presented as percent of input DNA are shown in supplemental Fig. S9. Corroborating the fact that the amounts of pSVED-A Tot transcripts were not altered in cells treated with CBP20 siRNA compared with the control siRNA (supplemental Fig. S8), the occupancy of total RNAPII at both minigene regions did not change either (Fig. 7B), suggesting that CBC depletion does not affect RNAPII elongation along the minigene. Because P-TEFb promotes alternative splicing via SF2/ASF (19), we examined its levels at the pSVED-A Tot minigene. Importantly, in CBP20-depleted cells, the SF2/ASF occupancy at the promoter and coding region decreased 2.9-fold and 2.7-fold, respectively, compared with the control siRNA-treated cells (Fig. 7E). The knockdown of CBP20 was substantial at both regions of the minigene as its levels decreased 3.5-fold and 2.5-fold, respectively, in cells treated with CBP20 siRNA compared with the control siRNA (Fig. 7F). In conclusion, depletion of CBC reduces the levels of P-TEFb at the promoter and coding region of the pSVED-A Tot minigene. Accordingly, the RNAPII CTD does not undergo efficient phosphorylation at Ser2, precluding a robust occupancy of SF2/ASF at the minigene regions, yielding lower levels of EDA-containing transcripts.
FIGURE 7.
Depletion of CBC reduces levels of Cdk9, S2-P RNAPII, and SF2/ASF at the pSVED-A Tot minigene. ChIP-qPCR analysis was performed at the promoter (Pr) and coding region (Cr) of pSVED-A Tot minigene and at the intergenic region (Ir) of the α-actin gene in HeLa cells. Cells were treated with the control (white bars) or CBP20 siRNA (black bars), and chromatin was immunoprecipitated with antibodies against the indicated proteins above the graphs. Results are presented as percent of input DNA or relative to RNAPII levels as indicated.
DISCUSSION
In this study, we provide evidence for a novel mechanism that connects pre-mRNA capping with two downstream transcriptional events, RNAPII elongation and alternative splicing. This coupling is mediated by CBC, which ensures optimal phosphorylation of the RNAPII CTD at Ser2 by interacting with P-TEFb and promoting its presence at target genes. Furthermore, our findings demonstrate a differential response of individual genes to reduced levels of P-TEFb and S2-P RNAPII at their loci in cells depleted of CBC. On one hand, the efficiency of transcription elongation is altered. On the other hand, alternative splicing but not the synthesis of transcript itself becomes affected. Thus, this work establishes a new role for CBC in gene regulation and suggests that different amounts of P-TEFb at target genes can modulate mRNA maturation without altering RNAPII elongation.
Given the interconnected nature of mRNA genesis and the involvement of CBC in multiple cotranscriptional and post-transcriptional gene expression pathways (1–3, 8–12), depletion of CBC may affect nearly every stage of the life of mRNA. For example, a decrease in the steady-state levels of HIV-1 Tat-regulated CAT or endogenous JTV1 and PEX1 mRNAs in CBC-depleted cells could stem from enhanced transcript degradation. However, by employing a combination of transcription-related ChIP and qNARIP techniques, our work is consistent with a conclusion that the synthesis of these transcripts, which precedes their potential degradation, is affected. The effects of CBC depletion on transcription were observed most directly by qNARIP assay, which detects nascent mRNA as it is being generated by RNAPII and not the steady-state level of mature mRNA. These qNARIP findings are complemented by ChIP results, which also reveal that CBC cooperates with P-TEFb to facilitate elongation phase of transcription. That CBC and P-TEFb interact with RNAPII engaged in transcription provides an additional biochemical evidence for the role of CBC in the synthesis of mRNA. Noteworthy, only a modest fraction of genes is expressed differently upon the knockdown of CBP80 (28), indicating that CBC affects transcription in a gene-specific manner. The study by Narita et al. also argues against the global role of CBC in ensuring transcript stability (28).
