Abstract
Human immunodeficiency virus-1 (HIV-1) exploits a number of host cellular factors for successful survival and propagation. The viral protein Nef plays an important role in HIV-1 pathogenesis by interacting with various cellular proteins. In the present work, we identified Cyclin K (CycK) as a novel Nef-interacting protein, and for the first time, we showed that CycK inhibits HIV-1 gene expression and replication in a Nef-dependent manner. The positive elongation factor b complex comprising cyclin-dependent kinase 9 (CDK9) and Cyclin T1 is a critical cellular complex required for viral gene expression and replication. Enhanced expression of CycK in the presence of Nef induced CycK-CDK9 binding, which prevented CDK9-Cyclin T1 complex formation and nuclear translocation of CDK9, resulting in inhibition of HIV-1 long terminal repeat-driven gene expression. Furthermore, this effect of CycK was not observed with Nef-deleted virus, indicating the importance of Nef in this phenomenon. Finally, silencing of CycK in HIV-1-infected cells resulted in increased translocation of CDK9 into the nucleus, leading to increased viral gene expression and replication. These data also suggest that endogenous CycK might act as an inhibitory factor for HIV-1 gene expression and replication in T-cells. Thus, our results clearly demonstrate that CycK utilizes HIV-1 Nef protein to displace CycT1 from the positive elongation factor b complex, resulting in inhibition of HIV-1 gene expression and replication.
Keywords: CDK (Cyclin-dependent Kinase), Cyclins, Gene Expression, Gene Regulation, HIV, CDK9, Cyclin K, Cyclin T1, HIV-1, Nef
Introduction
Human immunodeficiency virus type-1 (HIV-1) perpetuates its survival in the host by utilizing multiple strategies such as irreversible integration of its DNA into the host cell genome to establish the provirus (1), diversification of its genome to escape or tolerate adaptive immune responses (2), and use of the accessory genes (nef, vif, vpu, and vpr) to modulate host cell immune responses further (3–5). These viral accessory genes are frequently seen as dispensable in many in vitro cell culture systems but seem to be indispensable, in the context of natural infections in vivo. Among these accessory genes, pleiotropic functions of Nef have been studied extensively. It is a 27–30-kDa, N-terminally myristoylated protein having a well characterized CD4 down-regulatory activity (6). Nef does not possess any enzymatic activity, but it modulates diverse signaling pathways and the gene expression of host cell proteins (7, 8). Furthermore, Nef influences the activation state of the host cell by bringing together different host cell proteins, protein kinases, and components of the endocytic machinery and making the cells permissible to the virus (9, 10). All these functions of Nef are manifested by a number of events such as activation of signaling molecules, modulation of apoptosis, activation of transcription factors, mitigation of transcriptional repressors, and an increase in the infectivity of virions (11). Although these functions of Nef have been well studied, controversy exists on its role in viral infectivity and replication as both negative (12, 13) and positive (14, 15) roles have been attributed in the literature. Although a number of reports show that Nef increases viral replication by activating T-cells, the molecular basis of Nef-induced viral gene expression and replication remains to be clearly elucidated. Nef was also shown to physically interact with Tat and augment viral gene expression (16), although availability of Tat is critical for the elongation step of long terminal repeat (LTR)2-driven transcription by RNA polymerase II. Several cellular factors have been identified for their roles in this process, including positive transcription elongation factor b (P-TEFb) complex. Different P-TEFb complexes isolated from mammalian cells contain a common catalytic subunit, cyclin-dependent kinase 9 (CDK9), and one of the regulatory cyclins, CycT1, CycT2a, CycT2b, or CycK (17–19). However, CycT1-containing P-TEFb complex is the key complex responsible for HIV-1 transcription elongation (19). Tat is expressed early in the replicative cycle of HIV and is essential for viral gene expression from the LTR promoter. It recognizes the 5′-bulge in the transactivation response stem loop RNA, which is located at the 5′-end of all viral transcripts. Tat binds to CycT1, and together they form the combinatorial surface that interacts with the transactivation response RNA with high affinity and specificity. The obligate partner of CycT1, CDK9, then hyperphosphorylates the C-terminal domain (CTD) of RNA polymerase II, resulting in increased HIV-1 transcription elongation (20, 21).
HIV-1 gene expression is controlled by several cellular as well as viral proteins, including Nef. Evidences suggest that Nef enhances Tat-mediated viral transcription through a heterogeneous nuclear ribonucleoprotein K-nucleated signaling complex (22). In another study, Hsp40 was shown to interact and co-localize with Nef in the nucleus and thereby augment LTR-driven gene expression (23). In contrast, HEXIM1 and Hsp70 negatively regulate HIV-1 gene expression and replication (24, 25). HEXIM1 along with 7SK RNA was shown to be essential for inactivation of the P-TEFb complex (26, 27). Reportedly, the HEXIM1–7SK RNA complex physically competes with Tat for binding to P-TEFb (28). Recently, CycK has been also shown to inhibit viral replication in a T-cell line by inhibiting LTR-mediated transcription (29), although the mechanism of inhibition remains to be elucidated. CycK was first identified as a protein that could restore progression of the cell cycle and was most closely related to human Cyclins C and H (30). The kinase partner of CycK was identified as CDK9 (17), although a recent report also indicates CycK interaction with CDK12 and -13 (31). The complex between CycK and CDK9 was also shown to function as a CTD kinase in vitro (17).
Although Nef was earlier reported as a negative factor for viral replication (32), recent evidence has convincingly demonstrated Nef as an enhancer of viral replication (16, 22, 33–36). Nef performs most of its functions by interacting with cellular proteins. The majority of the functional studies of Nef to date have been performed with HIV-1 subtype B Nef protein, and there have been few studies using subtype C Nef protein. In the present study, we attempted to identify novel Nef-interacting host cell proteins using subtype C Nef as a bait in a yeast two-hybrid system. Our results show that both HIV-1 subtype C and B Nef interact with human Cyclin K in vitro and in vivo. Furthermore, CycK overexpression led to reduction in LTR-mediated gene expression and virus replication in the presence of Nef. Finally, CycK along with Nef disrupted CDK9-CycT1 interaction and thereby inhibited nuclear translocation of CDK9, consequently inhibiting viral gene expression and replication.
EXPERIMENTAL PROCEDURES
Plasmids, Cell Lines, and Antibodies
The nef gene from HIV-1 subtype C Indian isolate IN301904 (37) was cloned into pAS2-1 (Clontech) and pcDNA3.1 vector (Invitrogen), respectively, as described earlier (23). The sequence of cloned nef was confirmed by DNA sequencing using an ABI 310 Genetic Analyzer (Applied Biosystems). Immunoblotting was used to confirm the expression of Nef from both vectors. The pAS2-1NefC plasmid expressing subtype C Nef and Gal4 DNA binding domain as a fusion protein was used as the bait in yeast two-hybrid screening of a cDNA library of human leukocytes in the pACT2 vector obtained from Clontech. The NL4-3 molecular clone (pNL4-3) was obtained from the National Institutes of Health AIDS Reagent Program (38). The nef-deleted NL4-3 molecular clone (pNL4-3ΔNef) and glutathione S-transferase (GST)-Nef (B) plasmids were obtained from Dr. John C. Gautelli (39) and Dr. K. Saksela, respectively. The vectors expressing HA-Nef and HA-Nef mutants used in this study were obtained from Dr. W. C. Greene (40). The subtype C Nef-expressing GST-Nef plasmid was constructed by cloning subtype C nef gene into BamHI and EcoRI sites of pGEX-5X.1 (GE Healthcare). The sequence of cloned nef was confirmed by DNA sequencing as above and also by Western blotting with Nef- and GST-specific antibodies. EGFP-Nef plasmid was constructed by cloning the complete GFP-Nef fusion gene sequence from pEGFPN2-Nef in EcoRI and NotI sites of pcDNA6HisC (Invitrogen). Cyclin K (CCNK) gene encoding the 354-amino acid isoform (30) was cloned in-frame into BamHI and EcoRI sites of pcDNA6HisC to obtain pcDNA6HisC-Cyclin K (pc-CycK). The cloning of pc-CycK was confirmed by restriction digestion and by DNA sequencing in an ABI 310 Genetic Analyzer (Applied Biosystems). The expression was confirmed by Western blotting using anti-Xpress epitope and CycK antibodies. HEK-293T cells (human embryonic kidney cell line) and Jurkat cells (CD4+ human T cell line) were obtained from the National Centre for Cell Science Cell Repository in India. CEM-GFP, a CD4+ human T cell line, was obtained from the National Institutes of Health AIDS repository. Jurkat-1G5 cells (CD4+ human T cell line) having a copy of integrated LTR-Luc in the genome were obtained from the National Institutes of Health AIDS Reagent Program. Antibodies specific for Cyclin K, Cyclin T1, CDK9, GAPDH, and HA tag were obtained from Santa Cruz Biotechnology. Anti-Nef monoclonal antibody was obtained from Fitzgerald, anti-Nef polyclonal serum was obtained from Dr. Shahid Jameel, International Centre for Genetic Engineering and Biotechnology, New Delhi, India (41), and anti-p24 gag polyclonal serum was obtained from the National Institutes of Health AIDS repository.
