Abstract
Each segment of the influenza A virus (IAV) genome contains conserved sequences at the 5′- and 3′-terminal ends, which form the promoter region necessary for polymerase binding and initiation of RNA synthesis. Although several models of interaction have been proposed it remains unclear if these two short, partially complementary, and highly conserved sequences can form a stable RNA duplex at physiological temperatures. First, our time-resolved FRET analysis revealed that a 14-mer 3′-RNA and a 15-mer 5′-RNA associate in solution, even at 42 °C. We also found that a nonfunctional RNA promoter containing the 3′-G3U mutation, as well as a promoter containing the compensatory 3′-G3U/C8A mutations, was able to form a duplex as efficiently as wild type. Second, UV melting analysis demonstrated that the wild-type and mutant RNA duplexes have similar stabilities in solution. We also observed an increase in thermostability for a looped promoter structure. The absence of differences in the stability and binding kinetics between wild type and a nonfunctional sequence suggests that the IAV promoter can be functionally inactivated without losing the capability to form a stable RNA duplex. Finally, using uridine specific chemical probing combined with mass spectrometry, we confirmed that the 5′ and 3′ sequences form a duplex which protects both RNAs from chemical modification, consistent with the previously published panhandle structure. These data support that these short, conserved promoter sequences form a stable complex at physiological temperatures, and this complex likely is important for polymerase recognition and viral replication.
Keywords: Fluorescence Resonance Energy Transfer (FRET), Mass Spectrometry (MS), Promoters, RNA Folding, RNA Viruses
Introduction
Influenza A virus (IAV)2 belongs to the Orthomyxoviridae family and is the causative agent of both seasonal and pandemic influenza outbreaks. The IAV genome is composed of 8 segments of negative sense RNA and encodes an RNA-dependent RNA polymerase. IAV RNA polymerase is a trimeric complex, composed of two basic subunits, PB1 and PB2, as well as an acidic subunit, PA. This enzyme carries out both transcription of viral mRNAs and replication, producing (+) complementary RNAs from the incoming (−) viral genomes (vRNA) and then new (−) vRNAs (1). Although the IAV genome does not have a DNA stage, viral replication takes place in the host cell nucleus (2). During transcription, the virus uses host capped pre-mRNAs as primers for initiation of viral mRNA synthesis by binding the cap structure and cleaving a 10–13 nucleotide primer, which is then extended by the IAV polymerase complex (3). In contrast, replication of the genome occurs through a primer independent manner to generate a full length complement of the vRNA. Interestingly, though only the 3′-end of the genome serves as a template for initiation of RNA polymerization, both transcription and replication require the polymerase to be bound to both the 3′- and 5′-terminal ends of the vRNA segment, forming a looped structure. These terminal RNA sequences serve as a promoter for the initiation of RNA synthesis (4).
Influenza genomes are known to be highly variable as viral strains accumulate mutations over time and can also reassort. This facilitates viral host switch and adaptation, resulting in novel, possibly pandemic IAV strains (5). Despite this genetic variability, the sequences of the IAV promoter are highly conserved between strains (6). Indeed, the conserved 13 and 12 nucleotide promoter sequences are found at the 5′- and 3′-ends, respectively, of every vRNA segment in virtually every strain of the virus. The only known exception is a single variation in the 3′-sequence, U4C. It is found on segments encoding the polymerase proteins and neuraminidase in a few strains, and may play a role in regulating protein expression (7).
Although this viral promoter, composed of the short 5′- and 3′-end regions, is necessary for the initiation of RNA synthesis, the structure and mechanism of IAV polymerase recognition remain unclear and rather controversial. An NMR structure supports a panhandle-like duplex of the IAV promoter RNA, though this structure was obtained at 4 °C using the two promoter sequences connected by a tetraloop (8). This structure predicts base pairs between both the proximal and terminal ends of these sequences with a small internal loop. In contrast, other groups have evaluated the sequence and base pairing requirements for viral reporter gene expression from this promoter (9–13). Their work suggests that the sequences form a corkscrew like structure when bound by the polymerase, with only a small Watson-Crick paired region and hairpin loops forming in both the 3′- and 5′-sequences. This corkscrew conformation would be unlikely to form in solution as the helices leading to the hairpin loops are short (two basepairs). It has been postulated that the polymerase first binds to the 5′-promoter region and then binds the 3′-promoter region (12). This model is supported by a study showing that purified ribonucleoprotein particles no longer form a looped structure once stripped of the polymerase complex (15). This study however does not rule out more transient RNA interactions, which may have biological significance. It is also worth noting that the sequential model of interaction has been called into question by another study indicating that the polymerase is more active when the 5′- and 3′-promoter regions are added simultaneously rather than sequentially (14). This suggests that the RNA may form a structure in the absence of protein that is sufficient for polymerase recognition and may play a role in regulating polymerase activity.
