Abstract
In mammals, iron regulatory proteins (IRPs) 1 and 2 posttranscriptionally regulate expression of genes involved in iron metabolism, including transferrin receptor 1, the ferritin (Ft) H and L subunits, and ferroportin by binding mRNA motifs called iron responsive elements (IREs). IRP1 is a bifunctional protein that mostly exists in a non-IRE-binding, [4Fe-4S] cluster aconitase form, whereas IRP2, which does not assemble an Fe-S cluster, spontaneously binds IREs. Although both IRPs fulfill a trans-regulatory function, only mice lacking IRP2 misregulate iron metabolism. NO stimulates the IRE-binding activity of IRP1 by targeting its Fe-S cluster. IRP2 has also been reported to sense NO, but the intrinsic function of IRP1 and IRP2 in NO-mediated regulation of cellular iron metabolism is controversial. In this study, we exposed bone marrow macrophages from Irp1−/− and Irp2−/− mice to NO and showed that the generated apo-IRP1 was entirely responsible for the posttranscriptional regulation of transferrin receptor 1, H-Ft, L-Ft, and ferroportin. The powerful action of NO on IRP1 also remedies the defects of iron storage found in IRP2-null bone marrow macrophages by efficiently reducing Ft overexpression. We also found that NO-dependent IRP1 activation, resulting in increased iron uptake and reduced iron sequestration and export, maintains enough intracellular iron to fuel the Fe-S cluster biosynthetic pathway for efficient restoration of the citric acid cycle aconitase in mitochondria. Thus, IRP1 is the dominant sensor and transducer of NO for posttranscriptional regulation of iron metabolism and participates in Fe-S cluster repair after exposure to NO.
Keywords: Cell Metabolism, Iron, Iron Metabolism, Iron-Sulfur Protein, Nitric Oxide, IRP1, Macrophage
Introduction
Iron is a cofactor essential for fundamental metabolic processes and is therefore indispensable for cellular function. On the other hand, iron excess is toxic because of the ability of the metal to catalyze the formation of highly reactive hydroxyl radicals. Hence, cellular iron levels and bioavailability must be tightly controlled. Cellular iron homeostasis is coordinately regulated by iron regulatory protein (IRP)3 1 and 2, which posttranscriptionally modulate the expression of critical iron metabolism genes by interacting with conserved cis-regulatory iron responsive elements (IREs) present in the untranslated region (UTR) of target mRNAs (1). Either of the two IRPs inhibits translation when bound to the single 5′ UTR IRE of the mRNAs encoding, respectively, the H and L subunits of the iron storage protein ferritin (Ft) and the iron exporter ferroportin (Fpn). IRP binding to the multiple IREs within the 3′ UTR of the mRNA encoding the transferrin receptor 1 (TfR1) iron uptake molecule prevents its degradation. The IRE-binding activity of both IRPs responds to cellular iron levels, albeit via distinct mechanisms. Under iron-replete conditions, IRP1 assembles an iron-sulfur [4Fe-4S] cluster that precludes IRE binding, and the holo-protein functions as an aconitase. IRP1 is thus bifunctional. IRP2, which is not able to ligate an iron-sulfur cluster, is targeted for proteasomal degradation via the E3 ubiquitin ligase complex containing FBXL5 protein (2, 3). When iron and oxygen are high, FBXL5 assembles a di-iron-oxygen center in the hemerythrin domain, resulting in protein stabilization. The SKP1-CUL1-FBXL5 ubiquitin ligase complex then induces iron-dependent ubiquitination and degradation of IRP2. When iron is scarce, IRP1 loses its Fe-S cluster and aconitase activity and acquires its IRE-binding conformation. At the same time, FBXL5, which lacks its iron-oxygen center in the hemerythrin domain, is degraded, resulting in IRP2 stabilization. As a result, the translation of the H- and L-Ft and Fpn mRNAs is repressed, and the TfR1 mRNA is stabilized in iron-deficient cells, and vice versa when iron is plentiful. The opposite regulation of iron storage into Ft and export via Fpn on the one hand and TfR1-mediated iron uptake on the other hand maintains intracellular iron availability while preventing its excess.
The concerted regulation of both IRP1 and IRP2 by cellular iron levels raises the question of why cells possess two IRPs. Mice with targeted deletion of IRP2 display microcytic anemia and iron mismanagement in most tissues, showing that IRP2 is critical for maintaining the iron balance in vivo (4, 5). By contrast, IRP1-null mice exhibit no spontaneous iron misregulation (5, 6). This could be explained in part by the fact that IRP1 exists mostly in its non-IRE binding, [4Fe-4S] aconitase form in mouse tissues under standard laboratory conditions (6). Challenging cells to convert IRP1 to the cluster-free apo-form could therefore help to elucidate the specific functions of IRP1 and IRP2. The switch from the holo- to the apo-form of IRP1 can be rapidly driven by free radical molecules such as NO (7), a vital signaling and effector molecule whose interaction with the iron metabolism is well recognized (8, 9). NO is produced by NO synthases. Once released, it can directly target the [4Fe-4S] cluster of IRP1, promoting its gradual disassembly and complete removal, thus favoring the IRE-binding conformation of the protein (7, 10). The effect of NO on IRP2 has also been investigated, but seemingly contradictory conclusions have been reached, ranging from lack of regulation by NO (11–13) to NO-dependent IRP2 degradation (14–17) or activation (18). Moreover, both endogenous and exogenous NO has been shown to either increase (14, 19) or decrease (11, 20–22) Ft levels. Discrepant data on TfR1 regulation by NO have also been reported (15, 21–23). As a consequence, the role of the IRP/IRE system and, more particularly, the intrinsic function of IRP1 and IRP2 in NO-mediated regulation of cellular iron metabolism have remained elusive.
In this report, we took advantage of mouse models of IRP deficiency to define the respective role of IRP1 and IRP2 in the regulation of cellular iron metabolism by NO. Primary cultures of bone marrow macrophages (BMMs) lacking either of the two IRPs were exposed to NO, and the impact on cellular iron transport and storage was analyzed. The results show that NO imparts its effects on cellular iron metabolism via IRP1.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant mouse IFN-γ was purchased from R&D Systems (Minneapolis, MN). 5,6-Dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) and LPS from Escherichia coli serotype 055:B5 were from Sigma-Aldrich (Saint Louis, MO). Diethylenetriamine NONOate, L-NG-monomethyl arginine citrate, and N-((3-(aminomethyl)phenyl)methyl)-ethanimidamide dihydrochloride (1400W) were from Cayman Chemical (Ann Arbor, MI).
