Abstract
The amino acid sequence of collagen is composed of GlyXaaYaa repeats. A prevailing paradigm maintains that stable collagen triple helices form when (2S)-proline (Pro) or Pro derivatives that prefer the Cγ-endo ring pucker are in the Xaa position and Pro derivatives that prefer the Cγ-exo ring pucker are in the Yaa position. Anomalously, an amino acid sequence in an invertebrate collagen has (2S,4R)-4-hydroxyproline (Hyp), a Cγ-exo-puckered Pro derivative, in the Xaa position. In certain contexts, triple helices with Hyp in the Xaa position are now known to be hyperstable. Most intriguingly, the sequence (GlyHypHyp)n forms a more stable triple helix than does the sequence (GlyProHyp)n. Competing theories exist for the physicochemical basis of the hyperstability of (GlyHypHyp)n triple helices. By synthesizing and analyzing triple helices with different Cγ-exo-puckered proline derivatives in the Xaa and Yaa positions, we conclude that interstrand dipole-dipole interactions are the primary determinant of their additional stability. These findings provide a new framework for understanding collagen stability.
Keywords: Collagen, Hydroxyproline, Protein Conformation, Protein Stability, Protein Structure
Introduction
The abundance and importance of collagen has led to extensive research on the physicochemical basis of its structure and stability (1–3). Supramolecular collagen is composed of individual helices of three strands. To form a triple helix, every third amino acid in each strand must be a glycine (Gly) residue, resulting in a repeating GlyXaaYaa sequence. A second characteristic feature of natural collagen is that the residue in the Xaa position is often (2S)-proline (Pro), and that in the Yaa position is often (2S,4R)-4-hydroxyproline (Hyp)3 (4). A high percentage of Pro and Pro derivatives in the collagen sequence leads to the formation of triple helices with high thermostability.
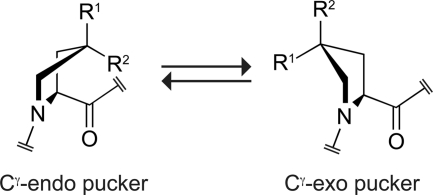
A paradigm for how Pro and Hyp stabilize the triple helix has emerged recently (5–13). This paradigm is based on preorganization. Briefly, it is known that two conformations of the pyrrolidine ring within Pro and Pro derivatives-Cγ-endo and Cγ-exo are dominant (Fig. 1). The peptide backbone torsion angles that accompany a Cγ-endo ring pucker are similar to those required in the Xaa position of the triple helix (9, 13). Likewise, the backbone torsion angles that accompany a Cγ-exo pucker are similar to those required in the Yaa position of the triple helix. Accordingly, Pro derivatives that prefer the Cγ-endo pucker stabilize the triple helix when substituted in the Xaa position, whereas Pro derivatives that prefer the Cγ-exo pucker stabilize the triple helix when substituted in the Yaa position (1, 14). As a corollary, Pro derivatives that prefer the Cγ-exo pucker are strongly destabilizing in the Xaa position, and those that prefer the Cγ-endo pucker are strongly destabilizing in the Yaa position (15–17). The preorganization paradigm explains the abundance of Pro (which prefers the Cγ-endo pucker) in the Xaa position and Hyp (which prefers the Cγ-exo pucker) in the Yaa position of stable, natural collagen (4). Likewise, this paradigm explains the instability of triple helices with Hyp in the Xaa position and Pro in the Yaa position (18). The preorganization paradigm enables the rational design of synthetic triple helices with virtually any thermostability, merely by using Pro derivatives with a particular Cγ-endo:Cγ-exo ratio (14).
FIGURE 1.
Preferred ring conformations of Pro and Pro derivatives. The Cγ-exo conformation is favored strongly by the gauche effect when R1 = OH (Hyp) or F (Flp), R2 = H, and by steric effects when R1 = H, R2 = CH3 (Mep). The Cγ-endo:Cγ-exo ratio is ∼2 when R1 = R2 = H (Pro) (13).
A recent discovery calls into question the generality of the preorganization paradigm. Bächinger and co-workers (19, 20) reported that Hyp is permissible, though not notably stabilizing, in the Xaa position of stable triple helices when the Yaa position is occupied by residues other than Pro. Although Hyp residues are found only in the Yaa position of the strands of vertebrate collagen, several invertebrates have Hyp residues in the Xaa position (21–24). Thus, the context-dependent stabilization of triple helices by Hyp in the Xaa position could be important for the structural integrity of invertebrates. Strikingly, several groups then showed that the instability of (HypProGly)n triple helices can be reverted by 4R hydroxylation of Pro residues in the Yaa position to form (HypHypGly)n (25–28). The (HypHypGly)n sequence actually forms a slightly more stable triple helix than does a (ProHypGly)n sequence, despite contradicting the previously absolute requirement for Pro derivatives that prefer the Cγ-endo conformation in the Xaa position of proline-rich hyperstable triple helices.
Various hypotheses have been put forth to explain the remarkable stability of (HypHypGly)n triple helices. The hyperstability could be due to the higher Ktrans/cis ratio of Hyp relative to Pro and the propensity of Hyp residues to adopt a PPII conformation (27, 29, 30), but this proposal is contravened by the instability of (HypProGly)n triple helices. Alternatively, if the Hyp residues in the Xaa position were to adopt the disfavored Cγ-endo pucker, then the hyperstability of (HypHypGly)n triple helices could be due to a hydrogen bond between the hydroxyl group of Hyp residues in the Xaa and Yaa position of different strands (26). Recent crystal structures of (HypHypGly)n triple helices revealed, however, that the Hyp residues adopt a Cγ-exo pucker (except when crystal-packing forces intervene), precluding such an interstrand hydrogen bond (31–33). These crystal structures also showed that a Cγ-exo pucker in the Xaa position leads to a small distortion of the backbone torsion angles of the triple helix from those adopted by a canonical triple helix. This distortion accounts for (HypHypGly)n triple helices being more flexible than (ProHypGly)n triple helices because of weaker interstrand hydrogen bonds (27, 34).