We first reveal that CBC is essential for HIV-1 Tat transactivation, a model system for stimulating RNAPII elongation by P-TEFb (14). Taking into account that treatment with CBP20 siRNA decreases mRNA levels of this gene but fails to affect the occupancy of initiated transcripts or RNAPII at the promoter, the observed accumulation of RNAPII and its associated elongated transcripts in the body of the gene indicates inefficient RNAPII elongation. In support of this notion, decreased levels of CBC at the promoter and coding region of Tat-activated gene lower the occupancy of P-TEFb significantly, leading to reduced levels of S2-P but not S5-P RNAPII. Moreover, that RNAPII accumulation within genes could result from hampered transcription elongation has been reported. For example, the human SWI/SNF subunit Brm induces accumulation of RNAPII on the variant region of CD44 gene by decreasing its elongation rate (30), and depletion of SC35 provokes RNAPII accumulation in the body of certain genes with a concomitant reduction in P-TEFb occupancy and levels of S2-P RNAPII (27). Besides the defective P-TEFb recruitment, the hampered transcription elongation along HIV-1 Tat-activated gene could also be attributed to suboptimal recruitment and performance of several positive transcription elongation factors, including DSIF, Tat-SF1, Paf1C, Spt6, and FACT, all of which act in cooperation downstream of P-TEFb (17, 31, 32). Overall, we propose that tethering of P-TEFb to the promoter-proximal region can be contingent upon pre-mRNA cap-bound CBC, which in effect enables productive RNAPII elongation. Therefore, cotranscriptional acquisition of CBC via the pre-mRNA cap structure is beneficial not only for transcript splicing, 3′ end processing, export, and translation, but also for its elongation. Noteworthy, similar yet distinct mechanisms linking transcription initiation or capping to elongation operate in other organisms. For example, in yeast Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe, a P-TEFb orthologue Bur1/Bur2 and P-TEFb complexed with the mRNA CE Pcm1, respectively, are recruited directly to S5-P RNAPII after promoter escape (33–36). It remains possible that P-TEFb could bind S5-P RNAPII directly or via CE in mammalian cells as well. Considering that neither S5-P RNAPII nor capping or CE activity/recruitment is affected by the depletion of CBC, our findings suggest that the interaction of P-TEFb with CBC is critical for coordinating capping with transcription elongation in human cells.
In addition, we report here that the necessity of CBC for transcription elongation is not limited to HIV-1 Tat transactivation. By examining JTV1 and PEX1, two endogenous genes that are down-regulated in response to the depletion of CBC (28), our results demonstrate that they also possess several hallmarks of inefficient transcription elongation after CBP20 knockdown. As is the case with the Tat-activated gene, depletion of CBC precludes optimal gene occupancy of P-TEFb and S2-P RNAPII. Consistently, we find accumulation of RNAPII and its associated elongated transcripts within the body of these genes, indicating transcription elongation defect. Of note, our qNARIP and ChIP analyses of gene bodies employ primer pairs that do not encompass splice sites or termination signals, known sites of RNAPII accumulation (37, 38). In addition, positions of the primers specific to the body of each gene differ between nascent RNA and DNA, suggesting that the observed accumulation of RNAPII and its associated transcripts is not a local phenomenon. Importantly, we note that JTV1 and PEX1 genes contain disproportionally high levels of RNAPII at their 5′ ends, implying that CBC and P-TEFb may be especially relevant for the expression of genes exhibiting promoter-proximal pausing of RNAPII. In contrast, the CYP26B1 gene, which belongs to a subset of CBC-regulated genes of which mRNA levels increase upon CBC depletion, displays a mirror image of the profile of RNAPII and its elongated transcripts, indicating that a reduction of RNAPII in the body of a gene can be compatible with enhanced RNAPII elongation. Taken together, these data highlight further that CBC and P-TEFb regulate gene transcription at the level of RNAPII elongation in a gene-specific manner.
Importantly, the present study is the first to disclose an involvement of CBC in modulating alternative splicing in human cells. A large body of evidence has revealed that transcription and alternative splicing could be connected by two major but not mutually exclusive mechanisms (39). These include the recruitment of splicing factors to the elongating RNAPII and the kinetic coupling, whereby the rate of transcription elongation determines the time in which splice sites are being displayed to the splicing machinery. Despite the requirement of CBC for maintaining the levels of P-TEFb and S2-P RNAPII at the pSVED-A Tot minigene, its depletion does not alter neither the amounts of mRNA produced from pSVED-A Tot nor the occupancy of the total RNAPII at the minigene. These findings are consistent with a previous work demonstrating the necessity of CBC for transcription of only a subset of human genes (28) and argue against the kinetic coupling of transcription and alternative splicing at the pSVED-A Tot minigene. Rather, our results are in agreement with the splicing factors recruitment model. Namely, CBC is critical for the loading of the stimulatory SF2/ASF to the S2-P RNAPII transcribing the minigene. By binding to the exonic splicing enhancer element on a nascent transcript, SF2/ASF could further attract spliceosomal U1 and U2 snRNPs and U2 auxiliary factor heterodimer and in doing so stimulate early steps of the spliceosome assembly (40). These events lead to recognition of weak splice sites, yielding an inclusion of the alternative exon into mRNA. In support of this scenario, a systematic proteomic analysis revealed that SR splicing factors and the components of U1 snRNP, P-TEFb, and CBC bound to the phosphorylated form of RNAPII (26). Given that depletion of CBC drastically affects splicing of a subset of pre-mRNAs (41, 42) and that it regulates cotranscriptional U1 snRNP binding to the cap-proximal 5′ splice site in yeast (8, 9), it remains plausible that CBC could also affect constitutive splicing in metazoans.