Mice
The generation of mouse lines carrying truncated Aco1 (herein designated Irp1−/−) or Ireb2 (Irp2−/−) alleles, respectively, has been described (5). These lines were backcrossed to C57BL6 mice for at least 10 generations. IRP1- and IRP2-null animals, respectively, and their corresponding wild-type littermates were obtained from heterozygous intercrosses. Mice were kept under a constant light/dark cycle on a standard diet and were sacrificed by cervical dislocation. All experimental procedures were approved by the Centre National de la Recherche Scientifique, Animal Care Committee (Gif-sur-Yvette, France) and the Third Local Ethical Commission (Warsaw, Poland).
Cell Culture and Treatment
Bone marrow cells were flushed out from the femur of adult mice with ice-cold Hanks' balanced salt solution and were differentiated into mature BMMs by cultivating them in RPMI 1640 + Glutamax (Invitrogen) supplemented with 10% fetal bovine serum (Lonza) and 10% L929 cell-conditioned medium as a source of macrophage colony-stimulating factor. Cells were grown at 37 °C in a 5% CO2 and 21% O2 atmosphere. After 4 days, non-adherent cells were removed by washing with Hanks' balanced salt solution, and the medium was subsequently replaced daily until cells were used. The homogeneity of the BMM population was assessed on day 5 of culture by flow cytometry analysis of the F4/80 macrophage-restricted cell surface antigen and was found to exceed 99%. On day 6, BMMs were exposed to the slowly releasing NO donor diethylenetriamine/NO (DETA/NO) (24) for the time indicated. This NONOate (X[N(O)NO−]) results from the combination of a nucleophilic molecule (X−) with NO and spontaneously releases 2 mol of NO/mol NONOate. The concentration and decomposition rate of DETA/NO in the culture medium were systematically determined by its characteristic absorbance at 252 nm (ϵ = 7640 M−1 × cm−1). The use of decomposed DETA/NO ascertained that the nucleophile molecule per se had no impact on the iron metabolism genes investigated (supplemental Fig. S1). In indicated experiments, BMMs were simultaneously treated with DRB (10 μg/ml) for 18 h to block transcription. In another set of experiments, wild-type and Irp1−/− BMMs were stimulated with IFN-γ and LPS for 18 h to induce the NO synthase 2 (NOS2). When indicated, cells were simultaneously incubated with the NOS inhibitors 1400W and L-NG-monomethyl arginine citrate.
Nitrite Measurement
The nitrite content was determined in cell medium using the Griess reagent (0.5% sulfanilamide and 0.05% N-1-napthylethylenediamine in 45% acetic acid). Nitrite reacts with the Griess reagent to give a red-violet diazo-dye, which is measured spectrophotometrically at 543 nm. Nitrite concentration was calculated from a calibration curve using sodium nitrite.
Preparation of Cell Extracts
Mitochondrial and cytosolic fractions and crude membrane extracts were performed as described previously (25). Protein concentration was determined spectrophotometrically using the Bio-Rad protein assay (Munich, Germany).
Aconitase Activity
Aconitase activity in cytosolic and mitochondrial extracts, respectively, was measured spectrophotometrically by monitoring the disappearance of cis-aconitate at 240 nm (ϵ = 3.6 mm−1 × cm−1) (25). Data are expressed as nmol of substrate consumed per minute per mg of protein.
Immunoblot Analysis
Ferritin expression was analyzed using cytosolic extracts and rabbit antisera raised against, respectively, the recombinant mouse ferritin H and L subunits (kindly provided by Dr. P. Santambrogio, Milan, Italy and Dr. Jeremy Brock, University of Glasgow, UK). Mitochondrial aconitase expression was determined using mitochondrial fractions and a rabbit polyclonal antibody against bovine mitochondrial aconitase (kindly provided by Dr. R. B. Franklin, University of Maryland, Baltimore, MD). Fpn and TfR1 levels in crude membrane extracts were detected using, respectively, a rabbit polyclonal anti-mouse metal transporter protein 1 (MTP-1, Alpha Diagnostics) antibody and a mouse monoclonal antibody against recombinant human TfR1 (Invitrogen). The loading controls β-actin, vinculin, and succinate dehydrogenase, subunit A in protein extracts were detected using murine monoclonal antibodies (from Sigma and Mitosciences, respectively). Protein detection and quantification were done using the Odyssey® infrared imaging system (LI-COR Biosciences).
Electromobility Shift Assay
Bandshift experiments were performed as described previously (26, 27) using 2 μg of cytosolic extract and a molar excess of [α-33P]UTP-labeled H-ferritin IRE in vitro transcribed from plasmid pSPT-fer (kindly provided by Dr. L. C. Kühn, Institut Suisse de Recherche Experimentale sur le Cancer, Switzerland). IRE-IRP complexes were resolved in 6% non-denaturing polyacrylamide gel. Samples were treated in parallel with 2% 2-mercaptoethanol prior the addition of the 33P-labeled IRE probe to fully activate IRP1 IRE-binding activity.
RNA Extraction and Real-Time Quantitative PCR
Total RNA was extracted by using the TRIzol reagent (Invitrogen). 1 μg of total RNA was reverse-transcribed using random hexamers and the Moloney murine leukemia virus reverse transcriptase (Promega). Real-time quantitative PCR was performed using the Roche Light Cycler system and the fast DNA Master SYBR Green I kit (Roche) together with the following primers: L-Ft, 5′-CGG AGG GTC AAC ATG CTA TAA-3′ (forward) and 5′-AAG AGA CGG TGC AGA CTG GT-3′ (reverse); H-Ft, 5′-GCT GAA TGC AAT GGA GTG TG-3′ (forward) and 5′-CAG GGT GTG CTT GTC AAA GA-3′ (reverse); TfR1, 5′-TCG CTT ATA TTG GGC AGA CC-3′ (forward) and 5′-CCA TGT TTT GAC CAA TGC TG-3′ (reverse); Fpn, 5′-GTT TGC AGG AGT CAT TGC TGC TA-3′ (forward) and 5′-TTA CAT TTT CTT GCA GCA ACT GTG T-3′ (reverse); NOS2, 5′-GCA ACT ACT GCT GGT GGT GA-3′ (forward) and 5′-GTT CGT CCC CTT CTC CTG TT-3′ (reverse); 18 S, 5′-CTG AGA AAC GGC TAC CAC ATC-3′ and 5′-CGC TCC CAA GAT CCA ACT AC-3′ (reverse). Data were analyzed with the Light Cycler 3.5 software. mRNA expression was standardized to 18 S ribosomal RNA levels. Identical data were obtained when using vimentin or actin mRNAs as standard.
Statistical Analysis
All experiments were performed at least in triplicate, and error bars indicate standard deviation. Statistical analysis was performed using a two-tailed Student's t test, with a p value of < 0.05 being considered statistically significant.