A consensus is developing that the hyperstability of (HypHypGly)n triple helices is due to differential hydration in the folded and unfolded states (32, 35, 36), although a possible role for interstrand dipole-dipole interactions has been proposed by one group based on theoretical calculations (37). Partial specific volume measurements and small-angle x-ray scattering data demonstrate the existence of different hydration levels in the folded and unfolded states. Still, we were skeptical that the hyperstability of (HypHypGly)n triple helices is due to the release of water upon triple-helix folding and hydroxyl group burial. We had shown previously that attributing differences in thermostability to hydration in the context of the collagen triple helix can be problematic (5, 7, 10, 38, 39).
Herein, we report on the physicochemical basis for the hyperstability of collagen triple helices with Hyp in the Xaa and Yaa positions. Recently, we established that both (2S,4R)-4-fluoroproline (Flp) and (2S,4S)-4-methylproline (Mep), like Hyp, strongly prefer the Cγ-exo pucker (13, 40). By incorporating these Pro derivatives, as well as Hyp, in the Xaa position of triple-helical collagen, we sought to reveal the determinants of triple-helix stability when a Pro derivative with a Cγ-exo pucker is in the Xaa position. We use FlpFlpGly triplets to test the differential hydration hypothesis, as they would likely be destabilizing if differential hydration is essential for triple helix stability. Triple helices formed with FlpHypGly and HypFlpGly triplets would likewise be unstable if differential hydration underlies hyperstability. Finally, we use HypMepGly triplets to provide further insight by retaining the conformational preferences of HypHypGly triplets but without an electronegative substituent in the Yaa position. For all of these experiments, ProHypGly and ProFlpGly triplets serve as controls. The ensuing data provide a compelling explanation for the stability of (HypHypGly)n triple helices.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
The peptides (GlyFlpFlp)7, (GlyFlpHyp)7, (GlyHypFlp)7, (GlyProFlp)7, and (HypMepGly)7 were synthesized by segment condensation of Fmoc-GlyFlpFlp-OH, Fmoc-GlyFlpHyp-OH, Fmoc-GlyHypFlp-OH, Fmoc-GlyProFlp-OH, and Fmoc-Hyp(tBu)MepGly-OH tripeptides, respectively. The synthesis of these tripeptides is described in supplemental information. Peptides (GlyProHyp)7 and (GlyHypHyp)7 were synthesized by condensation of Fmoc-Gly-OH, Fmoc-Hyp(tBu)-OH, and Fmoc-Pro-OH, as appropriate. Peptides were synthesized on a 2-chlorotrityl chloride resin using a Synergy 432A Peptide Synthesizer from Applied Biosystems (Carlsbad, CA) at the University of Wisconsin-Madison Biotechnology Center. For the peptides prepared by segment condensation, the first tripeptide was loaded onto the resin as described previously (40). Fmoc-deprotection was achieved by treatment with piperidine (20% v/v in DMF). The tripeptides (3 equiv) were converted to activated esters by treatment with HBTU, DIEA, and HOBt. Extended double couplings (60–90 min) were employed at room temperature for all segment condensations. Couplings of 2 h were employed at room temperature for single amino acid couplings. Peptides were cleaved from the resin and deprotected in 95:3:2 TFA/triisopropylsilane/H2O (2.5 ml), precipitated from methyl t-butyl ether at 0 °C, and isolated by centrifugation. After preheating solutions of the crude peptides to 70 °C to disassemble any oligomers, semi-preparative HPLC was used to purify the peptides using gradients of CH3CN/water containing TFA (0.1% v/v) at 60 °C. All peptides were >90% pure according to analytical HPLC and MALDI-TOF mass spectrometry (Applied Biosystems Voyager DE-Pro mass spectrometer) (m/z) [M + Na]+ calcd for C84H107F14N21O22Na 2050.8, found 2051.4 for (GlyFlpFlp)7; calcd for C84H114F7N21O29Na 2036.8, found 2038.1 for (GlyFlpHyp)7 and 2036.8 for (GlyHypFlp)7; calcd for C84H114F7N21O22Na 1924.8, found 1925.2 for (GlyProFlp)7; calcd for C91H135N21O29Na 2010.0, found 2010.1 for (HypMepGly)7; calcd for C84H121N21O29Na 1910.9, found 1912.2 for (GlyProHyp)7; calcd for C84H121N21O36Na 2023.8, found 2023.1 for (GlyHypHyp)7.
Circular Dichroism Spectroscopy
Peptides were dried under vacuum for ≥24 h before being weighed and dissolved to 0.2 mm in 50 mm sodium phosphate buffer (pH 7.0). Although we had previously used 50 mm acetic acid buffer (pH 2.9) for circular dichroism (CD) experimentss (10), we here elected to employ a phosphate buffer in our CD measurements to enable direct comparisons with analytical ultracentrifugation data. Analytical ultracentrifugation measurements using absorbance optics and performed on peptide species lacking side-chain chromophores are not reliable in an acetic acid buffer, which has a high absorbance near 230 nm. At pH 7.0, however, the C termini are at least partially unprotonated, decreasing Tm values by a few °C relative to values at pH 2.9. This pH-dependence is a hallmark of triple-helix formation and was part of the original evidence that triple helices form with a parallel arrangement of peptide strands (41, 42).
Solutions were incubated at 4 °C for ≥24 h before CD spectra were acquired with a 202SF CD spectrometer from Aviv Associates (Lakewood, NJ). Spectra were measured with a 1-nm band-pass in cuvettes with a 0.1-cm pathlength. The signal was averaged for 5 s during wavelength scans and unfolding experiments using a 0.6 °C temperature deadband. During unfolding experiments, CD spectra were acquired at 3 °C intervals using a 0.1-cm pathlength for all peptides except (GlyFlpFlp)7, whose thermal transition was monitored with a 1-cm pathlength because of its poor aqueous solubility. At each temperature, solutions were equilibrated for 5 min before data acquisition. Values of Tm were determined by fitting the molar ellipticity at 225 or 226 nm to a two-state model (43).