In conclusion, our study discloses a novel role of CBC in mediating the coupling of pre-mRNA capping to transcription elongation and alternative splicing through facilitating the tethering of P-TEFb. This mechanism illustrates the intertwined nature of gene expression processes and expands the complexity of P-TEFb recruitment mechanisms to specific genes for efficient transcription and cotranscriptional processing of pre-mRNA. Multiple manners of tethering this crucial transcriptional coactivator to target genes may allow fine tuning and/or regulation of transcription elongation and mRNA maturation at different levels under diverse circumstances.
Supplementary Material
Acknowledgments
We thank Peterlin laboratory members for discussions and Dr. Kalle Saksela for support. We also thank Dr. Elisa Izaurralde for providing the CBP80 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants AI058708 and GM082250 (to B. M. P.). This work was also supported by Academy of Finland FiDiPro programme Grant 1127313 (to Kalle Saksela and B. M. P.) and Academy of Finland Grants 137077 and 140996 (to M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9, Tables S1–S5, and control experiments.
- RNAPII
- RNA polymerase II
- CAT
- chloramphenicol acetyltransferase
- CBC
- cap-binding protein complex
- CBP
- cap-binding protein
- Cdk
- cyclin-dependent kinase
- CE
- capping enzyme
- CTD
- C-terminal domain
- Cyc
- cyclin
- F.
- FLAG-tagged
- MePCE
- γ-methylphosphate-capping enzyme
- P-TEFb
- positive transcription elongation factor b
- qNARIP
- quantitative nascent RNA immunoprecipitation
- qPCR
- quantitative PCR
- S5-P RNAPII
- phosphorylation of RNAPII CTD at Ser5
- SR
- Ser/Arg-rich
- WCE
- whole cell extract
- S2-P RNAPII
- phosphorylation of RNAPII CTD at Ser2
- SF2/ASF
- splicing factor 2/alternative splicing factor
- ChIP
- chromatin immunoprecipitation
- LTR
- long terminal repeat
- JTV1
- aminoacyl tRNA synthetase complex-interacting multifunctional protein 2; peroxisomal biogenesis factor 1 (PEX1)
- CYP26B1
- cytochrome P450, family 26, subfamily B, polypeptide 1.