RESULTS
Effect of NO on IRP1 and IRP2 Activity in BMMs
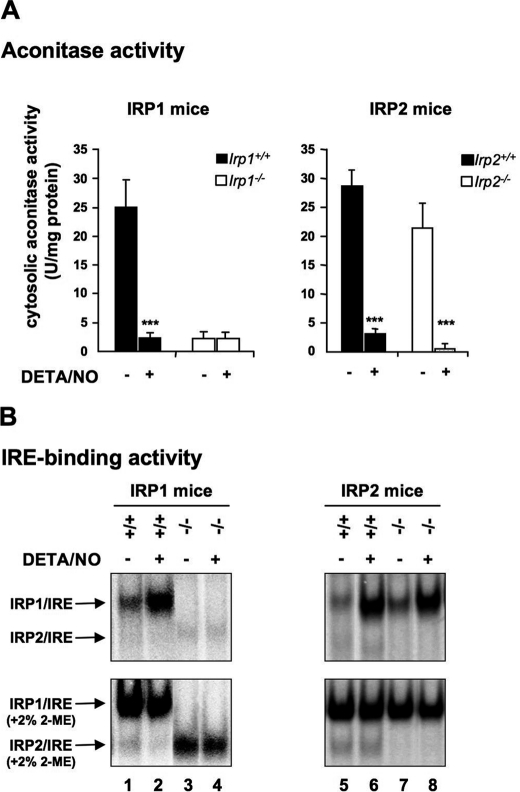
To delineate the respective contribution of IRP1 and IRP2 to the regulation of iron metabolism in response to NO, we first analyzed the modulation of IRP activity by NO in BMMs derived from Irp1−/− and Irp2−/− mice and their corresponding wild-type littermates. BMMs were exposed to a pure NO donor, DETA/NO. This compound has been shown to enhance IRP1 activity as efficiently as NO produced endogenously from NOS2 stimulation by, e.g., LPS and IFN-γ (28). Importantly, the use of an exogenous source of NO avoids potential confounding effects because of immunological stimuli (12). The analysis of aconitase and IRE-binding activities in cytosolic extracts from wild-type and IRP2-null BMMs reveals a clear switch from the aconitase to IRE-binding form of IRP1 in response to NO (Fig. 1, A and B, upper panels, lanes 2, 6, and 8). A dose response study shows that 250 μm DETA/NO suffice to fully activate IRP1 IRE-binding activity (supplemental Fig. S2). IRP1-null cells lack both activities, as expected (Fig. 1A, left panel, and B, lanes 3 and 4). Of note, we observed a slightly increased IRE-binding activity of IRP2 in Irp1−/− BMMs compared with Irp1+/+ (Fig. 1B, lanes 3 and 4 versus lanes 1 and 2) that becomes evident in the presence of 2-mercaptoethanol. Increased IRP2 activity in IRP1-null BMMs is reminiscent of the compensatory gain of IRP2 function in tissues of Irp1−/− mice (29, 30). By contrast, the lack of IRP2 has no major impact on the IRE binding of IRP1 under basal conditions or in response to NO (Fig. 1B, right panels). Importantly, both spontaneous and 2-mercaptoethanol-induced IRP2 activity remain largely insensitive to NO in wild-type and Irp1−/− cells (Fig. 1B, lanes 1–6). Taken together, these data show that NO selectively stimulates the IRE-binding activity of IRP1 in BMMs.
FIGURE 1.
IRP regulation in response to NO. BMMs derived from Irp1−/− and Irp2−/− mice and their corresponding wild-type littermates were exposed to 250 μm DETA/NO for 18 h. Cells were harvested and cytosolic extracts were used to determine IRP1 aconitase activity (mean ± S.D., n = 3) (A) and IRP IRE-binding activity in the presence (B, lower panel) or absence (upper panel) of 2% 2-mercaptoethanol. The experiments were performed at least three times. A representative autoradiogram is shown in B. ***, p < 0.001, Student's t test).
TfR1 Regulation in Response to NO Operates through Selective Activation of IRP1
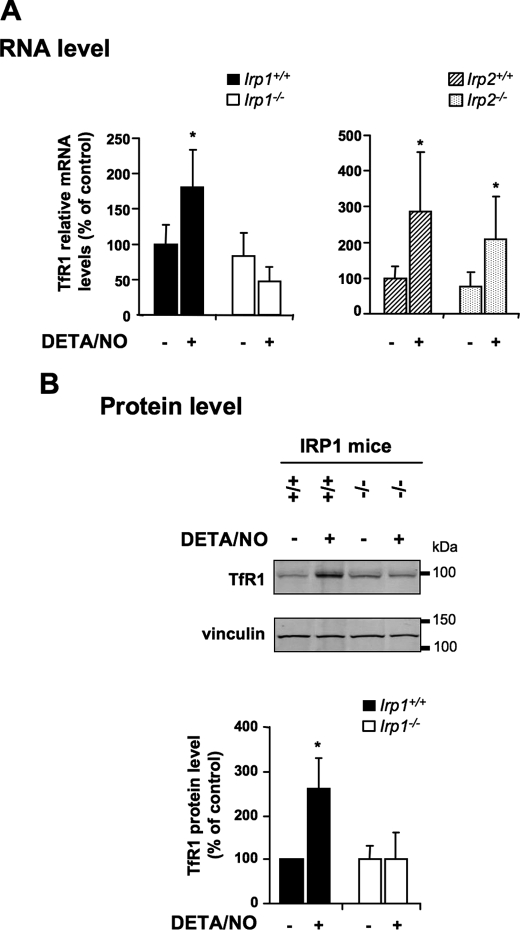
IRP binding to the 3′ UTR IREs of the TfR1 mRNA decreases its turnover. To determine the role of IRP1 and IRP2, respectively, in the regulation of IRP-target genes by NO, we analyzed TfR1 expression in wild-type and IRP-null BMMs exposed to DETA/NO. Real-time quantitative PCR shows that a 18-h exposure to 250 μm DETA/NO elicits a 2- to 3-fold increase in TfR1 mRNA levels in both wild-type and IRP2-null BMMs (Fig. 2A). By contrast, IRP1 deficiency abolishes TfR1 mRNA up-regulation in response to NO (Fig. 2A). Western blot analysis of membrane extracts shows that the 2- to 3-fold up-regulation of TfR1 mRNA expression in Irp1+/+ BMMs treated with NO (Fig. 2A) is associated with a commensurate (∼2.6-fold) increase at the protein level (B). Reflecting the lack of TfR1 mRNA up-regulation in IRP1 deficiency (Fig. 2A), IRP1-null BMMs fail to up-regulate TfR1 protein levels in response to NO (B). Therefore, TfR1 up-regulation by NO requires IRP1, whereas IRP2 is dispensable.
FIGURE 2.