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) measurements on peptides (GlyHypFlp)7, (GlyProFlp)7, (GlyFlpHyp)7, (GlyHypHyp)7, (GlyProHyp)7, and (HypMepGly)7 were conducted on a VP-DSC instrument from MicroCal (Northampton, MA). The (GlyFlpFlp)7 peptide could not be evaluated by DSC because of its poor solubility in aqueous solution. Instrument baselines were established by filling both the sample and the reference cells with degassed 50 mm sodium phosphate buffer (pH 7.0) and scanning from 5–98 °C at a scan rate of 6 °C/h with a 10-h pre-equilibration period at 5 °C until the baseline was stable. The final buffer-buffer scan was used as the baseline for subsequent peptide scans. Solutions of peptide (∼200 μm in 50 mm phosphate buffer; pH 7.0) were incubated at 4 °C for ≥24 h prior to their degassing and loading into the sample cell (the reference cell solution was not replaced) during the cooldown from the previous buffer-buffer scan (at ∼10 °C). Samples were scanned from 5–98 °C at a scan rate of 6 °C/h; the first scan of each sample was used in the analysis. Subsequent scans of the same sample after cooling to 4 °C overnight and without reloading the chamber showed high reversibility with >85% recovery. After DSC measurements, peptide concentrations were determined by quantitative amino acid analysis (Protein Chemistry Core, Biomolecular Resource Facility, University of Texas Medical Branch). Peptide concentrations of 138, 136, 130, 289, 176, and 478 μm were discerned for (GlyFlpHyp)7, (GlyHypFlp)7, (GlyProFlp)7, (HypMepGly)7, (GlyProHyp)7, and (GlyHypHyp)7, respectively.
DSC data were processed with the MicroCal software in the Origin 7 program from OriginLab (Northampton, MA). The appropriate reference scan was subtracted from the first sample scan and the data were normalized to monomer concentrations. Progress baselines were subtracted from the data, yielding the traces shown in Fig. 4. Values of ΔH (per mol of monomer) were obtained by direct integration of the baseline-subtracted DSC endotherms. The value of ΔS at the Tm was calculated by using the equation: Tm = ΔH/(ΔS + R·ln(0.75c2)), where c is the concentration of monomeric peptide determined by amino acid analysis and Tm is the maximum of the DSC endotherm (44, 45). Because our model assumes ΔCp = 0 for unfolding (39, 44, 46), ΔH and ΔS are independent of T. Values of ΔG and –TΔS (Table 1) were calculated by using the equation: ΔG = ΔH − TΔS. Thermodynamic parameters determined by DSC measurements are most valid at temperatures near the Tm. Accordingly, parameters were calculated at T = 31.7 °C, which is the mean Tm value for the five triple helices evaluated by DSC.
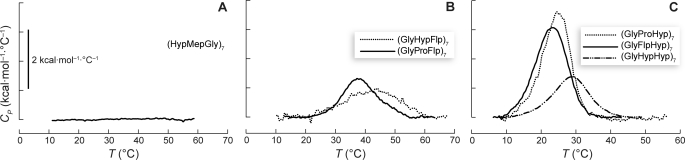
FIGURE 4.
Differential scanning calorimetry of peptides. A, (HypMepGly)7 (289 μm). B, (GlyHypHyp)7 (478 μm), (GlyFlpHyp)7 (138 μm), and (GlyProHyp)7 (176 μm). C, (GlyProFlp)7 (130 μm) and (GlyHypFlp)7 (136 μm). Data were collected with a scan rate of 6 °C/h.
TABLE 1.
Thermodynamic parameters for the unfolding of collagen triple helices
The largest source of error (±5%) was in the determination of [peptide].
| Peptide | Tma | ΔHb | −TΔSb | ΔGb |
|---|---|---|---|---|
| kcal/mol | kcal/mol | kcal/mol | ||
| (GlyFlpFlp)7 | ≥43c | NDd | ND | ND |
| (GlyHypFlp)7 | 43 | 10.1 | 1.3 | 11.3 |
| (GlyProFlp)7 | 41 | 9.4 | 1.8 | 11.2 |
| (GlyHypHyp)7 | 29 | 9.2 | 0.1 | 9.4 |
| (GlyProHyp)7 | 27 | 18.2 | −8.0 | 10.2 |
| (GlyFlpHyp)7 | 25 | 17.9 | −7.5 | 10.4 |
| (HypMepGly)7 | No helix | – | – | – |
a Values (±1 °C) determined by CD spectroscopy at pH 7.0.
b Values at T = 31.7 °C, which is the average of the Tm values determined by DSC at pH 7.0 with a scan rate of 6 °C/h.
c Collagen triple-helix formation by (GlyFlpFlp)7 was evaluated only by CD spectroscopy at low [peptide] because of its poor aqueous solubility.
d Not determined.
Analytical Ultracentrifugation
The self-assembly of (GlyFlpHyp)7, (GlyHypFlp)7, (GlyProFlp)7, (HypMepGly)7, and (GlyHypHyp)7 was evaluated by sedimentation equilibrium experiments using a Beckman XL-A analytical ultracentrifuge. Samples were diluted to ∼0.1 mm in 50 mm potassium phosphate buffer (pH 7.0) and equilibrated at 4 °C for ≥24 h. Equilibrium data were collected at multiple speeds at 4 °C and either 37 or 40 °C. Gradients were monitored at 230 nm. Solvent densities of 1.0048 and 0.99699 g/ml at 4 and 40 °C, respectively, were calculated based on buffer composition and temperature. Partial specific volumes (PSVs) for the peptides were calculated based on amino acid content, with corrections for fluorine, hydroxyl, or methyl content (47). PSVs of 0.654, 0.654, 0.684, 0.713, and 0.660 cm3/g were used for (GlyFlpHyp)7, (GlyHypFlp)7, (GlyProFlp)7, (HypMepGly)7, and (GlyHypHyp)7, respectively. Data at multiple speeds at each temperature were fitted globally as either a single species or as a mixture of monomer and trimer by using programs written for IgorPro (Wavemetrics) by Dr. Darrell R. McCaslin (University of Wisconsin-Madison Biophysics Instrumentation Facility). A small baseline of A ≤ 0.1 at 230 nm was employed at both temperatures for each peptide, but improved the fit only slightly and did not alter any conclusions.