Yeast Two-hybrid Screening
A human leukocyte cDNA library in pACT2 vector (Clontech) was screened for Nef-interacting proteins by co-transformation with the pAS2-1NefC bait plasmid into yeast strain AH109 (Clontech) as described previously (23). Positive clones were selected based on growth in medium lacking adenine, histidine, tryptophan, and leucine and also by expression of β-galactosidase. Co-transformants in AH109 yeast strain were screened three times for growth on these selection plates and also for β-galactosidase activity to exclude false positive clones. A liquid β-galactosidase assay was finally performed to confirm the interaction using the yeast β-galactosidase assay kit (Pierce) according to the manufacturer's protocol. The interacting protein in the positive clone was identified by rescuing the gene fragment using PCR amplification with pACT2-specific primers followed by DNA sequencing and BLAST analysis.
HIV-1 Infection and Virus Quantitation
CEM-GFP cells (2 × 106) were infected with HIV-1NL4-3 virus at a multiplicity of infection of 0.5 in the presence of Polybrene (1 μg/ml) as described earlier (23). 24 h postinfection, cells were transfected with 200 nm siRNA, and then 48 h later, cells were harvested and lysed for preparation of whole cell lysate. The culture supernatants from infected and molecular clone-transfected cells were used to determine virus production by p24 gag antigen capture ELISA (Perkin Elmer Life Sciences).
Transient Transfection and Luciferase Assay
HEK-293T cells were transfected with HIV-1 LTR-reporter vector (pLTR-Luc) along with other expression vectors using the calcium phosphate precipitation method and were harvested 36 h post-transfection for luciferase assay. Jurkat-1G5 cells were transfected with pNL4-3 and pc-CycK using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and cells were harvested 48 h post-transfection for luciferase assay. The cells were then lysed in cell lysis reagent (Promega), and luciferase assays were performed using Luclite substrate (PerkinElmer Life Sciences). Normalization of transfection efficiency was done using enhanced green fluorescent protein reporter (pEGFP-N1) co-transfection and quantitation as described earlier (42). For RNAi experiments, Jurkat-1G5 cells were transfected with 100 nm siRNAs along with expression vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The culture supernatants were also collected at the time of luciferase assay from HIV-1 NL4-3 molecular clone-transfected cells to determine virus production.
GST Pulldown, Immunoprecipitation, and Immunoblotting
Escherichia coli BL21(DE3) cells expressing either GST or GST-Nef were induced with isopropyl β-d-thiogalactoside followed by purification of proteins using glutathione-Sepharose beads (Amersham Biosciences). Jurkat cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 5 mm EDTA, 0.12 m NaCl, 0.5% Nonidet P-40, 0.5 mm NaF, 1 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride) with a protease inhibitor mixture (Roche Applied Bioscience) on ice for 45 min. The clarified lysates were incubated with either GST or GST-Nef protein immobilized on glutathione-Sepharose beads at 4 °C and subjected to five washes with lysis buffer. The complexes were resuspended in Laemmli sample buffer, boiled, and resolved by 12% SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride (PVDF) membrane, and the membrane was probed with anti-CycK antibody.
Infected and uninfected CEM-GFP cells and co-transfected HEK-293T cells were lysed in lysis buffer as mentioned above. The clarified lysate was incubated with polyclonal antibody, and the antigen-antibody complex was pulled down by an equal mixture of Protein A and G beads followed by resolution by 12% SDS-PAGE. The proteins were transferred onto PVDF membrane, and then the membrane was probed with specific antibodies mentioned above. The blots were developed by using the ECL Plus system (Amersham Biosciences). Furthermore, equal amounts of protein were taken from cell lysates and were resolved by SDS-PAGE followed by immunoblotting for CycK and other proteins.
Immunofluorescence Microscopy
HEK-293T cells grown on coverslips were co-transfected with expression vectors using Lipofectamine (Invitrogen). Cells were harvested 24 h post-transfection and stained with CycK and Nef antibodies after fixing with 2% paraformaldehyde. The secondary antibodies used were Cy3-conjugated, Cy5-conjugated, and fluorescein isothiocyanate-conjugated specific antibodies (Chemicon). After washing, cells were mounted in mounting medium containing antifade reagent (Santa Cruz Biotechnology) on the slide, and the samples were analyzed with a confocal microscope (Zeiss LSM 510).
Reverse Transcription-PCR
RNA was prepared from HIV-1 NL4-3-infected and uninfected CEM-GFP cells or from Jurkat-1G5 cells using TRIzol reagent (Invitrogen). The cDNA was made using Moloney murine leukemia virus reverse transcriptase (Invitrogen) followed by amplification by PCR for CCNK, β-ACTIN, and HIV-p24 gene with Taq polymerase (Bangalore Genei) using standard conditions and gene-specific oligonucleotide primers. The primers used were as follows: CCNK: forward primer, 5′-TGCAAAAGCAACTCAAAGGTG-3′; reverse primer, 5′-AACAAACGCTCCCACCCTC-3′; β-ACTIN: forward primer, 5′-ATCTACCCGTGTCACACCCACTGGGGCAGT-3′; reverse primer, 5′-GGAGGTAGCAGGTGGCGTTTACGAAGATC3′; and HIV-p24: forward primer, 5′-ATAATCCACCTATCCCAGTAGGAGAAAT-3′; reverse primer, 5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′.
Preparation of Nuclear and Cytoplasmic Extracts
HIV-1-infected CEM-GFP and transfected HEK-293T cells were harvested and washed with ice-cold phosphate-buffered saline (PBS). Nuclear and cytoplasmic extracts were prepared using ProteoJET reagent (Fermentas) according to the manufacturer's protocol. Equal amounts of nuclear and cytoplasmic extracts were used for immunoblotting.
Statistical Analysis
Each individual experiment was repeated at least three times. The error bars in the figures represent the mean ± S.D. of three independent experiments. Statistical analysis of the experimental data was performed using Student's t test with the levels of significance defined as p < 0.05 (*) and p < 0.01 (**).
RESULTS
HIV-1 Nef protein interacts with a number of cellular proteins, most of which perform critical functions in signaling pathways. Although the role of Nef in HIV-1 pathogenesis has been well studied, the molecular mechanism of how Nef utilizes these interacting cellular factors to modulate viral replication and gene expression is still a conundrum. The functions attributed to Nef are believed to be conserved across various HIV-1 subtypes, and to date, the majority of the functional studies have been performed with subtype B Nef protein. As subtype C has become the most prevalent subtype in the world at present, we wanted to identify novel cellular factors, if any, that may interact with subtype C Nef protein. The subtype C Nef from Indian isolate IN301904 (37) used in the yeast two-hybrid screening was shown to have 84% similarity with subtype B NL4-3 Nef (23).