Effect of IRP1 and IRP2 deficiency on TfR1 regulation by NO. IRP1- and IRP2-null BMMs and their corresponding wild-type counterparts were exposed to 250 μm DETA/NO for 18 h. A, real-time quantitative PCR analysis of TfR1 mRNA expression. The histograms display TfR1 mRNA levels (mean ± S.D., n = 3) as a percentage of untreated wild-type cells after normalization to 18 S ribosomal RNA levels. B, Western blot analysis of TfR1 protein levels in membrane fractions. A representative immunoblot is shown (upper panel). Vinculin was used as a loading control. The histograms show TfR1 protein levels (mean ± S.D., n = 3) as a percentage of untreated wild-type cells after normalization to vinculin *, p < 0.05, Student's t test.
Role of IRP1 in Ft and Fpn Regulation by NO
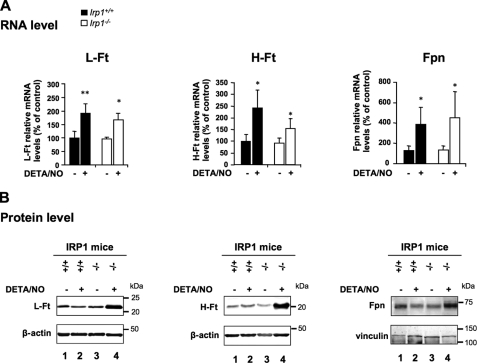
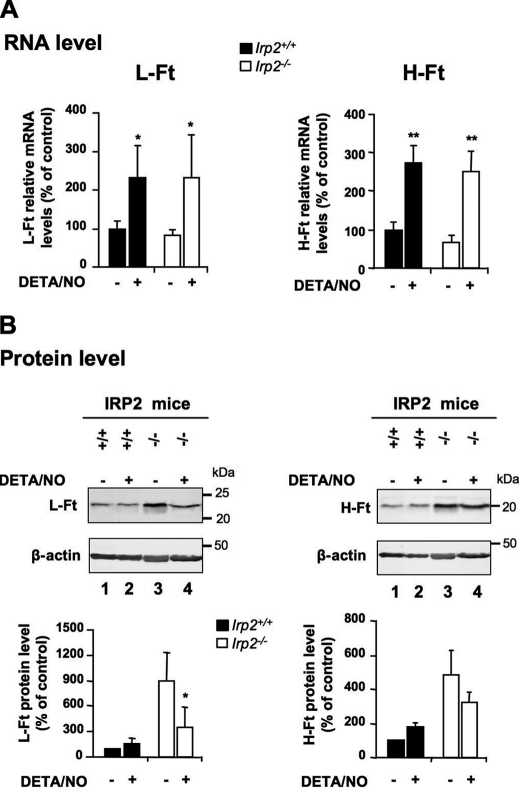
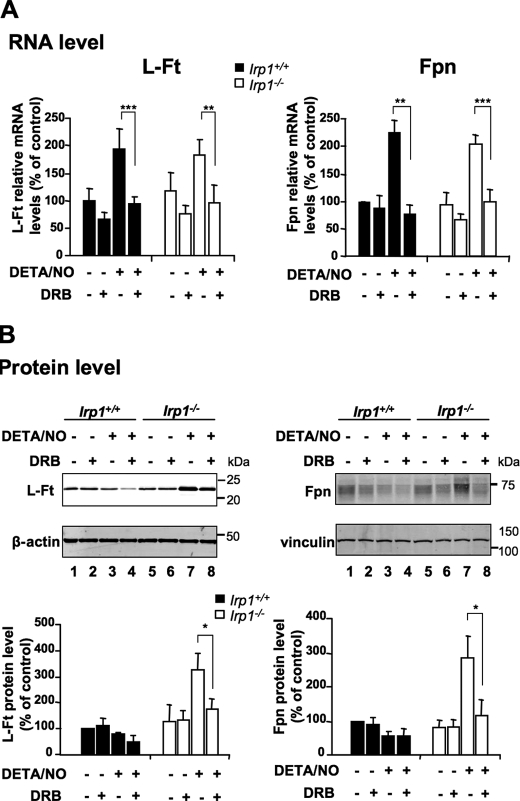
To further explore each IRP unique function in the modulation of the IRP regulon by NO in BMMs, we analyzed the expression of Ft and Fpn, which are translationally silenced by IRPs. NO treatment was found to up-regulate the H-Ft, L-Ft, and Fpn mRNAs (Figs. 3A and 4A). Importantly, neither IRP1 nor IRP2 deficiency alters the up-regulation. Despite up-regulation at the mRNA level, NO does not increase the protein levels of H-Ft, L- Ft, and Fpn in wild-type BMMs, as assessed by Western blot analysis (Fig. 3B, lane 2 versus lane 1). The lack of Ft and Fpn protein up-regulation by NO in the context of increased mRNA expression (Fig. 3) could be explained by translational repression of the Ft and Fpn mRNAs resulting from the concomitant increase in IRP1 IRE-binding activity (Fig. 1). Supporting this hypothesis, IRP1-deficient cells respond to NO by increasing the expression of H-Ft, L-Ft, and Fpn at both mRNA (Fig. 3A) and protein (B, lane 4 versus lane 2) levels, regardless of the dose of DETA/NO used (supplemental Fig. S3). In addition, BMMs were stimulated with IFN-γ and LPS to produce endogenous NO. As shown in supplemental Fig. S4D, Irp1−/− BMMs also respond to a physiological source of NO by increasing both H- and L-Ft levels. This effect strictly depends on endogenous NO, as confirmed by the use of NOS inhibitors (supplemental Fig. S4, A, B and D, compare lanes 5 and 6). In contrast, NO-producing wild-type BMMs maintain a low L-Ft level by increasing IRE-binding activity of IRP1 (supplemental Fig. S4, C and D). Interestingly, in IRP2-null BMMs that display increased H- and L-Ft protein expression under basal conditions (Fig. 4B, lane 3 versus lane 1), NO-treatment prevents H-Ft protein up-regulation and even triggers a nearly 60% decrease in L-Ft protein levels (Fig. 4B, lane 4 versus lane 3) despite a marked increase in the expression of the corresponding mRNAs (Fig. 4A). Indeed, the ∼9-fold excess of L-Ft protein resulting from IRP2 deficiency (Fig. 4B, lane 3 versus lane 1) was reduced to about 3-fold after NO exposure (Fig. 4B, lane 4 versus lanes 1 and 2), showing that IRP1 activation by NO counteracts IRP2 deficiency and tends to restore the iron storage capacity of the cell. Treatment of BMMs with the DRB transcriptional inhibitor abrogates L-Ft and Fpn mRNA up-regulation by NO both in Irp1+/+ and Irp1−/− cells (Fig. 5A), showing that NO enhances L-Ft and Fpn mRNA expression transcriptionally. In IRP1-null cells, L-Ft and Fpn protein levels mirror the expression of their corresponding mRNAs and are no longer increased by NO in the presence of DRB (Fig. 5B, lane 8 versus lane 7), reflecting the sole contribution of transcriptional regulation by NO. In Irp1+/+ cells, the lack of transcriptional stimulation of L-Ft seems to potentiate the inhibitory effect of NO-dependent activation of IRP1 on L-Ft protein synthesis (Fig. 5B, lane 4 versus lane 3). Altogether, these data show that NO enhances Ft and Fpn transcription in BMMs but that parallel stimulation of IRP1 IRE-binding activity prevents a rise at the protein level. In conclusion, although IRP2 is important to set the basal expression of the IRP regulon, IRP1 appears to be the key player in the fine tuning of IRP-target genes in response to NO.