RESULTS
We began our analysis by testing the possibility that the hyperstability of (HypHypGly)n triple helices results merely from a Pro derivative with a high PPII propensity and Ktrans/cis ratio being in the Xaa and Yaa positions simultaneously. To do so, we synthesized (HypMepGly)7 by segment condensation of Fmoc-Hyp(tBu)MepGly-OH tripeptides. Because Mep strongly prefers the Cγ-exo pucker, favors a trans peptide bond, and stabilizes triple helices in the Yaa position (40), we anticipated that a (HypMepGly)7 triple helix would, like a (HypHypGly)n triple helix, be hyperstable if the stability of (HypHypGly)n triple helices is due to the strong preference for a Cγ-exo pucker and high Ktrans/cis ratio of Hyp.
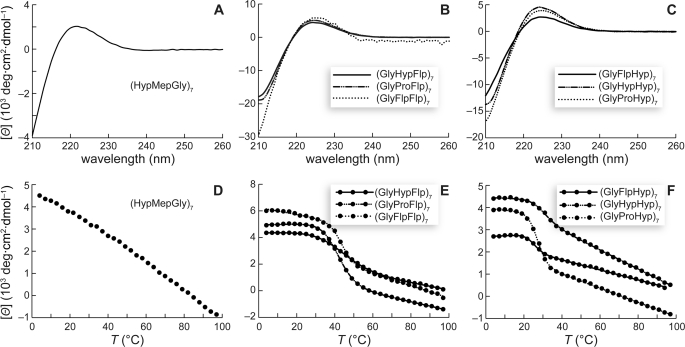
The CD spectrum of a collagen triple helix is characterized by a PPII-like maximal ellipticity near 225 nm that upon heating undergoes a cooperative transition. Our data for the (HypMepGly)7 peptide are shown in Fig. 2, A and D. Although the CD spectrum is PPII-like, the absence of a cooperative transition demonstrates that (HypMepGly)7 does not form a stable triple helix. We confirmed that (HypMepGly)7 is monomeric, even at 4 °C, by sedimentation equilibrium experiments (Fig. 3A). Thus, the mere presence in the Yaa position of a Pro derivative with a Cγ-exo ring pucker is not sufficient to confer hyperstability on triple helices with Hyp in the Xaa position.
FIGURE 2.
Conformational analysis of peptides. A–C, circular dichroism spectra of peptide solutions (∼0.2 mm in 50 mm sodium phosphate buffer, pH 7.0; except (GlyFlpFlp)7, which was ∼0.01 mm) at 4 °C after incubating at 4 °C for ≥24 h. D–F, effect of temperature on the molar ellipticity at 225 nm. Data were recorded at 3-°C intervals after a 5-min equilibration.
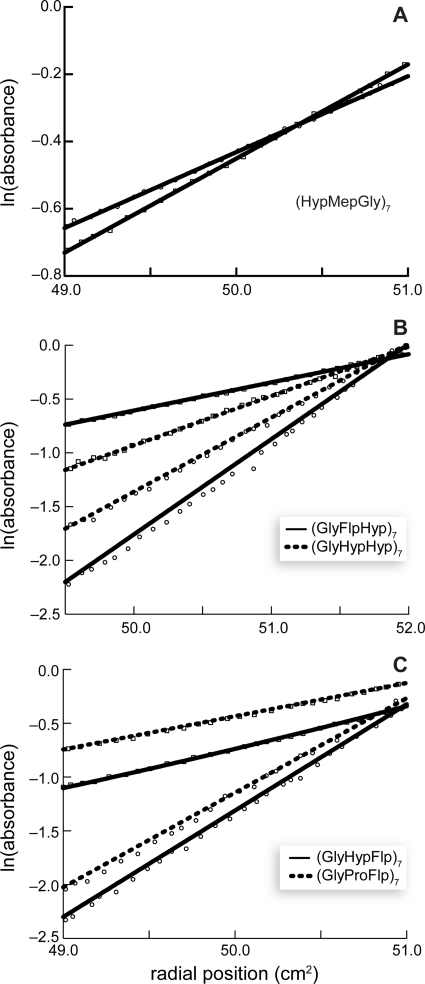
FIGURE 3.
Sedimentation equilibrium analysis of peptides. Circles, 4 °C; squares, 37 or 40 °C. For clarity, only every third data point is shown. Best fits are shown as solid or dashed lines. A, (HypMepGly)7 (40,000 rpm at 4 °C and 50,000 rpm at 37 °C). Fits shown are for monomer at both 4 and 37 °C. B, (GlyFlpHyp)7 (45,000 rpm at 4 °C and 42,000 rpm at 40 °C) and (GlyHypHyp)7 (40,000 rpm at 4 °C and 50,000 rpm at 37 °C). Fits shown are for trimer at 4 °C and monomer at 37 or 40 °C. C, (GlyHypFlp)7 (45,000 rpm at 4 °C and 42,000 rpm at 40 °C) and (GlyProFlp)7 (45,000 rpm at 4 °C and 42,000 rpm at 40 °C). Fits shown are for trimer at 4 °C, and a mixture of monomer and trimer at 40 °C.
Our findings with (HypMepGly)7 suggested that differential hydration indeed might underlie the hyperstability of (HypHypGly)n triple helices, as proposed by others (32, 35, 36), because the hydration of a (HypMepGly)7 peptide will more closely resemble a (HypProGly)7 than a (HypHypGly)7 peptide. To test the differential hydration hypothesis more directly, we designed triple helices with the conformational characteristics and substitution patterns of (HypHypGly)n triple helices, but with vastly altered hydration tendencies. Specifically, we sought to replace the hydroxyl groups in (HypHypGly)n triple helices with fluorine atoms (that is, replace the Hyp residues with Flp). Flp, like Hyp, strongly prefers the Cγ-exo ring pucker (12, 13) and strongly stabilizes the triple helix when substituted in the Yaa position (7). Organic fluorine does not, however, form substantive hydrogen bonds (48, 49). Flp residues are, therefore, poorly hydrated in the folded and unfolded states, and cannot stabilize triple helices by the differential hydration mechanism proposed for (HypHypGly)n, which relies on hydrogen bonds between water and proline substituents in the Xaa and Yaa positions (32, 35, 36). These hydrogen bonds can form with Hyp residues in (HypHypGly)n, but not when those residues are replaced with Flp. Also, Flp residues are known not to stabilize the triple helix by the hydrophobic effect (38).