Identification of Cyclin K as Subtype C Nef-interacting Protein
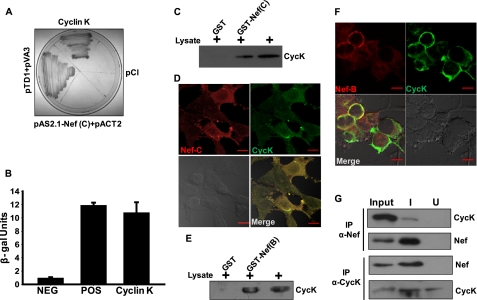
To identify cellular factors interacting with subtype C Nef, we performed a yeast-two hybrid screening of a human leukocyte cDNA library using full-length subtype C Nef as bait. Positive clones were identified after several rounds of screening and culture on X-gal-containing dropout plates (Fig. 1A). The gene expressed in the positive clone was amplified by PCR using pACT2-specific primers followed by DNA sequencing. Sequence analysis and database comparison revealed that the clone expressed a gene encoding for Cyclin K (GenBankTM Gene ID: 8812). To validate this interaction, a liquid β-galactosidase assay was performed, and the clone expressing Cyclin K and Nef showed β-galactosidase activity comparable with the positive control (Fig. 1B). To further confirm our finding, a GST-Nef (subtype C) pulldown assay was performed using Jurkat cell lysate as described under “Experimental Procedures.” The result of the pulldown clearly indicates that Cyclin K (CycK) interacts with subtype C Nef but not with GST (Fig. 1C). To see whether these proteins interact in vivo, immunofluorescence staining was performed in subtype C Nef-transfected 293T cells. Interestingly, these proteins co-localized primarily in the cytoplasm with some localization in the nucleus (Fig. 1D).
FIGURE 1.
Cyclin K physically interacts with HIV-1 Nef protein. A, Cyclin K interacts with HIV-1 subtype C Nef in the yeast two-hybrid assay. A human leukocyte cDNA library was screened using subtype C Nef as bait in the yeast two-hybrid system. One of the clones co-expressing Cyclin K and Nef was found to be positive after repeated screening and was grown on a quadruple knock-out plate with appropriate controls. B, β-galactosidase activity of Cyclin K-Nef yeast co-transformant in liquid β-galactosidase activity assay as described in the text. C, GST-Nef (subtype C) pulls down Cyclin K from Jurkat lysate. GST and GST-Nef (subtype C) were incubated with Jurkat cell lysate followed by SDS-PAGE and immunoblotting with CycK antibody. D, co-localization of Cyclin K and subtype C Nef in co-transfected HEK-293T cells. HEK-293T cells were grown on coverslips and transfected with pcDNA-NefC and pc-CycK. After 24 h, transfected cells were stained with Nef and Cyclin K antibodies as described in the text. The immunofluorescence was visualized by confocal microscopy. E, GST-Nef (subtype B) also pulls down Cyclin K from Jurkat lysate. GST and GST-Nef (subtype B) were incubated with Jurkat cell lysate followed by SDS-PAGE and immunoblotting with CycK antibody. F, co-localization of Cyclin K and subtype B Nef in co-transfected HEK-293T cells. HEK-293T cells were grown on coverslips and transfected with pNL4-3 and pc-CycK. The immunofluorescence staining and visualization were performed as described above. G, Nef and CycK interact in HIV-1-infected CEM-GFP cells. CEM-GFP cells were infected with HIV-1NL4-3 virus as described in the text. The cells were lysed on day 5 postinfection. The lysates of uninfected (U) and infected (I) CEM-GFP cells were co-immunoprecipitated (IP) with either Nef or CycK antibody followed by immunoblotting with CycK or Nef antibody, respectively, as presented in the figure. NEG, negative; POS, positive. The red lines in panels D and F indicate 10 μm scale.
Cyclin K Also Interacts with Subtype B Nef Both in Vitro and in Vivo
To test whether CycK also interacts with subtype B Nef, a GST pulldown assay was performed as described above. Jurkat cell lysate was incubated with bead-bound GST and GST-Nef (subtype B) followed by immunoblotting with Cyclin K antibody, which showed that CycK specifically interacts with GST-Nef but not with GST alone (Fig. 1E). Immunofluorescence staining was also performed in pNL4-3- and pc-CycK-co-transfected HEK-293T cells to confirm in vivo interaction of CycK with Subtype B Nef. Both proteins were found to be co-localized extensively in the cytoplasm as shown in Fig. 1F. We further validated this interaction in vivo by co-immunoprecipitation from uninfected and HIV-1NL4-3-infected CEM-GFP cells. Uninfected and HIV-1-infected cell lysates were immunoprecipitated with either Nef or Cyclin K antibody followed by immunoblotting with CycK or Nef antibody. The analyses of immunoprecipitates clearly suggest that CycK and Nef specifically interact in HIV-1-infected cells (Fig. 1G). The uninfected cell lysate served as a control in co-immunoprecipitation experiments. These results clearly suggest that Nef interacts with Cyclin K both in vitro and in vivo.
Cyclin K Inhibits HIV-1 LTR-driven Gene Expression in Presence of Nef
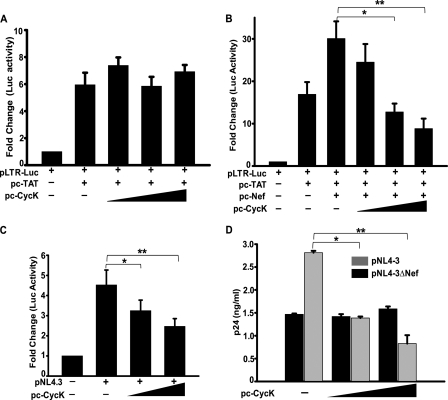
Cyclin K was first identified as a novel C type cyclin that is associated with an in vitro kinase activity toward CTD of RNA polymerase II (30). As this activity is important for HIV-1 gene expression, we wanted to examine the effect of CycK on HIV-1 LTR-driven viral gene expression. First, HEK-293T cells were co-transfected with the pLTR-Luc construct along with Tat and CycK expression vectors. As shown in Fig. 2A, overexpression of CycK in the presence of Tat alone did not cause any significant change in LTR-driven reporter activity. However, in the presence of Nef, Cyclin K overexpression led to inhibition of both Tat- and Nef-induced LTR-driven gene expression in 293T cells in dose-dependent manner (Fig. 2B). This result clearly shows that CycK inhibits HIV-1 LTR-driven gene expression in the presence of Nef. To explore this finding further, we then used Jurkat-1G5 cells, a human CD4+ reporter T-cell line having a copy of “integrated” LTR-Luc in the genome (43). Jurkat-1G5 cells were transfected with increasing amounts of CycK along with an HIV-1 molecular clone (pNL4-3) followed by a luciferase activity assay 48 h post-transfection. There was a significant reduction in luciferase activity upon CycK overexpression in a dose-dependent manner (Fig. 2C). Concurrently, Western blotting indicated that p24 expression in these cells was also reduced in a dose-dependent manner with increasing expression of CycK (data not shown). To further elucidate the effect of CycK on viral gene expression and replication, a p24 antigen capture ELISA of culture supernatant of these cells was also performed. Virus production in Cyclin K-overexpressing cells analyzed by p24 antigen release in the culture supernatant was significantly reduced in a dose-dependent manner as compared with pNL4-3 only-transfected cells (Fig. 2D, gray bars). Moreover, when Jurkat-1G5 cells were transfected with a Nef-deleted molecular clone (pNL4-3ΔNef), CycK-mediated reduction in virus production was not observed at all (Fig. 2D, black bars). These data clearly suggest that Cyclin K acts as a cellular inhibitory factor against HIV-1 by reducing viral gene expression and replication in a Nef-dependent manner.
FIGURE 2.