FIGURE 3.
Impact of IRP1 deficiency on NO-mediated regulation of Ft and Fpn. Wild-type and IRP1-null BMMs were exposed to 250 μm DETA/NO for 18 h. A, L-Ft, H-Ft, and Fpn mRNA levels were assayed by real-time quantitative PCR. The histograms display mRNA expression (mean ± S.D., n = 3) as a percentage of untreated wild-type cells after calibration to 18 S ribosomal RNA levels. *, p < 0.05, Student's t test. B, cytosolic levels of L- and H-Ft were analyzed by Western blotting. β-actin was used as a loading control. Fpn expression was analyzed using membrane fractions and vinculin as a loading control. The experiments were performed at least three times, and representative immunoblots are shown.
FIGURE 4.
NO regulation of L- and H-Ft in IRP2-deficient BMMs. Wild-type and IRP2-null BMMs were exposed to 250 μm DETA/NO for 18 h. A, L- and H-Ft mRNA levels were analyzed by real-time quantitative PCR. The histograms display mRNA expression (mean ± S.D., n = 3) as a percentage of untreated wild-type cells after normalization to 18 S ribosomal RNA levels. B, Western blot analysis of L-Ft (left panels) and H-Ft (right panels) levels in cytosolic extracts. Representative immunoblots are shown. β-actin was used as a loading control. The histograms show L- and H-Ft expression (mean ± S.D., n = 3) as a percentage of untreated wild-type cells after normalization to β-actin. *, p < 0.05, **, p < 0.01, Student's t test.
FIGURE 5.
Transcriptional and IRP1-dependent posttranscriptional regulation of L-Ft and Fpn by NO. Wild-type and IRP1-null BMMs were exposed to 250 μm DETA/NO for 18 h alone or in the presence of the DRB transcriptional inhibitor, as indicated. A, L-Ft and Fpn mRNA levels were assayed by real-time quantitative PCR. The histograms shows mRNA expression (mean ± S.D., n = 3) as a percentage of untreated (no DETA/NO, no DRB) wild-type cells after normalization to 18 S ribosomal RNA levels. B, Western blot analysis of L-Ft (left panels) and Fpn (right panels) levels. Representative immunoblots are shown. β-actin and vinculin, respectively, were used as loading controls. The histograms show L-Ft and Fpn levels (mean ± S.D., n = 3) as a percentage of untreated wild-type cells. *, p < 0.05; **, p < 0.01; ***, p < 0.01; Student's t test.
Role of IRP1 in NO Modulation of Intracellular Iron Availability
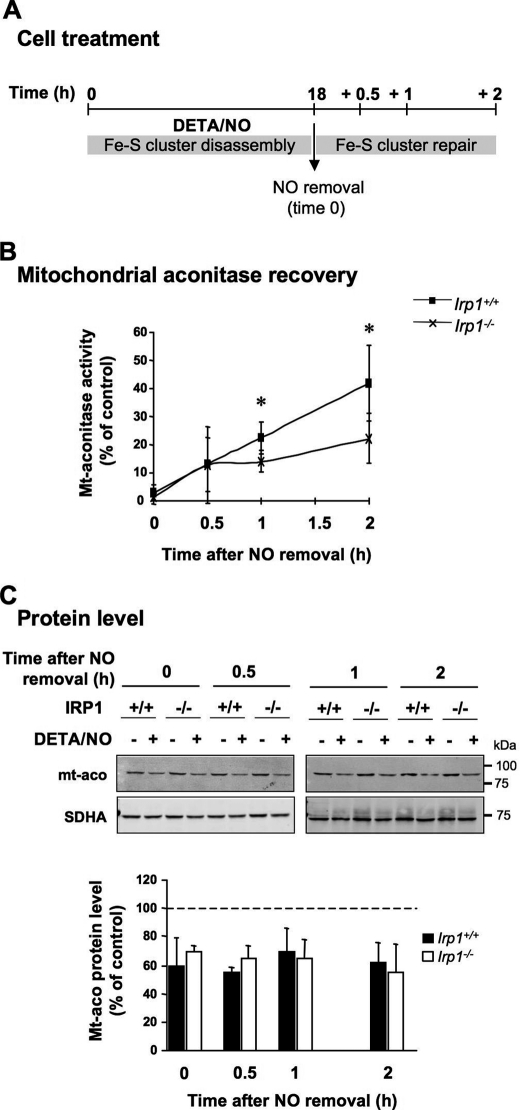
We have shown that IRP1 plays a critical role in the regulation of iron transport and storage molecules by NO. Next we analyzed the impact of NO-dependent activation of IRP1 on intracellular iron availability. Wild-type BMMs exposed to NO exhibit TfR1 up-regulation (Fig. 2) associated with unchanged Ft and Fpn protein levels (Fig. 3). Increased iron uptake in the context of steady iron storage and export would be predicted to augment intracellular iron availability. By contrast, IRP1-deficient cells fail to up-regulate TfR1 (Fig. 2) and display increased Ft and Fpn expression (Fig. 3), a pattern expected to result in intracellular iron depletion. Intracellular iron availability is typically determined by measuring the level of the chelatable iron pool (CIP) using the fluorescent iron sensor dye calcein (31). However, NO reacts with the CIP and forms stable dinitrosyl iron complexes that are resistant to the iron chelator used in the calcein method (32). Hence, availability of the dynamic pool of iron was determined indirectly, on the basis of the dependence of aconitase 2, mitochondrial aconitase (ACO2), whose activity strictly relies on the assembly of an iron-limiting [4Fe-4S] cluster. BMMs from Irp1+/+ and Irp1−/− mice were first challenged with DETA/NO for 18 h to induce their respective regulation of iron-related genes. At that time point (Fig. 6A, time zero hour), ACO2 activity is almost completely abolished both in wild-type and IRP1-null cells (B). This is most likely due to the disruption of the enzyme Fe-S cluster, as Western blotting reveals only a 30 to 40% reduction in ACO2 expression (Fig. 6C). NO was then removed, and the recovery of ACO2 activity, which reflects Fe-S cluster repair (21) and is strongly dependent on cellular iron bioavailability (33), was analyzed at the times indicated (Fig. 6A). In wild-type BMMs, ACO2 activity increases linearly and reaches 40% of control levels 2 h after NO removal (Fig. 6B). Compared with the wild type, IRP1-deficient cells display significantly lower ACO2 activity recovery (Fig. 6B) although ACO2 protein levels are similar (C). This suggests that IRP1 deficiency limits intracellular iron availability, impeding efficient repair of the ACO2 Fe-S cluster. In conclusion, IRP1 activation by NO and the ensuing TfR1 up-regulation and translational inhibition of Ft and Fpn increases intracellular iron bioavailability.