For this experiment, we first attempted to synthesize (FlpFlpGly)n and (FlpHypGly)n, as well as (HypFlpGly)n, by segment condensation of the corresponding Fmoc-XaaYaaGly-OH tripeptides. We were not successful in doing so, presumably because of the low nucleophilicity of the N-terminal Flp residue. We suspected that segment condensation using Fmoc-GlyXaaYaa-OH tripeptides would be more efficacious. Extensive experimental data demonstrates that (GlyXaaYaa)n and (XaaYaaGly)n triple helices have nearly identical biophysical and structural properties (27, 32). We were successful in using Fmoc-GlyXaaYaa tripeptides to synthesize (GlyFlpFlp)7, (GlyFlpHyp)7, and (GlyHypFlp)7. We also synthesized (GlyProFlp), (GlyProHyp)7, and (GlyHypHyp)7 as controls for our analyses.
CD spectroscopy indicated that all six of these peptides formed collagen triple helices. At 4 °C, each exhibited a signature PPII-like CD spectrum (Fig. 2, B and C), and each underwent a cooperative thermal transition at 225 nm upon heating (Fig. 2, E and F). The linear change in ellipticity observed after the cooperative transition is consistent with the presence of residual PPII secondary structure (30, 50, 51). Values of Tm are listed in Table 1.
The ability to form a triple helix at 4 °C was confirmed by sedimentation equilibrium experiments for five of the six peptides (Fig. 3, B and C). Unfortunately, the (GlyFlpFlp)7 was only slightly soluble (<10 μm) in aqueous solution, even after extensive sonication. Because the Tm value for the (GlyFlpFlp)7 triple helix was determined at low concentration and triple-helix stability is concentration-dependent, the Tm value in Table 1 underestimates the actual Tm value for a (GlyFlpFlp)7 triple helix relative to those for other triple helices. Although we could not confirm the self-association of (GlyFlpFlp)7 by sedimentation equilibrium experiments, we note that CD data are widely recognized as sufficient evidence for collagen triple-helix formation (26, 39, 52), and we conclude with confidence that (GlyFlpFlp)7 forms a hyperstable triple helix.
We used thermal scans in a differential scanning calorimeter (DSC) to extract thermodynamic parameters for the six peptides with sufficient solubility for data collection. We used a heating rate of 0.1 °C/min for these scans (10, 32), finding that slower rates increased the noise substantially. Subsequent scans of the same sample showed high reversibility, with >85% endotherm recovery upon re-equilibration at 4 °C. After collecting the DSC data, we determined peptide concentrations by quantitative amino acid analysis. To calculate thermodynamic parameters, we assumed that ΔCp = 0 for triple-helix folding, and analyzed the data by progress baseline subtraction and direct integration of the resultant endotherms (Fig. 4). The thermodynamic parameters thus obtained are listed in Table 1.
The absence of a DSC endotherm for (HypMepGly)7 (Fig. 4A) provided further evidence that (HypMepGly)7 does not form a stable triple helix. The other important finding from our DSC data is that all the other peptides display endotherms near the Tm values determined by CD spectroscopy, as expected for triple-helix formation. Further interpretation of the DSC data must be performed with caution, because we cannot be sure of having obtained equilibrium at each temperature (53, 54). Nonetheless, consistent with previous reports from the Kobayashi and Bächinger groups (27, 32), we observed that triple-helix formation by (GlyHypHyp)7 is favored weakly by both enthalpy and entropy. This result is reiterated in (GlyHypFlp)7, the other stable triple helix with Hyp in the Xaa position. In contrast, triple-helix formation by (GlyProHyp)7 appears to be favored by enthalpy but disfavored by entropy. A (GlyFlpHyp)7 triple helix has a highly favorable enthalpy of formation but a highly unfavorable entropy, yielding thermodynamic parameters similar to those of a (GlyProHyp)7 triple helix.
DISCUSSION
A peptide-by-peptide comparison of our results provides substantial insight into the mechanism of stabilization for triple helices with Cγ-exo-puckered proline derivatives in the Xaa position. We begin by comparing (GlyHypPro)n triple helices to (GlyHypHyp)n triple helices. Differential hydration could underlie the hyperstability of the (GlyHypHyp)n triple helix. There is substantial evidence that the folded (GlyHypHyp)n and (GlyProHyp)n triple helices have similar hydration, whereas the single coil form of (GlyHypHyp)n is more heavily hydrated than the single coil form of (GlyProHyp)n (32, 35). (Alternatively, the hydration number of (GlyProHyp)n is increased and the hydration number of (GlyHypHyp)n is decreased upon triple-helix formation.) Does this differential hydration actually contribute to triple-helix stability? Enthalpy-entropy compensation, in particular, often undermines any such contribution (55–57). Indeed, the hydration of Hyp residues is known to be deleterious to the stability of (GlyProHyp)n triple helices (39). We sought to address this issue directly for (GlyHypHyp)n triple helices.
To assess the role of differential hydration, we studied analogous peptides with altered hydration properties. Because (GlyHypPro)n and (GlyFlpPro)n peptides both fail to form stable triple helices (15, 16, 18), we surmised that if differential hydration affects the stability of triple helices with a Cγ-exo-puckered proline in the Xaa position, then its contribution to stability could depend merely on the presence of a Cγ-exo-puckered Pro derivative in the Yaa position. We synthesized a peptide to test this hypothesis. (HypMepGly)7 has a Cγ-exo-puckered Pro derivative in the Yaa position but lacks the hydroxyl moiety of Hyp in the Yaa position. We found that this peptide does not fold into a stable triple helix, much less a hyperstable one. Thus, simply installing a Cγ-exo-puckered proline derivative in the Yaa position does not enable differential hydration to stabilize the collagen triple helix.