Cyclin K inhibits HIV-1 LTR-driven gene expression and replication in Nef-dependent manner. A, Cyclin K does not modulate LTR-driven luciferase expression in the presence of Tat alone. HEK-293T cells were transfected with HIV-1 LTR-Luc reporter vector along with pc-CycK in increasing amounts (0.3–1.2 μg) and with pcDNA-Tat (0.25 μg) followed by a luciferase assay 36 h post-transfection. The total amount of DNA transfected was equal in all lanes. B, Cyclin K down-regulates Tat-induced LTR-driven luciferase expression in the presence of Nef. HEK-293T cells were transfected with HIV-1 LTR-Luc reporter vector along with pc-CycK in increasing amounts (0.3–1.2 μg) and with pcDNA-Nef (0.5 μg) and pcDNA-Tat (0.25 μg). 36 h post-transfection, cells were lysed in Promega cell lysis reagent, and a luciferase assay was performed. C, Cyclin K inhibits integrated LTR-driven gene expression in T-cells. Jurkat-1G5 cells were transfected with pNL4-3 (1 μg) and increasing amounts of pc-CycK (0.25–0.5 μg). 48 h post-transfection, cells were lysed in Promega cell lysis reagent followed by a luciferase assay. D, Nef is necessary for Cyclin K-mediated inhibition of HIV-1 replication. Jurkat-1G5 cells were transfected with either pNL4-3 (gray bars) or pNL4-3ΔNef (black bars) and with increasing amounts of pc-CycK (0.25–0.5 μg) as presented in the figure. 48 h post-transfection, the culture supernatant was collected, and the amount of virus present in the culture supernatant was estimated by a p24 gag antigen capture ELISA.
Endogenous Cyclin K Reduces Viral Gene Expression and Replication in T-cells
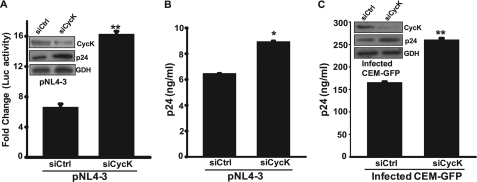
To identify the role of endogenous CycK in HIV-1 gene expression and replication, Jurkat-1G5 cells were co-transfected with pNL4-3 and specific siRNA to silence CycK expression as described under “Experimental Procedures.” After 72 h of transfection, cells were harvested, and culture supernatants were collected for estimation of virus production by a p24 antigen capture ELISA. The harvested cells were used for cell lysate preparation. The efficacy of siRNA was checked by immunoblotting, and the cell lysate was also used for a luciferase activity assay. As compared with control siRNA-transfected Jurkat-1G5 cells, transfection of CycK siRNA showed significant silencing of CycK expression (Fig. 3A, inset). The luciferase activity in CycK siRNA-transfected cells was significantly increased as compared with the control siRNA-transfected cells (Fig. 3A). The viral expression was also checked by immunoblotting for p24 gag protein. The expression level of p24 was higher in CycK siRNA-transfected cells as compared with control (Fig. 3A, inset). Furthermore, a similar result was obtained when virus production from siRNA-transfected cells was determined by a p24 antigen capture ELISA. The amount of virus in the culture supernatant of CycK siRNA-transfected cells was significantly increased as compared with control siRNA-transfected cells (Fig. 3B). These results clearly indicate that CycK down-regulation leads to an increase in HIV-1 gene expression and virus production.
FIGURE 3.
Cyclin K silencing increases LTR-mediated gene expression and virus production in T-cells. A, inhibition of Cyclin K expression by siRNA leads to an increase in LTR-driven luciferase activity in Jurkat-1G5 cells. Jurkat-1G5 cells were transfected with 100 nm control (siCtrl) siRNA or CycK-specific siRNA (siCycK), and 24 h post-transfection, cells were transfected with pNL4-3 vector (1 μg). 48 h post-transfection, cells were harvested, and a luciferase assay was performed. The inset shows the efficiency of siRNA-mediated Cyclin K silencing by immunoblotting. B, inhibition of Cyclin K expression by siRNA leads to an increase in virus production from Jurkat-1G5 cells. The virus released in the culture supernatant of cells in the experiment described in A was estimated by a p24 gag antigen capture ELISA. C, Cyclin K silencing increases virus production in HIV-1NL4-3-infected CEM-GFP cells. CEM-GFP cells were first transfected with 200 nm control or CycK-specific siRNA. 24 h post-transfection, cells were infected with HIV-1NL4-3 at 0.5 multiplicity of infection, and on day 3, the culture supernatant was used for a p24 antigen capture ELISA. The inset shows the efficacy of siRNA-mediated silencing by immunoblotting. GDH, Glyceraldehyde-3-phosphate dehydrogenase loading control.
To assess the role of Cyclin K in a more physiological scenario, we then performed the above experiment in an HIV-1-infected CEM-GFP reporter T-cell line. The CEM-GFP cells were first transfected with siRNA, and after 24 h, they were infected with HIV-1NL4-3 at 0.5 multiplicity of infection. On day 3 postinfection, cells were harvested to monitor progression of infection by flow cytometry. The cells transfected with CycK siRNA showed a higher number of GFP-positive cells, indicating increased infection, as compared with control siRNA-transfected cells (data not shown). Analysis of the virus present in culture supernatant by a p24 antigen capture ELISA showed significantly more virus production by CycK siRNA-transfected cells as compared with the control siRNA-transfected cells (Fig. 3C). A similar pattern was obtained by immunoblotting for p24 (Fig. 3C, inset), thereby indicating that CycK acts as an inhibitory factor in HIV-1-infected cells.
Cyclin Box 1 Is Required for CycK-mediated Reduction of LTR-driven Gene Expression
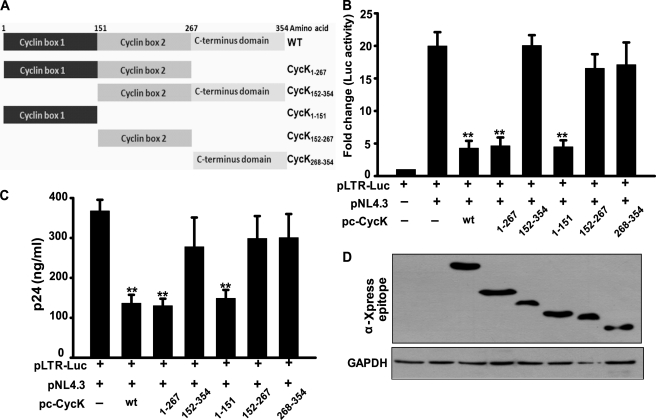
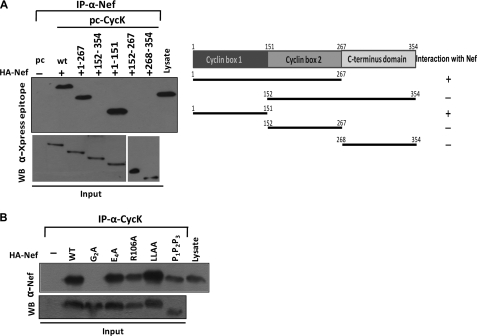
Human CycK is a 354-amino acid protein; however, a less abundant isoform of CycK consisting of 580 amino acids also exists. The overall structure of CycK is similar to other cyclin structures. It consists of 10 helical domains, which are arranged in two cyclin boxes along with two short helices (HNa and HNb) preceding the N-terminal cyclin box. The five helices comprising the N- and C-terminal cyclin domains are designated as H1–H5 and H1′–H5′, respectively. The N-terminal domain (amino acids 1–151) of CycK is responsible for binding to CDK9; however, the two N-terminal helices HNa and HNb are not essential (44). The region from amino acids 152 to 267 forms cyclin box 2, and the region comprising amino acids 268–354 can be considered as the C terminus of the protein (Fig. 4A). Different deletion mutants of Cyclin K, as shown in Fig. 4A, were cloned in pcDNA6HisC.