FIGURE 6.
Iron bioavailability and restoration of mitochondrial aconitase activity after DETA/NO removal in wild-type versus IRP1-deficient BMMs. A, wild-type and IRP1-null BMMs were exposed to 250 μm DETA/NO for 18 h. NO was then removed by extensive washing (time 0), and cells were further incubated in fresh medium for up to 2 h. Crude mitochondrial extracts were collected 0, 0.5, 1, and 2 h after NO removal to analyze the recovery of mitochondrial aconitase activity. B, mitochondrial aconitase activity in wild-type versus IRP1-null BMMs expressed as a percentage of control, i.e. wild-type and IRP1-null cells incubated in DETA/NO-free medium during the first 18 h but otherwise processed in an identical manner. Data are presented as mean ± S.D., n = 5. C, Western blot analysis of mitochondrial aconitase levels in crude mitochondrial extracts. Representative immunoblots are shown. The succinate dehydrogenase, subunit A complex II subunit was used as a loading control. The histograms show mitochondrial aconitase levels (mean ± S.D., n = 4) as a percentage of control (no DETA/NO) cells after normalization to succinate dehydrogenase, subunit A. *, p < 0.05, Student's t test.
DISCUSSION
NO is widely recognized as a regulator of cellular iron metabolism (8). Among its various effects, it has been proposed to regulate iron storage, transport, and utilization by targeting the IRP/IRE regulatory network (34). Although IRP1 can compensate for IRP2 deficiency (and vice versa) in regulating iron homeostasis in response to iron fluctuations under normoxia (29), the respective contribution of each IRP to the regulation of cellular iron metabolism by NO is not clear. Most studies focusing on the regulation of IRPs in macrophages reported that NO, in contrast to iron, conversely regulates IRP1 and IRP2. Although the IRP1 IRE binding activity is strongly activated by NO, IRP2 is down-regulated in NO-treated (14, 17) or IFN-γ LPS-stimulated macrophages (12, 13). Under these conditions, discrepancies mostly concern not only the intrinsic contribution of each IRP to regulate downstream IRP targets but also the mechanism of NO-dependent IRP2 down-regulation (12–14, 17, 35). An explanation for the effect of NO has been proposed, suggesting that S-nitrosylation at Cys-178 leads to proteasome-dependent IRP2 degradation (17). However, this mechanism, once believed in, was not confirmed by other authors and is still a matter of controversy (35).
Animal and cellular models of IRP deficiency have proven useful for defining the specific as well as redundant functions of IRP1 and IRP2 under basal conditions (4, 5) or in response to iron fluctuation (6), local inflammation (36), and oxidative stress (30, 37). In our study, primary cultures of BMMs derived from IRP1- and IRP2-null mice were exposed to the chemical NO donor DETA/NO that mimics NO release from NOS2 and recapitulates the aconitase/IRP1 switch induced by IFN-γ/LPS-stimulated macrophages (28). We found that 1) NO indeed converts the vast reservoir of cytosolic aconitase into its IRE-binding form but has no major impact on IRP2, 2) that TfR1 up-regulation by NO is abolished in IRP1-null cells but is unaffected by IRP2 deficiency, and 3) that the compensatory increase in IRP2 activity in IRP1-null cells does not alter TfR1 regulation by NO. This shows that the regulation of cellular iron uptake by NO is mediated by IRP1 and cannot be compensated by IRP2. Further highlighting the importance of IRP1 for NO-dependent regulation of iron metabolism, we found that IRP1 controls the NO-dependent translational inhibition of mRNAs containing IRE in their 5′ UTR. Although NO stimulates Ft and Fpn transcription regardless of the IRP genotype, only IRP1-deficient cells display a proportional increase in Ft and Fpn protein levels. Furthermore, we characterized the effect of endogenously produced NO on Ft chain levels by stimulating Irp1−/− and Irp1+/+ BMMs with IFN-γ and LPS in the presence or absence of NOS inhibitors. We showed that, similarly to exogenous NO, endogenous NO is the signal leading to both increased H- and L-Ft protein levels in IRP1-null macrophages. These data show that NO controls iron export and storage both at the transcriptional and posttranscriptional level through an incoherent feed-forward loop. On the one hand, NO up-regulates Ft and Fpn transcription, possibly by stimulating the binding of Nrf2 (38) to the antioxidant response elements present in the promoter of the Ft (39) and Fpn genes (40). On the other hand, IRP1 activation by NO represses Ft and Fpn translation and antagonizes the transcriptional stimulation of those genes (Fig. 7). Our study thus indicates that bifunctional IRP1 is a key player in the complex network of interactions between NO and cellular iron transport and storage, whereas IRP2, which is important for steady-state control of iron metabolism, is dispensable.
FIGURE 7.
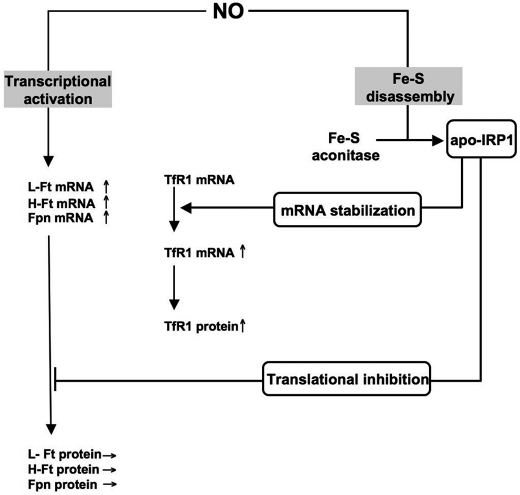
Scheme of dual transcriptional and IRP1-dependent regulation of cellular iron metabolism in response to NO. In wild-type BMMs, NO is responsible for the transcriptional activation of the L-, H-Ft, and Fpn genes (left side). Nonetheless, up-regulation of the L-, H-Ft, and Fpn mRNAs does not finally lead to increased L, H-Ft, and Fpn protein levels. This is explained by the fact that NO mediates posttranslational modification of the aconitase/IRP1 system (right side). By rapidly converting the constitutively expressed cytosolic Fe-S aconitase into an iron regulatory factor (apo-IRP1), NO in fine inhibits translation of the L-, H-Ft, and Fpn mRNAs. This latter action of NO predominates on the former one because IRP1 Fe-S disassembly by NO is a fast process that does not require protein synthesis and that generates sufficient amount of apo-IRP1 to efficiently prevent translation of elevated levels of L-, H-Ft, and Fpn mRNAs. In an opposite way, TfR1 mRNA is stabilized by the NO dependent-IRP1 activation, leading to the increase in TfR1 protein levels.