There is an alternative hypothesis: the stabilization due to differential hydration requires a Cγ-exo puckered Pro derivative with hydration properties similar to those of Hyp to be in the Yaa position. This hypothesis is consistent with the instability of a (GlyHypMep)n triple helix. We sought confirmatory evidence for this hypothesis by installing a Flp residue in the Yaa position. Because organic fluoro groups do not form strong hydrogen bonds, a Flp residue in the Yaa position cannot participate in a hydration network accessible to Hyp. Thus, a (GlyHypFlp)n triple helix cannot be stabilized by the differential hydration effect proposed for (GlyHypHyp)n triple helices. Note that our data for the (GlyHypMep)n triple helix revealed that a Cγ-exo puckered proline derivative in the Yaa position is not sufficient to induce stabilizing differential hydration. Further, we showed previously that the stabilization conferred on triple helices by incorporating Flp instead of Hyp in the Yaa position is due not to the hydrophobic effect but to preorganization (5, 7, 10, 38). Surprisingly, we found that a (GlyHypFlp)n triple helix is not only hyperstable, but also more stable than even a (GlyHypHyp)n triple helix (Table 1). Apparently, the sequence-specific benefit conferred by a Hyp residue in the Xaa position is linked somehow to having a Cγ-exo-puckered proline in the Yaa position, even when that Yaa position residue cannot participate in any hydration network. Yet, because a (HypMepGly)7 triple helix is unstable, more than the mere presence of a Cγ-exo-puckered proline in the Yaa position is required to induce beneficial stabilization. Does hydration of a Hyp residue in the Xaa position have any role in that benefit?
To probe the importance of differential hydration of the residue in the Xaa position, we replaced the Hyp residue in that position of (GlyHypHyp)n with Flp. Because a fluoro group cannot form strong hydrogen bonds with water, (GlyFlpHyp)n should be unstable if differential hydration of a Cγ-exo-puckered residue in the Xaa position has a role in the hyperstability of (GlyHypHyp)n triple helices. We note too that (GlyFlpPro)n strands do not form a stable triple helix (15, 16), so a hydrophobic effect arising from a Flp residue in the Xaa position is insubstantial. Nonetheless, we found that a (GlyFlpHyp)n triple helix is quite stable (Table 1). This result is counter to the idea that differential hydration of the residue in the Xaa position has a role in the hyperstability of (GlyHypHyp)n triple helices.
Finally, we considered the (GlyFlpFlp)n peptide, which is hydrated poorly in both the single coil and triple-helical state and should not endow stability via the hydrophobic effect (38). The parent (GlyFlpPro)n peptide cannot form a triple helix (15, 16), whereas a (GlyProPro)n triple helix is stable. We found that having Flp residues in the Xaa and Yaa positions yields a triple helix that is more stable than any other in this series (Table 1). This result is inexplicable by considerations of differential hydration.
Thus, the evidence is formidable against the differential hydration hypothesis as an explanation for the unusual stability of triple helices with Cγ-exo-puckered proline derivatives in the Xaa position. Is there a more parsimonious and general principle underlying our findings? Preorganization and cis:trans ratios of the Pro derivatives in our peptides cannot explain the data, as Flp, Hyp, and Mep all strongly prefer the Cγ-exo ring pucker and trans peptide bond, yet the (HypMepGly)7 triple-helix is unstable.
A computational analysis by Improta, Berisio, and Vitagliano provided key insight. They suggested that interstrand dipole-dipole interactions could more than compensate for the energetic penalty of triple-helix distortion caused by the incorporation of a Pro derivative with a Cγ-exo pucker in the Xaa position of a folded triple helix (37). (GlyFlpFlp)7, (GlyHypFlp)7, (GlyFlpHyp)7, and (GlyHypHyp)7 all form hyperstable triple helices (Table 1), and all can engage in just such stabilizing interstrand dipole-dipole interactions (Fig. 5). In contrast, (HypMepGly)7 retains the pyrrolidine ring pucker of the other six peptides, but cannot engage in a stabilizing dipole-dipole interaction due to the absence of a 4R electron-withdrawing group (Fig. 5). It is noteworthy that our natural bond orbital calculations (58) at the 6–311+G*(2d,p) level of theory on models of triple helices with Flp and Hyp in both the Xaa and Yaa positions indicate that there is not a significant covalent component to this interstrand interaction (e.g. n → σ* electron donation), but that it is instead a genuine electrostatic interaction (data not shown).
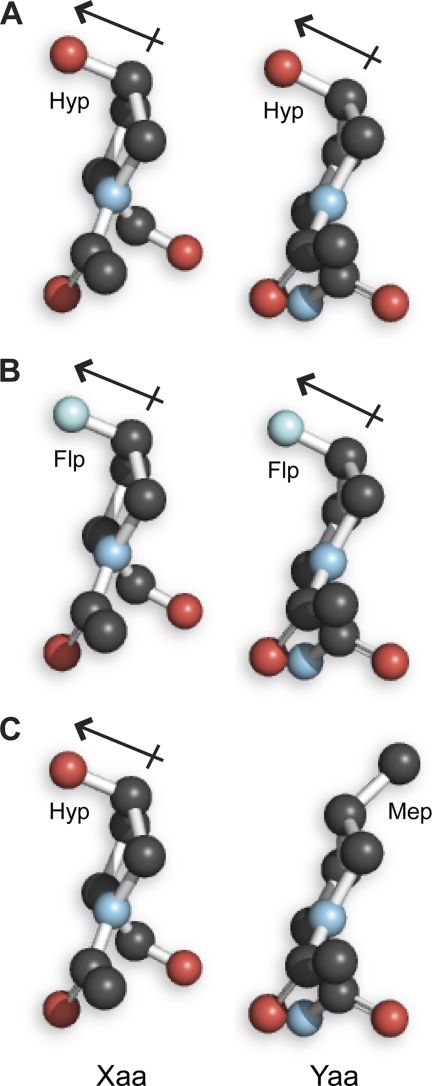
FIGURE 5.
Illustration of dipoles in the side chains of proximal Xaa and Yaa residues in a (GlyXaaYaa)n triple helix. Images were created by modifying Hyp residues in panel A, which shows a cross section of a (HypHypGly)10 triple helix (32), with the program PyMOL (Delano Scientific, Palo Alto, CA). The dipole-dipole interaction in panel A could contribute 0.6 kcal/mol to the stability of the triple helix (37), and likely more in panel B.
In summary, the one constant theme across the ensemble of collagen-related peptides that we (and others) have prepared is the presence of a favorable dipole-dipole interaction whenever proline-rich triple helices containing Cγ-exo-puckered proline derivatives in the Xaa position are hyperstable and its absence when they are unstable. Accordingly, dipole-dipole interactions provide the simplest explanation for the hyperstability of such triple helices.