FIGURE 4.
Cyclin box 1 domain of Cyclin K is necessary for its inhibitory activity on LTR-driven gene expression and HIV-1 virus production. A, schematic representation of Cyclin K mutants generated and used in the present study. B, cyclin box 1 domain is required for the inhibition of HIV-1 LTR-driven luciferase activity. HEK-293T cells were transfected with LTR-Luc reporter and pNL4-3 molecular clone along with various CycK mutant expression vectors as indicated in the figure. 24 h post-transfection, cells were harvested followed by a luciferase assay of the lysate. C, cyclin box 1 domain is required for the inhibition of HIV-1 virus production. HEK-293T cells were transfected with LTR-Luc reporter and pNL4-3 molecular clone along with various CycK mutant expression vectors as indicated in the figure. 24 h post-transfection, cell supernatants were used to analyze the virus production by a p24 antigen capture ELISA. D, expression of different Cyclin K mutant proteins in transfected HEK-293T cells. HEK-293T cell lysates were examined for the expression of various CycK mutants using Xpress epitope antibody and GAPDH by Western blot analysis in the experiments in B and C.
To investigate the effect of different CycK mutants on LTR-mediated gene expression, HEK-293T cells were transfected with pLTR-Luc reporter plasmid along with pNL4-3 and wild type or different CycK mutants followed by a luciferase activity assay. Upon wild type CycK overexpression (Fig. 4B, third bar), the luciferase activity was reduced by ∼5-fold as compared with that of vector control (second bar). A similar pattern was observed when CycK(1–267) (fourth bar) and CycK(1–151) (sixth bar) mutants were used in the experiment (Fig. 4B). However, no significant change in luciferase activity was observed when cyclin box 1 deletion mutants like CycK(152–354), CycK(152–267), and CycK(268–354) were co-transfected with LTR-Luc and pNL4-3 vectors (Fig. 4B), indicating that the N-terminal end of Cyclin K was important for CycK activity, whereas the C-terminal end was not required for this activity. The amount of virus produced in culture supernatant of these transfected cells was also estimated, and a similar pattern was observed, indicating that amino acids 1–151 of CycK were important for CycK-mediated reduction in virus production (Fig. 4C). These results suggest that cyclin box 1 is responsible for the inhibitory activity of Cyclin K. The expression levels of Cyclin K wild type and the mutants in the above experiment were confirmed by immunoblotting with anti-Xpress epitope antibody (Fig. 4D).
Domains/Motifs Responsible for CycK-Nef Interaction
To find the region of Cyclin K important for interaction with Nef, immunoprecipitation of wild type or CycK mutants and Nef-transfected HEK-293T cell lysate was performed with anti-Nef antibody followed by immunoblotting of the complex with anti-Xpress epitope antibody. Our results clearly show that Nef specifically interacts with only wild type CycK and the two mutants having the region comprising amino acids 1–151 (Fig. 5A). These data suggest that domain 1–151 is responsible for the Nef binding property of CycK, which correlates well with the inhibitory activity of CycK delineated in the earlier figure (Fig. 4C). It has been reported that Nef has some signature motifs responsible for binding to various cellular proteins (45). To explore the motif of Nef important for binding to CycK, we used a set of Nef point mutants (40). HEK-293T cells were co-transfected with HA-Nef mutant expression vectors along with CycK-WT expression vector. The cell lysates were then subjected to immunoprecipitation with CycK antibody followed by immunoblotting with Nef antibody. The co-immunoprecipitation analysis indicates that myristoylation of Nef is required for CycK interaction as complete loss of interaction was observed when the glycine at the second position, which is critical for myristoylation, was replaced with alanine in Nef (Nef-G2A mutant) (Fig. 5B).
FIGURE 5.
Mutational analysis of CycK-Nef interaction. A, cyclin box 1 of Cyclin K is important for its interaction with Nef. HEK-293T cells were transfected with HA-Nef expression vector along with various CycK mutant vectors as indicated in the figure. 24 h post-transfection, immunoprecipitation (IP) was performed with Nef antibody followed by Western blotting (WB) with Xpress tag antibody for Cyclin K. The right side shows a schematic representation of CycK mutants and the results of their interaction with HIV-1 Nef protein. The lower panels show the immunoblotting of input samples. B, G2A mutation in Nef leads to abrogation of Nef-Cyclin K interaction. HEK-293T cells were transfected with HA-Nef mutant expression vector along with CycK vector as indicated in the figure. 24 h post-transfection, immunoprecipitation was performed with Cyclin K antibody followed by Western blot analysis with Nef antibody. The lower panel shows the immunoblotting of input samples.
Nef Induces Cyclin K Interaction with CDK9
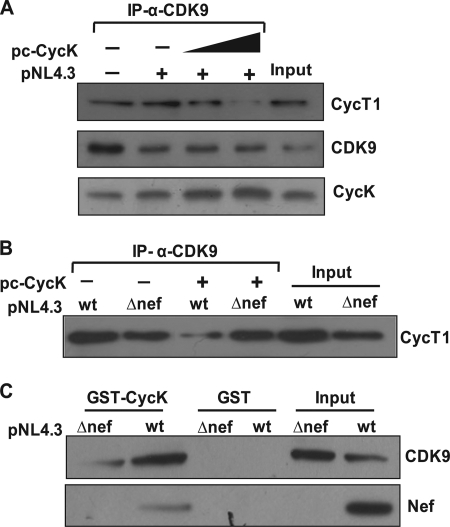
The CDK9-CycT1 complex (P-TEFb complex) plays a central role in HIV-1 LTR-driven gene expression (46). Both CycK and CycT1 are known to bind CDK9 through their N-terminal cyclin box of which 53 of 80 residues are identical (17, 44). Based on this report, we initiated studies to elucidate the mechanism of CycK-mediated reduction in viral gene expression. To test whether the presence of CycK alters the physical association between CycT1 and CDK9, we performed a co-immunoprecipitation experiment by transfecting pNL4-3 along with increasing amounts of CycK expression vector. The co-immunoprecipitation experiment revealed that in the presence of increasing levels of CycK in pNL4-3-transfected cells CycK interaction with CDK9 was increased, whereas CycT1 association with CDK9 was reduced in a dose-dependent manner (Fig. 6A). This result suggests that the increase in CycK levels in the cell reduces the formation of the CDK9-CycT1 complex, which is an active protein complex required for HIV-1 LTR-driven gene expression.
FIGURE 6.
Cyclin K overexpression leads to increased binding with CDK9 and inhibition of CDK9-Cyclin T1 interaction in Nef-dependent manner. A, Cyclin K binds to CDK9 and inhibits CDK9-Cyclin T1 interaction in pNL4-3-transfected cells. HEK-293T cells were co-transfected with pNL4-3 and CycK expression vector as shown in the figure. 200 μg of lysate was immunoprecipitated (IP) with CDK9 antibody followed by immunoblotting with CycT1, CycK, and CDK9 antibodies. B, Cyclin K inhibits CDK9-Cyclin T1 interaction in a Nef-dependent manner. HEK-293T cells were transfected with pNL4-3 or pNL4-3ΔNef along with pc-CycK. 150 μg of cell lysate was immunoprecipitated with CDK9 antibody followed by immunoblotting with CycT1 antibody. C, Cyclin K binding to CDK9 is increased in the presence of Nef. HEK-293T cells were transfected with pNL4-3 or pNL4-3ΔNef. 150 μg of lysate was incubated with an equal amount of bead-bound GST and GST-CycK protein for a GST pulldown assay as described in the text. The pulldown was detected by immunoblotting with Nef and CDK9 antibodies.