REFERENCES
- 1. Perales R., Bentley D. (2009) Mol. Cell 36, 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buratowski S. (2009) Mol. Cell 36, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egloff S., Murphy S. (2008) Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 4. Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shuman S. (2001) Prog. Nucleic Acids Res. Mol. Biol. 66, 1–40 [DOI] [PubMed] [Google Scholar]
- 6. Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I. W. (1994) Cell 78, 657–668 [DOI] [PubMed] [Google Scholar]
- 7. Kataoka N., Ohno M., Moda I., Shimura Y. (1995) Nucleic Acids Res. 23, 3638–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Görnemann J., Kotovic K. M., Hujer K., Neugebauer K. M. (2005) Mol. Cell 19, 53–63 [DOI] [PubMed] [Google Scholar]
- 9. Lewis J. D., Izaurralde E. (1997) Eur. J. Biochem. 247, 461–469 [DOI] [PubMed] [Google Scholar]
- 10. Raczynska K. D., Simpson C. G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J. W., Jarmolowski A. (2010) Nucleic Acids Res. 38, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flaherty S. M., Fortes P., Izaurralde E., Mattaj I. W., Gilmartin G. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11893–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hosoda N., Kim Y. K., Lejeune F., Maquat L. E. (2005) Nat. Struct. Mol. Biol. 12, 893–901 [DOI] [PubMed] [Google Scholar]
- 13. Nechaev S., Adelman K. (2010) Biochim. Biophys. Acta 1809, 43–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peterlin B. M., Price D. H. (2006) Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 15. Wei P., Garber M. E., Fang S. M., Fischer W. H., Jones K. A. (1998) Cell 92, 451–462 [DOI] [PubMed] [Google Scholar]
- 16. Brès V., Yoh S. M., Jones K. A. (2008) Curr. Opin. Cell Biol. 20, 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lenasi T., Barboric M. (2010) RNA Biol. 7, 145–150 [DOI] [PubMed] [Google Scholar]
- 18. Ahn S. H., Kim M., Buratowski S. (2004) Mol. Cell 13, 67–76 [DOI] [PubMed] [Google Scholar]
- 19. Barboric M., Lenasi T., Chen H., Johansen E. B., Guo S., Peterlin B. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7798–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ni Z., Schwartz B. E., Werner J., Suarez J. R., Lis J. T. (2004) Mol. Cell 13, 55–65 [DOI] [PubMed] [Google Scholar]
- 21. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., Young R. A. (2010) Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Q., Yik J. H. (2006) Microbiol. Mol. Biol. Rev. 70, 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Felber B. K., Pavlakis G. N. (1988) Science 239, 184–187 [DOI] [PubMed] [Google Scholar]
- 24. Yik J. H., Chen R., Nishimura R., Jennings J. L., Link A. J., Zhou Q. (2003) Mol Cell 12, 971–982 [DOI] [PubMed] [Google Scholar]
- 25. Caputi M., Casari G., Guenzi S., Tagliabue R., Sidoli A., Melo C. A., Baralle F. E. (1994) Nucleic Acids Res. 22, 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das R., Yu J., Zhang Z., Gygi M. P., Krainer A. R., Gygi S. P., Reed R. (2007) Mol. Cell 26, 867–881 [DOI] [PubMed] [Google Scholar]
- 27. Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X. D. (2008) Nat. Struct. Mol. Biol. 15, 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narita T., Yung T. M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. (2007) Mol. Cell 26, 349–365 [DOI] [PubMed] [Google Scholar]
- 29. Cramer P., Cáceres J. F., Cazalla D., Kadener S., Muro A. F., Baralle F. E., Kornblihtt A. R. (1999) Mol. Cell 4, 251–258 [DOI] [PubMed] [Google Scholar]
- 30. Batsché E., Yaniv M., Muchardt C. (2006) Nat. Struct. Mol. Biol. 13, 22–29 [DOI] [PubMed] [Google Scholar]
- 31. Chen Y., Yamaguchi Y., Tsugeno Y., Yamamoto J., Yamada T., Nakamura M., Hisatake K., Handa H. (2009) Genes Dev. 23, 2765–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoh S. M., Lucas J. S., Jones K. A. (2008) Genes Dev. 22, 3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guiguen A., Soutourina J., Dewez M., Tafforeau L., Dieu M., Raes M., Vandenhaute J., Werner M., Hermand D. (2007) EMBO J. 26, 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pei Y., Du H., Singer J., Stamour C., Granitto S., Shuman S., Fisher R. P. (2006) Mol. Cell. Biol. 26, 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qiu H., Hu C., Hinnebusch A. G. (2009) Mol. Cell 33, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Viladevall L., St. Amour C. V., Rosebrock A., Schneider S., Zhang C., Allen J. J., Shokat K. M., Schwer B., Leatherwood J. K., Fisher R. P. (2009) Mol. Cell 33, 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alexander R. D., Innocente S. A., Barrass J. D., Beggs J. D. (2010) Mol. Cell 40, 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Glover-Cutter K., Kim S., Espinosa J., Bentley D. L. (2008) Nat. Struct. Mol. Biol. 15, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kornblihtt A. R. (2007) Adv. Exp. Med. Biol. 623, 175–189 [DOI] [PubMed] [Google Scholar]
- 40. Chen M., Manley J. L. (2009) Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Colot H. V., Stutz F., Rosbash M. (1996) Genes Dev. 10, 1699–1708 [DOI] [PubMed] [Google Scholar]
- 42. Fortes P., Kufel J., Fornerod M., Polycarpou-Schwarz M., Lafontaine D., Tollervey D., Mattaj I. W. (1999) Mol. Cell. Biol. 19, 6543–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.