CONCLUSION
In the last few years, the unexpected finding that peptides composed of GlyHypHyp triplets form hyperstable collagen triple helices has generated great interest (25–28, 31–34, 37). We have synthesized a diverse set of peptides that could, in principle, form stable triple helices, despite having Pro derivatives with Cγ-exo puckers in the Xaa position. We find that the sole essential requirement for triple-helix formation when Pro derivatives with a Cγ-exo pucker are in the Xaa and Yaa positions is that a favorable interstrand dipole-dipole interaction be established between electronegative substituents on Cγ of Pro derivatives in the Xaa and Yaa positions. Thus, we explain the ability of (GlyFlpFlp)7 to form a hyperstable triple helix, and the failure of (HypMepGly)7 to form even a slightly stable one. We conclude that triple helices can violate the preorganization paradigm (that is, Cγ-endo pucker in the Xaa position and Cγ-exo pucker in the Yaa position) and still be highly stable if a dipole-dipole interaction provides recompense. Analogous strategies could be applicable in other contexts, such as coiled-coils.
We speculate that the context-dependent stabilization conferred upon invertebrate collagen triple helices when Hyp is in the Xaa position and certain residues other than Pro are in the Yaa position is due to a combination of interstrand dipole-dipole interactions (observed most clearly when an O-galactosylated (2S,3R)-threonine residue is in the Yaa position) and the less stringent structural requirements for triple-helix formation when Pro derivatives do not occupy both the Xaa and the Yaa positions (19, 20).
Supplementary Material
Acknowledgments
We thank Dr. D. R. McCaslin for both experimental aid and helpful discussions. CD spectroscopy, DSC, analytical ultracentrifugation, and MALDI-TOF mass spectrometry were performed at the University of Wisconsin-Madison Biophysics Instrumentation Facility, which was established with Grants BIR-9512577 (NSF) and S10 RR13790 (National Institutes of Health). NMR spectroscopy was performed at the National Magnetic Resonance Facility at Madison, which is supported by Grant P41RR02301 (National Institutes of Health).
This work was supported, in whole or in part, by Grant R01 AR044276 from the National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental information.
- Hyp
- (2S,4R)-4-hydroxyproline
- DIEA
- N,N-diisopropylethylamine
- DMF
- N,N-dimethylformamide
- DSC
- differential scanning calorimetry
- Flp
- (2S,4R)-4-fluoroproline
- HBTU
- O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- HOBt
- N-hydroxybenzotriazole
- HPLC
- high-performance liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time-of-flight
- Mep
- (2S,4S)-4-methylproline
- Pro
- (2S)-proline.
REFERENCES
- 1. Shoulders M. D., Raines R. T. (2009) Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brodsky B., Thiagarajan G., Madhan B., Kar K. (2008) Biopolymers. 89, 345–353 [DOI] [PubMed] [Google Scholar]
- 3. Fields G. B. (2010) Org. Biomol. Chem. 8, 1237–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramshaw J. A., Shah N. K., Brodsky B. (1998) J. Struct. Biol. 122, 86–91 [DOI] [PubMed] [Google Scholar]
- 5. Holmgren S. K., Bretscher L. E., Taylor K. M., Raines R. T. (1999) Chem. Biol. 6, 63–70 [DOI] [PubMed] [Google Scholar]
- 6. Improta R., Mele F., Crescenzi O., Benzi C., Barone V. (2002) J. Am. Chem. Soc. 124, 7857–7865 [DOI] [PubMed] [Google Scholar]
- 7. Holmgren S. K., Taylor K. M., Bretscher L. E., Raines R. T. (1998) Nature 392, 666–667 [DOI] [PubMed] [Google Scholar]
- 8. Bretscher L. E., Jenkins C. L., Taylor K. M., DeRider M. L., Raines R. T. (2001) J. Am. Chem. Soc. 123, 777–778 [DOI] [PubMed] [Google Scholar]
- 9. Vitagliano L., Berisio R., Mazzarella L., Zagari A. (2001) Biopolymers. 58, 459–464 [DOI] [PubMed] [Google Scholar]
- 10. Shoulders M. D., Satyshur K. A., Forest K. T., Raines R. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Umashankara M., Babu I. R., Ganesh K. N. (2003) Chem. Commun. 2606–2607 [DOI] [PubMed] [Google Scholar]
- 12. Improta R., Benzi C., Barone V. (2001) J. Am. Chem. Soc. 123, 12568–12577 [DOI] [PubMed] [Google Scholar]
- 13. DeRider M. L., Wilkens S. J., Waddell M. J., Bretscher L. E., Weinhold F., Raines R. T., Markley J. L. (2002) J. Am. Chem. Soc. 124, 2497–2505 [DOI] [PubMed] [Google Scholar]
- 14. Shoulders M. D., Raines R. T. (2009) Adv. Exp. Med. Biol. 611, 251–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hodges J. A., Raines R. T. (2003) J. Am. Chem. Soc. 125, 9262–9263 [DOI] [PubMed] [Google Scholar]
- 16. Doi M., Nishi Y., Uchiyama S., Nishiuchi Y., Nakazawa T., Ohkubo T., Kobayashi Y. (2003) J. Am. Chem. Soc. 125, 9922–9923 [DOI] [PubMed] [Google Scholar]
- 17. Shoulders M. D., Guzei I. A., Raines R. T. (2008) Biopolymers. 89, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inouye K., Kobayashi Y., Kyogoku Y., Kishida Y., Sakakibara S., Prockop D. J. (1982) Arch. Biochem. Biophys. 219, 198–203 [DOI] [PubMed] [Google Scholar]