To demonstrate the role of Nef in the context of CycK-mediated inhibition of CycT1-CDK9 interaction, we tested whether the presence of Nef is essential for this CycK function. We performed a co-immunoprecipitation experiment with wild type and a Nef-deleted NL4-3 molecular clone (pNL4-3ΔNef). Our results clearly show that CycK overexpression leads to inhibition of CycT1-CDK9 interaction only in the presence of Nef, and there was no significant change in CycT1-CDK9 interaction in the cells transfected with Nef-deleted molecular clone (Fig. 6B). Thus, our co-immunoprecipitation experiments indicate that CycK associates with CDK9 in the presence of Nef and thereby prevents association of CDK9 with CycT1. We further validated this finding by performing a GST pulldown assay using bead-bound GST-CycK to pull down CDK9 from pNL4-3- and pNL4-3ΔNef-transfected HEK-293T cell lysate. Western blot analysis of the GST-CycK pulldown complex clearly indicates that in the presence of Nef CDK9 pulldown was significantly greater as compared with that in the absence of Nef (Fig. 6C). All these results clearly suggest that CycK physically interacts with CDK9 in the presence of Nef, and this interaction prevents CDK9-CycT1 complex formation.
Cyclin K Binding to CDK9 Inhibits Its Nuclear Translocation in Presence of Nef
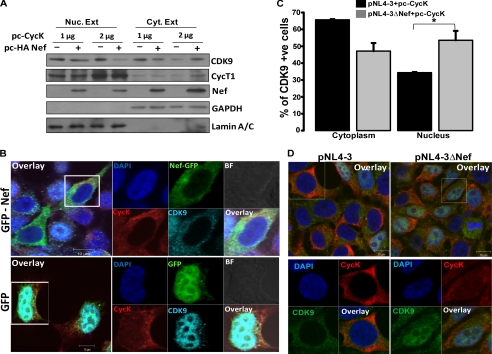
Previous studies have indicated that CDK9 shuttles between the nucleus and cytoplasm, and enhanced expression of CycT1 increases CDK9 nuclear translocation, suggesting a role for CycT1 in transporting CDK9 into the nucleus (47). The localization of CycK has not been studied in detail. However, when the sequences of CDK9, CycK, and CycT1 were analyzed to predict the presence of a nuclear localization signal, we found that only CycT1 has a nuclear localization signal sequence. So based on the above information, we hypothesized that inhibition of CDK9-CycT1 complex formation by CycK could be responsible for restricting CDK9 from translocating into the nucleus to form active P-TEFb complex and thus causing reduction in viral gene expression. We therefore analyzed whether ectopic expression of CycK inhibits CDK9 nuclear translocation. We performed Western analysis of nuclear and cytoplasmic extracts of HEK-293T cells transfected with Nef and different amounts of CycK. It was clearly evident that the presence of Nef and CycK significantly reduced CDK9 translocation into the nucleus as analyzed by immunoblotting (Fig. 7A). This experiment also showed that with increasing CycK expression there was a substantial decrease in CDK9 levels in the nucleus (Fig. 7A). To further substantiate this finding, we transfected HEK-293T cells with EGFP-Nef and CycK expression vectors to perform an immunofluorescence study. In EGFP-Nef-transfected cells, CDK9 was evidently co-localized in the cytoplasm with CycK and EGFP-Nef (Fig. 7B). When EGFP alone was used as a control, CDK9 was visible mostly in the nucleus. Thus, the CycK-Nef complex may act as an inhibitory complex, which may restrict viral replication by binding to CDK9 and sequestering it in the cytoplasm. To further confirm the above mentioned finding, we directly examined the localization of CDK9 in pNL4-3- and pNL4-3ΔNef-transfected cells in the presence of CycK by immunofluorescence. Cells were counted, and the percentage of cells with CDK9 localized in the cytoplasm and nucleus was calculated for three independent experiments. The results presented in Fig. 7C clearly show that CDK9 was localized more in the nucleus of pNL4-3ΔNef-transfected cells as compared with pNL4-3-transfected cells. Representative immunofluorescence data are shown in Fig. 7D. Put together, all these results clearly indicate that CycK inhibits nuclear translocation of CDK9 in the presence of Nef.
FIGURE 7.
Cyclin K restricts CDK9 nuclear translocation in presence of Nef. A, Cyclin K and Nef overexpression restricts CDK9 from nuclear translocation. HEK-293T cells were co-transfected with increasing amounts of pc-CycK with or without HA-Nef expression vector as indicated in the figure. Equal amounts of nuclear (Nuc. Ext) and cytoplasmic extracts (Cyt. Ext) were used for immunoblotting with CDK9, CycT1, and Nef antibodies as described in the text. The same blot was probed for Lamin A/C and GAPDH to serve as loading controls. B, Cyclin K and Nef overexpression inhibits CDK9 nuclear translocation as visualized by immunofluorescence. HEK-293T cells were grown on a coverslip and co-transfected with pc-CycK and EGFP-Nef or EGFP control vectors. After 24 h, cells were stained for CycK and CDK9 as described in the text. Cells were then mounted onto a glass slide and visualized by a confocal microscope. C, CDK9 translocation into the nucleus is increased in the cells transfected with the Nef-deleted molecular clone. HEK-293T cells were grown on a coverslip and were transfected with pNL4-3 or pNL4-3ΔNef along with pc-CycK. Immunofluorescence staining was performed with CDK9 and CycK antibodies. Cells stained for CDK9 in the nucleus or cytoplasm were counted and plotted as a bar diagram with data from three independent experiments. Black and gray bars indicate CDK9-stained cells transfected with pNL4-3 and pNL4-3ΔNef, respectively. D, representative immunofluorescence images of CDK9 localization in the experiment described in C. Left and right panels are immunofluorescence images of cells transfected with pNL4-3 and pNL4-3ΔNef along with pc-CycK, respectively. BF, bright field; +ve, positive.
Endogenous Cyclin K Restricts CDK9 Nuclear Translocation in HIV-1-infected Cells
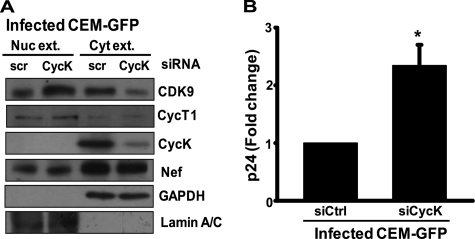
For further confirmation of the above findings, we then investigated the localization of CDK9 in HIV-1-infected T-cells in which Cyclin K expression was silenced. We made nuclear and cytoplasmic extracts of CycK-silenced and HIV-1-infected CEM-GFP cells and performed immunoblotting. The infected CEM-GFP cells that were transfected with CycK-specific siRNA had more CDK9 in the nucleus as compared with control siRNA-transfected cells (Fig. 8A). Simultaneously, we checked for virus production in the culture supernatant of these cells by a p24 antigen capture ELISA, and as shown in Fig. 8B, the virus produced from CycK-silenced infected cells was significantly increased as compared with control siRNA-transfected cells. These data clearly suggest that CycK in the presence of Nef restricts viral gene expression and replication by binding to CDK9 and inhibiting its translocation into the nucleus. The inhibition of nuclear translocation of CDK9 seems to prevent formation of active P-TEFb complex necessary for HIV-1 LTR-driven gene expression and replication.
FIGURE 8.
Cyclin K silencing leads to increased CDK9 nuclear translocation and virus production in HIV-1-infected CEM-GFP T-cells. A, Cyclin K down-regulation leads to increased CDK9 translocation into the nucleus of HIV-1-infected T cells. CEM-GFP cells were transfected with 200 nm control (siCtrl) and CycK-specific siRNA (siCycK). 24 h post-transfection, cells were infected with HIV-1NL4-3 at 0.5 multiplicity of infection; on day 3, cells were harvested; and nuclear and cytoplasmic fractions were prepared as mentioned under “Experimental Procedures.” Equal amounts of extracts were used for immunoblotting with CDK9, CycT1, CycK, and Nef antibodies. GAPDH and Lamin A/C were used as loading controls. B, Cyclin K down-regulation leads to increased virus production from HIV-1-infected T-cells. The amount of virus present in the culture supernatant of the cells described in A was estimated by a p24 antigen capture ELISA. Nuc. Ext, nuclear extract; Cyt. Ext, cytoplasmic extract; scr, scrambled.