Most tissues of IRP2-null mice overexpress ferritin under standard laboratory conditions (4, 5). We found that basal expression of L-Ft is also posttranscriptionally increased in IRP2-deficient BMMs ex vivo. Interestingly, NO treatment of IRP2-null BMMs substantially reduces L-Ft protein expression despite a concomitant up-regulation of the L-Ft mRNA. This shows that IRP1 activation by NO is not only able to override the transcriptional stimulation of L-Ft but can also partially remedy the defects of iron storage in IRP2 deficiency. This is reminiscent of the recently reported ability of Tempol, a stable nitroxide, to trigger the conversion of cytosolic aconitase into apo-IRP1 and reduce Ft expression in the brain of IRP2-deficient mice (41). However, Tempol acts as a superoxide scavenger (42) and is used mostly for antioxidant therapy (43). Unlike NO, its mechanism of action on the aconitase/IRP1 switch remains unclear. Hence, pharmacological NO donors that are able to release physiological fluxes of NO in specific tissues (44) might represent interesting alternatives to Tempol to restore the cellular iron balance in IRP2 deficiency.
Our data show that IRP1 activation by NO increases TfR1 expression while neutralizing the transcriptional up-regulation of the Ft and Fpn genes via translational repression of the corresponding mRNAs. Therefore, modulation of the cellular iron transport and storage capacities by NO is predicted to increase cellular iron availability in wild-type BMMs and have the opposite effect in IRP1-deficient cells (Fig. 2 and 3). In this context, a critical question is how NO-dependent activation of IRP1 impacts on intracellular iron availability and how this may affect cellular function. Rapidly available iron sources serve for use in synthesis of heme, Fe-S centers, and other non-heme iron proteins (45). This dynamic source of iron is constituted by the CIP. However, in cells producing (or exposed to) NO, the CIP is in part converted into stable dinitrosyl-iron complexes (32). Determination of iron in a readily available form in NO-treated BMMs was therefore based on the strict dependence of ACO2 on the flux of iron from the CIP to the Fe-S cluster biosynthetic pathway (45). We found that IRP1 deficiency impairs the restoration of ACO2 activity after disruption of the enzyme Fe-S cluster by NO. This indicates that the failure of IRP1-null cells to increase cellular iron uptake and/or limit iron export and storage in response to NO curtails the iron supply from the CIP to the Fe-S cluster biogenesis machinery. This is reminiscent of the widespread impairment of Fe-S cluster enzymes (including ACO2) in the liver of mice with coablation of IRP1 and IRP2 in hepatocytes, which display hepatocytic iron depletion under steady-state conditions because of abnormally high Ft and Fpn expression and down-regulation of iron import molecules (46). Hence, activation of IRP1 by NO and the resulting increase in iron uptake and down-regulation of iron storage and export, respectively, may serve as a stratagem to maintain adequate levels of chelatable iron in the cell to fuel the Fe-S biosynthetic pathway for efficient repair of Fe-S cluster proteins. On the other hand, IRP1 activation by NO and the ensuing increase in the CIP could promote iron toxicity in inflammation-related pathologies, such as inflammatory bowel disease, Parkinson's disease, and acute lung injury (47, 48) that are characterized by continuous production of reactive oxygen and nitrogen species. IRP1-mediated modulation of cellular iron metabolism by NO could also be important for macrophage cellular immune functions (49).
In conclusion, our work shows that IRP1 is critically important for the regulation of cellular iron transport and storage by NO and uncovers aspects of iron metabolism that cannot be compensated by IRP2. The situation may differ under hypoxic conditions because IRP2 is then stabilized (2, 3), and hypoxia-inducible factor 1-α is activated (50). The prominence of IRP1 over IRP2 in NO regulation of the cellular iron metabolism lies in the ability of the radical to promote the conversion of the cytosolic aconitase to the IRE-binding form of IRP1 independently of iron-mediated control. It is noteworthy that NO can also fully activate IRP1 under hypoxia (51). Therefore, future in vivo studies will be devoted to elucidate the intricate interplay between IRP1, IRP2, and hypoxia-inducible factor 1-α in the regulation of cellular iron metabolism by NO under hypoxia. This approach will help pinpoint the impact of the NO/IRP1/IRE system among the wide-ranging actions of NO in physiology and inflammation.
Supplementary Material
Acknowledgments
We thank Prof. M. W. Hentze for provision of the Irp1- and Irp2-deficient mice. We also thank Dr. J. Bignon (Institut de Chimie des Substances Naturelles, Gif-sur-Yvette) for helping to determine the homogeneity of the BMM population using flow cytometry.
This work was supported by a French government scholarship (Thèse en Cotutelle) granted by the Embassy of France in Poland, Regional Council of Ile-de-France Grant 09SETCI019, Polish State Committee for Scientific Research Grant N N303 373636 (cofinanced within the framework of Integrated Regional Development Operational Programme (PhD student scholarship no. 1365/1).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- IRP
- iron regulatory protein
- IRE
- iron response element
- UTR
- untranslated region
- Ft
- ferritin
- Fpn
- ferroportin
- TfR1
- transferrin receptor 1
- BMM
- bone marrow-derived macrophage
- DRB
- 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside
- DETA
- diethylenetriamine
- NOS
- NO synthase
- CIP
- chelatable iron pool
- ACO2
- aconitase 2, mitochondrial aconitase.