- 19. Bann J. G., Bächinger H. P. (2000) J. Biol. Chem. 275, 24466–24469 [DOI] [PubMed] [Google Scholar]
- 20. Mizuno K., Hayashi T., Bächinger H. P. (2003) J. Biol. Chem. 278, 32373–32379 [DOI] [PubMed] [Google Scholar]
- 21. Goldstein A., Adams E. (1970) J. Biol. Chem. 245, 5478–5483 [PubMed] [Google Scholar]
- 22. Muir L., Lee Y. C. (1970) J. Biol. Chem. 245, 502–509 [PubMed] [Google Scholar]
- 23. Gaill F., Mann K., Wiedemann H., Engel J., Timpl R. (1995) J. Mol. Biol. 246, 284–294 [DOI] [PubMed] [Google Scholar]
- 24. Mann K., Mechling D. E., Bächinger H. P., Eckerskorn C., Gaill F., Timpl R. (1996) J. Mol. Biol. 261, 255–266 [DOI] [PubMed] [Google Scholar]
- 25. Buechter D. D., Paolella D. N., Leslie B. S., Brown M. S., Mehos K. A., Gruskin E. A. (2003) J. Biol. Chem. 278, 645–650 [DOI] [PubMed] [Google Scholar]
- 26. Berisio R., Granata V., Vitagliano L., Zagari A. (2004) J. Am. Chem. Soc. 126, 11402–11403 [DOI] [PubMed] [Google Scholar]
- 27. Mizuno K., Hayashi T., Peyton D. H., Bächinger H. P. (2004) J. Biol. Chem. 279, 38072–38078 [DOI] [PubMed] [Google Scholar]
- 28. Doi M., Nishi Y., Uchiyama S., Nishiuchi Y., Nishio H., Nakazawa T., Ohkubo T., Kobayashi Y. (2005) J. Pept. Sci. 11, 609–616 [DOI] [PubMed] [Google Scholar]
- 29. Panasik N., Jr., Eberhardt E. S., Edison A. S., Powell D. R., Raines R. T. (1994) Int. J. Pept. Protein Res. 44, 262–269 [DOI] [PubMed] [Google Scholar]
- 30. Horng J. C., Raines R. T. (2006) Protein Sci. 15, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schumacher M., Mizuno K., Bächinger H. P. (2005) J. Biol. Chem. 280, 20397–20403 [DOI] [PubMed] [Google Scholar]
- 32. Kawahara K., Nishi Y., Nakamura S., Uchiyama S., Nishiuchi Y., Nakazawa T., Ohkubo T., Kobayashi Y. (2005) Biochemistry 44, 15812–15822 [DOI] [PubMed] [Google Scholar]
- 33. Okuyama K., Morimoto T., Narita H., Kawaguchi T., Mizuno K., Bächinger H. P., Wu G., Noguchi K. (2010) Acta. Crystallogr. D Biol. Crystallogr. 66, 88–96 [DOI] [PubMed] [Google Scholar]
- 34. Radmer R. J., Klein T. E. (2006) Biophys. J. 90, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terao K., Mizuno K., Murashima M., Kita Y., Hongo C., Okuyama K., Norisuye T., Bächinger H. P. (2008) Macromolecules 41, 7203–7210 [Google Scholar]
- 36. Okuyama K., Hongo C., Wu G., Mizuno K., Noguchi K., Ebisuzaki S., Tanaka Y., Nishino N., Bächinger H. P. (2009) Biopolymers. 91, 361–372 [DOI] [PubMed] [Google Scholar]
- 37. Improta R., Berisio R., Vitagliano L. (2007) Protein Sci. 17, 955–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shoulders M. D., Kamer K. J., Raines R. T. (2009) Bioorg. Med. Chem. Lett. 19, 3859–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kotch F. W., Guzei I. A., Raines R. T. (2008) J. Am. Chem. Soc. 130, 2952–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shoulders M. D., Hodges J. A., Raines R. T. (2006) J. Am. Chem. Soc. 128, 8112–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berg R. A., Olsen B. R., Prockop D. J. (1970) J. Biol. Chem. 245, 5759–5763 [PubMed] [Google Scholar]
- 42. Venugopal M. G., Ramshaw J. A., Braswell E., Zhu D., Brodsky B. (1994) Biochemistry 33, 7948–7956 [DOI] [PubMed] [Google Scholar]
- 43. Becktel W. J., Schellman J. A. (1987) Biopolymers 26, 1859–1877 [DOI] [PubMed] [Google Scholar]
- 44. Engel J., Bächinger H. P. (2005) Top. Curr. Chem. 247, 7–33 [Google Scholar]
- 45. Breslauer K. J. (1994) Methods Mol. Biol. 26, 347–372 [DOI] [PubMed] [Google Scholar]
- 46. Engel J., Chen H. T., Prockop D. J., Klump H. (1977) Biopolymers 16, 601–622 [DOI] [PubMed] [Google Scholar]
- 47. Durschlag H., Zipper P. (1994) Prog. Colloid Polym. Sci. 94, 20–39 [Google Scholar]
- 48. Dunitz J. D., Taylor R. (1997) Chem. Eur. J. 3, 89–98 [Google Scholar]
- 49. O'Hagan D. (2008) Chem. Soc. Rev. 37, 308–319 [DOI] [PubMed] [Google Scholar]
- 50. Ruzza P., Siligardi G., Donella-Deana A., Calderan A., Hussain R., Rubini C., Cesaro L., Osler A., Guiotto A., Pinna L. A., Borin G. (2006) J. Pept. Sci. 12, 462–471 [DOI] [PubMed] [Google Scholar]
- 51. Kümin M., Sonntag L. S., Wennemers H. (2007) J. Am. Chem. Soc. 129, 466–467 [DOI] [PubMed] [Google Scholar]
- 52. Gauba V., Hartgerink J. D. (2007) J. Am. Chem. Soc. 129, 2683–2690 [DOI] [PubMed] [Google Scholar]
- 53. Mizuno K., Boudko S. P., Engel J., Bächinger H. P. (2010) Biophys. J. 98, 3004–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Persikov A. V., Xu Y., Brodsky B. (2004) Protein Sci. 13, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lumry R., Rajender S. (1970) Biopolymers 9, 1125–1227 [DOI] [PubMed] [Google Scholar]
- 56. Dunitz J. D. (1995) Chem. Biol. 2, 709–712 [DOI] [PubMed] [Google Scholar]
- 57. Liu L., Guo Q. X. (2001) Chem. Rev. 101, 673–695 [DOI] [PubMed] [Google Scholar]
- 58. Glendening E. D., Badenhoop J. K., Reed A. E., Carpenter J. E., Bohmann J. A., Morales C. M., Weinhold F. (2001) NBO 5.0, Theoretical Chemistry Institute, University of Wisconsin-Madison, Madison, WI [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.