DISCUSSION
The dependence of HIV-1 on host cellular factors is well established. The multifunctional protein Nef is capable of imparting extensive aid for HIV-1 pathogenesis by making a niche in the form of an interactome. The Nef interactome consists of multiple branches leading to critical cellular functions like receptor down-modulation, cell signaling, cell migration, apoptosis, and transcription regulation. To date, Nef has been shown to interact with several important cellular factors, and new proteins are being added frequently, unraveling its novel functions. In the present study, we show for the first time that HIV-1 Nef interacts with Cyclin K, which was identified by yeast two-hybrid screening and was further confirmed by immunoprecipitation and co-localization studies. This interaction is not subtype-specific as both subtypes B and C Nef bind to CycK. Our data seem to corroborate previous reports (48), suggesting that despite significant sequence differences in the Nef gene from various subtypes functional properties remain relatively well conserved.
CycK was first identified as a protein that could restore progression through the cell cycle and was most closely related to human cyclins C and H (30). The kinase partner of CycK was identified as CDK9 and was reported to function as a CTD kinase in vitro (17). Recently Cyclin K was also shown to interact with CDK12 and -13 (31), but its functional implication remains to be clearly elucidated. CycT1 on the other hand is an obligate cyclin partner of CDK9, and this complex (P-TEFb) plays a key role in HIV-1 gene expression (19). As both CycK and CycT1 bind to CDK9, it was reasonable to speculate a possible role of Nef-CycK interaction in HIV-1 LTR-driven gene expression. Our results clearly show that CycK reduces LTR-driven gene expression in a dose-dependent manner. Moreover, CycK was able to reduce LTR activity only in the presence of Nef, which suggests an apparent role of Nef-CycK interaction in this phenomenon. Furthermore, CycK ectopic expression in Jurkat-1G5 cells was able to reduce LTR-mediated gene expression and thus virus production. Additionally, when endogenous CycK was knocked down using specific siRNA, the viral gene expression was enhanced, and virus particle release was also augmented in Jurkat-1G5 cells. Hence, our findings convincingly indicate that CycK might possess an HIV restricting property and that it is not a cell type-dependent phenomenon.
Endogenous CycK seems to substantially reduce HIV-1 gene expression and virus production as was evident in RNAi experiments performed with HIV-1NL4-3-infected CEM-GFP cells. In a report in 2008, Urano et al. (29) demonstrated that CycK overexpression leads to reduction in HIV-1 production from T-cell lines; however, the mechanism behind CycK-mediated reduction of HIV-1 gene expression remained unexplored. Our co-immunoprecipitation experiment along with immunofluorescence studies confirmed that CycK not only binds to CDK9 in the presence of Nef but also reduces binding of CycT1 to CDK9 and thereby restricts CDK9 translocation into the nucleus. It is worth mentioning here that our mutational studies indicate CycK binding to only myristoylated Nef. Thus, it might be hypothesized that membrane-associated Nef-CycK complex only interacts with CDK9 and thereby restricts it from translocating into the nucleus. Moreover, immunofluorescence studies with wild type and a nef-deleted HIV-1 molecular clone demonstrated that Nef is essential for CycK-mediated restriction of CDK9 nuclear translocation. Furthermore, immunoblotting analyses of nuclear and cytoplasmic extracts of CycK-specific siRNA-treated HIV-1-infected CEM-GFP cells showed increased translocation of CDK9 into the nucleus and increased virus production.
Although CDK9-CycK forms an active kinase complex that possesses CTD phosphorylating activity in vitro (17), its role in HIV-1 transcription has not been studied. CycK and CycT1 have 53 identical residues in the cyclin homology box domain, which is responsible for binding to CDK9 (44); hence, the possibility of competition between CycK and CycT1 for binding to CDK9 cannot be ruled out. The Tat-interacting region, also known as the Tat-responsive motif, located outside the cyclin homology box of CycT1, is not present in CycK (49, 50), and thus a possible role of the CDK9-CycK complex in modulating HIV-1 transcription can be ruled out. To function as a part of an active P-TEFb complex for HIV-1 gene transcription, CDK9 needs to be translocated into the nucleus, and it shuttles between the nucleus and cytoplasm under the influence of CycT1 levels in the cell (47). However, if CDK9 is restricted in the cytoplasm, the amount of active P-TEFb complex in the nucleus will be reduced, leading to inhibition of transcription. The present work in the context of Nef-CycK interaction has shown convincingly that CDK9 binding to CycK increases in the presence of Nef, thereby reducing its binding to the obligate partner CycT1. Hence, it may be hypothesized that binding to Nef is responsible for the increased affinity of CycK toward CDK9.
Thus, the present study provides insight into the molecular mechanism of CycK-mediated reduction in HIV-1 gene expression and replication. In summary, we conclude that Nef interacts with CycK, and the resulting complex leads to reduction in HIV-1 LTR-driven gene expression by interfering with CDK9 binding to CycT1 and its translocation into the nucleus. Our data also indicate that CycK function is not cell type-specific, and endogenous CycK is capable of reducing viral gene expression. Finally, our results also suggest that Nef-CycK interaction leads to reduction in the formation of active P-TEFb complex, consequently inhibiting HIV-1 LTR-driven viral gene expression and replication. Based upon our findings, we propose a model depicting how Nef-Cyclin K interaction leads to restriction of CDK9 in the cytoplasm, thereby reducing the formation of active CDK9-CycT1 complex and consequently reducing HIV-1 gene expression and virus production (Fig. 9). It is interesting to note that a recent report finds that CycK along with CDK9 binds to checkpoint proteins and thus plays an important role in replication-induced stress (51). Finally, identification of CycK as a novel Nef-interacting host factor and the consequences of this interaction provide another example for utilization of a viral protein by the host in combating the pathogen.
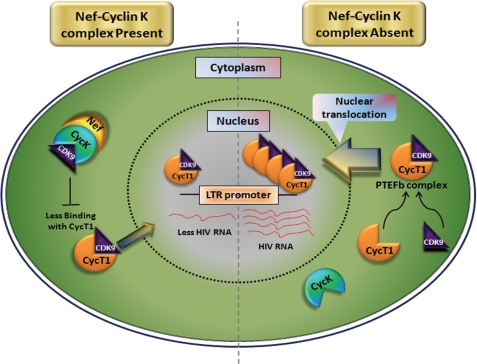
FIGURE 9.
Proposed mechanistic model for Cyclin K-mediated inhibition of HIV-1 gene expression and replication. The results obtained in the present study indicate that in the presence of the Nef-Cyclin K complex CDK9-CycT1 interaction is abrogated, causing a reduction in active P-TEFb complex in the nucleus and resulting in inhibition of HIV-1 LTR-driven gene expression and replication. However, in the absence of the Nef-Cyclin K complex, CDK9 is free to bind to CycT1 and translocate into the nucleus, consequently leading to enhanced viral gene expression and replication.
Acknowledgments
We thank Dr. G. C. Mishra, Director, National Centre for Cell Science, for support and constant encouragement. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: pNL4-3 plasmid, p24 antibody, and CEM-GFP and Jurkat-1G5 cell lines. We thank Dr. J. C. Guatelli, Dr. K. Saksela, Dr. W. C. Greene, and Dr. S. Jameel for providing different reagents as detailed in the text. We also thank Dr. M. Kumar for help with the yeast two-hybrid assay.
This work was supported by the Department of Biotechnology, Government of India.