REFERENCES
- 1. Hentze M. W., Muckenthaler M. U., Galy B., Camaschella C. (2010) Cell 142, 24–38 [DOI] [PubMed] [Google Scholar]
- 2. Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LaVaute T., Smith S., Cooperman S., Iwai K., Land W., Meyron-Holtz E., Drake S. K., Miller G., Abu-Asab M., Tsokos M., Switzer R., 3rd, Grinberg A., Love P., Tresser N., Rouault T. A. (2001) Nat. Genet. 27, 209–214 [DOI] [PubMed] [Google Scholar]
- 5. Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schümann K., Hentze M. W. (2005) Blood 106, 2580–2589 [DOI] [PubMed] [Google Scholar]
- 6. Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T. A. (2004) EMBO J. 23, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soum E., Drapier J. C. (2003) J. Biol. Inorg. Chem. 8, 226–232 [DOI] [PubMed] [Google Scholar]
- 8. Watts R. N., Ponka P., Richardson D. R. (2003) Biochem. J. 369, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill B. G., Dranka B. P., Bailey S. M., Lancaster J. R., Jr., Darley-Usmar V. M. (2010) J. Biol. Chem. 285, 19699–19704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouton C., Raveau M., Drapier J. C. (1996) J. Biol. Chem. 271, 2300–2306 [DOI] [PubMed] [Google Scholar]
- 11. Phillips J. D., Kinikini D. V., Yu Y., Guo B., Leibold E. A. (1996) Blood 87, 2983–2992 [PubMed] [Google Scholar]
- 12. Bouton C., Oliveira L., Drapier J. C. (1998) J. Biol. Chem. 273, 9403–9408 [DOI] [PubMed] [Google Scholar]
- 13. Mulero V., Brock J. H. (1999) Blood 94, 2383–2389 [PubMed] [Google Scholar]
- 14. Recalcati S., Taramelli D., Conte D., Cairo G. (1998) Blood 91, 1059–1066 [PubMed] [Google Scholar]
- 15. Kim S., Ponka P. (1999) J. Biol. Chem. 274, 33035–33042 [DOI] [PubMed] [Google Scholar]
- 16. Kim S., Ponka P. (2000) J. Biol. Chem. 275, 6220–6226 [DOI] [PubMed] [Google Scholar]
- 17. Kim S., Wing S. S., Ponka P. (2004) Mol. Cell. Biol. 24, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J., Chen G., Pantopoulos K. (2005) Mol. Cell. Biol. 25, 1347–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., Ponka P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12214–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weiss G., Goossen B., Doppler W., Fuchs D., Pantopoulos K., Werner-Felmayer G., Wachter H., Hentze M. W. (1993) EMBO J. 12, 3651–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bouton C., Chauveau M. J., Lazereg S., Drapier J. C. (2002) J. Biol. Chem. 277, 31220–31227 [DOI] [PubMed] [Google Scholar]
- 22. Pantopoulos K., Hentze M. W. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1267–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drapier J. C., Hirling H., Wietzerbin J., Kaldy P., Kühn L. C. (1993) EMBO J. 12, 3643–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keefer L. K., Nims R. W., Davies K. M., Wink D. A. (1996) Methods Enzymol. 268, 281–293 [DOI] [PubMed] [Google Scholar]
- 25. Drapier J. C., Hibbs J. B., Jr. (1996) Methods Enzymol. 269, 26–36 [DOI] [PubMed] [Google Scholar]
- 26. Leibold E. A., Munro H. N. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2171–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Müllner E. W., Neupert B., Kühn L. C. (1989) Cell 58, 373–382 [DOI] [PubMed] [Google Scholar]
- 28. Oliveira L., Drapier J. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6550–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. (2004) Science 306, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 30. Corna G., Galy B., Hentze M. W., Cairo G. (2006) J. Mol. Med. 84, 551–560 [DOI] [PubMed] [Google Scholar]
- 31. Epsztejn S., Kakhlon O., Glickstein H., Breuer W., Cabantchik I. (1997) Anal. Biochem. 248, 31–40 [DOI] [PubMed] [Google Scholar]
- 32. Toledo J. C., Jr., Bosworth C. A., Hennon S. W., Mahtani H. A., Bergonia H. A., Lancaster J. R., Jr. (2008) J. Biol. Chem. 283, 28926–28933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paradkar P. N., Zumbrennen K. B., Paw B. H., Ward D. M., Kaplan J. (2009) Mol. Cell. Biol. 29, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bouton C., Drapier J. C. (2003) Sci STKE 2003, pe17. [DOI] [PubMed] [Google Scholar]
- 35. Wang J., Fillebeen C., Chen G., Andriopoulos B., Pantopoulos K. (2006) Mol. Cell. Biol. 26, 1948–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Viatte L., Gröne H. J., Hentze M. W., Galy B. (2009) J. Mol. Med. 87, 913–921 [DOI] [PubMed] [Google Scholar]
- 37. Wang W., Di X., D'Agostino R. B., Jr., Torti S. V., Torti F. M. (2007) J. Biol. Chem. 282, 24650–24659 [DOI] [PubMed] [Google Scholar]
- 38. Liu X. M., Peyton K. J., Ensenat D., Wang H., Hannink M., Alam J., Durante W. (2007) Cardiovasc. Res. 75, 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hintze K. J., Katoh Y., Igarashi K., Theil E. C. (2007) J. Biol. Chem. 282, 34365–34371 [DOI] [PubMed] [Google Scholar]
- 40. Marro S., Chiabrando D., Messana E., Stolte J., Turco E., Tolosano E., Muckenthaler M. U. (2010) Haematologica 95, 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghosh M. C., Tong W. H., Zhang D., Ollivierre-Wilson H., Singh A., Krishna M. C., Mitchell J. B., Rouault T. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soule B. P., Hyodo F., Matsumoto K., Simone N. L., Cook J. A., Krishna M. C., Mitchell J. B. (2007) Free Radic. Biol. Med. 42, 1632–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Caldero H., Rodriguez-Vilarrupla A., Gracia-Sancho J., Divi M., Lavina B., Bosch J., Garcia-Pagan J. C. (2011) J. Hepatol. 54, 660–665 [DOI] [PubMed] [Google Scholar]
- 44. Inami K., Nims R. W., Srinivasan A., Citro M. L., Saavedra J. E., Cederbaum A. I., Keefer L. K. (2006) Nitric Oxide 14, 309–315 [DOI] [PubMed] [Google Scholar]
- 45. Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 46. Galy B., Ferring-Appel D., Sauer S. W., Kaden S., Lyoumi S., Puy H., Kölker S., Gröne H. J., Hentze M. W. (2010) Cell Metab. 12, 194–201 [DOI] [PubMed] [Google Scholar]
- 47. Cross R. K., Wilson K. T. (2003) Inflamm. Bowel Dis. 9, 179–189 [DOI] [PubMed] [Google Scholar]
- 48. Andersen J. K. (2004) J. Alzheimer's Dis. 6, S47–52 [DOI] [PubMed] [Google Scholar]
- 49. Nairz M., Schroll A., Sonnweber T., Weiss G. (2010) Cell Microbiol. 12, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 50. Majmundar A. J., Wong W. J., Simon M. C. (2010) Mol. Cell 40, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Soum E., Brazzolotto X., Goussias C., Bouton C., Moulis J. M., Mattioli T. A., Drapier J. C. (2003) Biochemistry 42, 7648–7654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.