- LTR
- long terminal repeat
- CDK9
- cyclin-dependent kinase 9
- HEXIM1
- hexamethylene bisacetamide-inducible protein 1
- CTD
- C-terminal domain
- Cyc
- Cyclin
- P-TEFb
- positive elongation factor b
- Luc
- luciferase
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Colin L., Van Lint C. (2009) Retrovirology 6, 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Streeck H., Brumme Z. L., Anastario M., Cohen K. W., Jolin J. S., Meier A., Brumme C. J., Rosenberg E. S., Alter G., Allen T. M., Walker B. D., Altfeld M. (2008) PLoS Med. 5, e100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boutwell C. L., Rolland M. M., Herbeck J. T., Mullins J. I., Allen T. M. (2010) J. Infect. Dis. 202, Suppl. 2, S309–S314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson W. E., Desrosiers R. C. (2002) Annu. Rev. Med. 53, 499–518 [DOI] [PubMed] [Google Scholar]
- 5. Kamp W., Berk M. B., Visser C. J., Nottet H. S. (2000) Eur. J. Clin. Invest. 30, 740–746 [DOI] [PubMed] [Google Scholar]
- 6. Geyer M., Fackler O. T., Peterlin B. M. (2001) EMBO Rep. 2, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saksela K. (1997) Front. Biosci. 2, d606–d618 [DOI] [PubMed] [Google Scholar]
- 8. Simmons A., Aluvihare V., McMichael A. (2001) Immunity 14, 763–777 [DOI] [PubMed] [Google Scholar]
- 9. Doms R. W., Trono D. (2000) Genes Dev. 14, 2677–2688 [DOI] [PubMed] [Google Scholar]
- 10. Fackler O. T., Baur A. S. (2002) Immunity 16, 493–497 [DOI] [PubMed] [Google Scholar]
- 11. Arora V. K., Fredericksen B. L., Garcia J. V. (2002) Microbes Infect. 4, 189–199 [DOI] [PubMed] [Google Scholar]
- 12. Ahmad N., Venkatesan S. (1988) Science 241, 1481–1485 [DOI] [PubMed] [Google Scholar]
- 13. Fackler O. T., d'Aloja P., Baur A. S., Federico M., Peterlin B. M. (2001) J. Virol. 75, 6601–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collette Y., Olive D. (1999) Virology 265, 173–177 [DOI] [PubMed] [Google Scholar]
- 15. Miller M. D., Warmerdam M. T., Gaston I., Greene W. C., Feinberg M. B. (1994) J. Exp. Med. 179, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joseph A. M., Ladha J. S., Mojamdar M., Mitra D. (2003) FEBS Lett. 548, 37–42 [DOI] [PubMed] [Google Scholar]
- 17. Fu T. J., Peng J., Lee G., Price D. H., Flores O. (1999) J. Biol. Chem. 274, 34527–34530 [DOI] [PubMed] [Google Scholar]
- 18. Peng J., Marshall N. F., Price D. H. (1998) J. Biol. Chem. 273, 13855–13860 [DOI] [PubMed] [Google Scholar]
- 19. Price D. H. (2000) Mol. Cell. Biol. 20, 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mancebo H. S., Lee G., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Flores O. (1997) Genes Dev. 11, 2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M. B., Price D. H. (1997) Genes Dev. 11, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolf D., Witte V., Clark P., Blume K., Lichtenheld M. G., Baur A. S. (2008) Cell Host Microbe 4, 398–408 [DOI] [PubMed] [Google Scholar]
- 23. Kumar M., Mitra D. (2005) J. Biol. Chem. 280, 40041–40050 [DOI] [PubMed] [Google Scholar]
- 24. Shimizu S., Urano E., Futahashi Y., Miyauchi K., Isogai M., Matsuda Z., Nohtomi K., Onogi T., Takebe Y., Yamamoto N., Komano J. (2007) AIDS 21, 575–582 [DOI] [PubMed] [Google Scholar]
- 25. Iordanskiy S., Zhao Y., DiMarzio P., Agostini I., Dubrovsky L., Bukrinsky M. (2004) Blood 104, 1867–1872 [DOI] [PubMed] [Google Scholar]
- 26. Yik J. H., Chen R., Pezda A. C., Samford C. S., Zhou Q. (2004) Mol. Cell. Biol. 24, 5094–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barboric M., Kohoutek J., Price J. P., Blazek D., Price D. H., Peterlin B. M. (2005) EMBO J. 24, 4291–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulte A., Czudnochowski N., Barboric M., Schönichen A., Blazek D., Peterlin B. M., Geyer M. (2005) J. Biol. Chem. 280, 24968–24977 [DOI] [PubMed] [Google Scholar]
- 29. Urano E., Shimizu S., Futahashi Y., Hamatake M., Morikawa Y., Takahashi N., Fukazawa H., Yamamoto N., Komano J. (2008) AIDS 22, 1081–1083 [DOI] [PubMed] [Google Scholar]
- 30. Edwards M. C., Wong C., Elledge S. J. (1998) Mol. Cell. Biol. 18, 4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. (2010) Genes Dev. 24, 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niederman T. M., Thielan B. J., Ratner L. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glushakova S., Grivel J. C., Suryanarayana K., Meylan P., Lifson J. D., Desrosiers R., Margolis L. (1999) J. Virol. 73, 3968–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joseph A. M., Kumar M., Mitra D. (2005) Curr. HIV Res. 3, 87–94 [DOI] [PubMed] [Google Scholar]
- 35. Lundquist C. A., Zhou J., Aiken C. (2004) J. Virol. 78, 6287–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papkalla A., Münch J., Otto C., Kirchhoff F. (2002) J. Virol. 76, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., Ingersoll R., Sheppard H. W., Ray S. C. (1999) J. Virol. 73, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chowers M. Y., Spina C. A., Kwoh T. J., Fitch N. J., Richman D. D., Guatelli J. C. (1994) J. Virol. 68, 2906–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stoddart C. A., Geleziunas R., Ferrell S., Linquist-Stepps V., Moreno M. E., Bare C., Xu W., Yonemoto W., Bresnahan P. A., McCune J. M., Greene W. C. (2003) J. Virol. 77, 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chaudhry A., Das S. R., Hussain A., Mayor S., George A., Bal V., Jameel S., Rath S. (2005) J. Immunol. 175, 4566–4574 [DOI] [PubMed] [Google Scholar]
- 42. Dandekar D. H., Kumar M., Ladha J. S., Ganesh K. N., Mitra D. (2005) Anal. Biochem. 342, 341–344 [DOI] [PubMed] [Google Scholar]
- 43. Aguilar-Cordova E., Chinen J., Donehower L., Lewis D. E., Belmont J. W. (1994) AIDS Res. Hum. Retrovir. 10, 295–301 [DOI] [PubMed] [Google Scholar]
- 44. Baek K., Brown R. S., Birrane G., Ladias J. A. (2007) J. Mol. Biol. 366, 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Das S. R., Jameel S. (2005) Indian J. Med. Res. 121, 315–332 [PubMed] [Google Scholar]
- 46. Jones K. A., Peterlin B. M. (1994) Annu. Rev. Biochem. 63, 717–743 [DOI] [PubMed] [Google Scholar]
- 47. Napolitano G., Licciardo P., Carbone R., Majello B., Lania L. (2002) J. Cell. Physiol. 192, 209–215 [DOI] [PubMed] [Google Scholar]
- 48. Jubier-Maurin V., Saragosti S., Perret J. L., Mpoudi E., Esu-Williams E., Mulanga C., Liegeois F., Ekwalanga M., Delaporte E., Peeters M. (1999) AIDS Res. Hum. Retrovir. 15, 23–32 [DOI] [PubMed] [Google Scholar]
- 49. Bieniasz P. D., Grdina T. A., Bogerd H. P., Cullen B. R. (1998) EMBO J. 17, 7056–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garriga J., Peng J., Parreño M., Price D. H., Henderson E. E., Graña X. (1998) Oncogene 17, 3093–3102 [DOI] [PubMed] [Google Scholar]
- 51. Yu D. S., Zhao R., Hsu E. L., Cayer J., Ye F., Guo Y., Shyr Y., Cortez D. (2010) EMBO Rep. 11, 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]