The native, physiologic structure of the influenza promoter remains unclear. As noted above, formation of a panhandle-like duplex was observed when the promoter regions were linked by synthetic loop sequences considerably smaller than viral genomic segments (8, 16). This, along with low temperatures, could have facilitated the interaction of the promoter sequences. We therefore used multiple biophysical methodologies to investigate the interaction of the conserved short 5′- and 3′-promoter RNAs in solution, under more physiologic conditions. We also evaluated the impact of two different mutations in the 3′-sequence on the promoter duplex: (i) a 3′-G3U mutation that inactivates IAV promoter function in reporter gene assays and (ii) a compensatory mutation, C8A that restores wild-type levels of gene expression (12).
EXPERIMENTAL PROCEDURES
RNA Sequences and Labels
RNA oligonucleotides were synthesized by Integrated DNA Technologies. For time-resolved Förster Resonance Energy Transfer (trFRET) experiments, the 5′-vRNA sequence was synthesized with a Cy3 label on the 5′-end. The 3′-vRNA sequences were synthesized with a Cy5 label on the 3′-end. The sequences used are shown in Figs. 1A and 2A. The sequences are shown and numbered starting from the terminal ends of the promoter such that the 5′-vRNA sequence is written 5′ to 3′ while the 3′-vRNA sequence is written 3′ to 5′. For optical melting, a looped RNA was also used that contained the 5′- and 3′-sequences connected by a UUCG tetraloop.
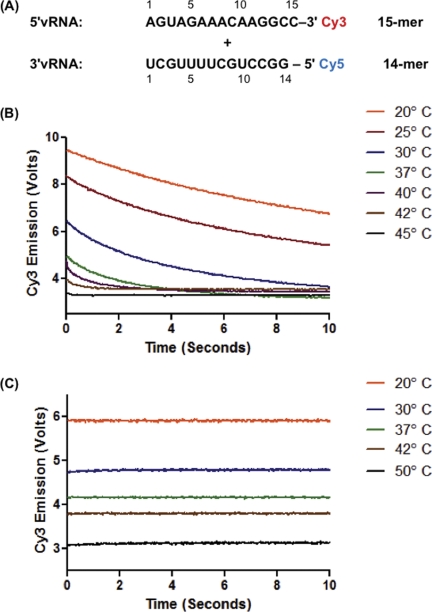
FIGURE 1.
Conserved influenza A promoter RNA sequences interact in solution at biological temperatures. A, sequences of the 5′-vRNA and 3′-vRNA promoters with Cy3 donor and Cy5 acceptor fluorophores. Numbering is from the terminal end of the promoter such that the 5′-vRNA sequence is written 5′ to 3′ while the 3′-vRNA sequence is written 3′ to 5′. B, 5′-vRNA and 3′-vRNA (100 nm each) were mixed at indicated temperatures and the donor (Cy3) emission was measured over 10 s. C, poly(rA) control oligonucleotide labeled with Cy3 was mixed with the 3′-vRNA using the same conditions as in B.
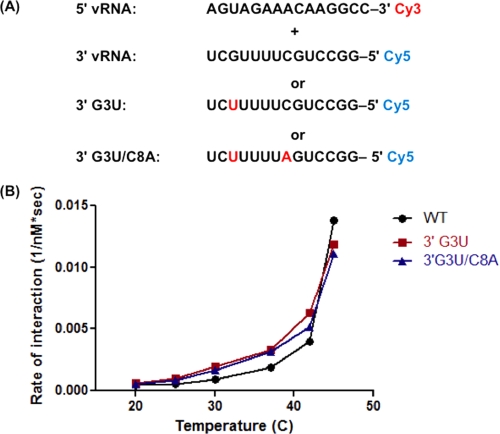
FIGURE 2.
Mutations in the 3′-vRNA sequence do not prevent interaction of promoter RNAs. A, sequences and fluorophores used. B, sequences shown in A were mixed, and the Cy3 emission was recorded as shown in Fig. 1B. Four replicates of Cy3 emission were averaged and fit to a single exponential curve to determine the rate of interaction. Rates from three independent experiments were averaged and plotted versus temperature.
Time-resolved FRET Measurements
The fluorophore-labeled 5′-vRNA or 3′-vRNA was diluted to 200 nm in 50 mm HEPES pH 7.8 containing 5 mm MgCl2 and 150 mm KCl. This buffer was used for all subsequent experiments as well. All trFRET measurements were carried out using a SF2004 Stopped Flow Fluorimeter (Kintek Inc.). The two RNAs were loaded into independent syringe chambers. The RNAs were then pushed into a temperature controlled observation cell where the Cy3 donor fluorophore was excited with 545 nm light. The emission of Cy3 donor and Cy5 acceptor were detected with two photomultiplier tubes using 580 and 680 ± 10 nm bandpass filters, respectively. The emission of both fluorophores was measured at 1000 time points over 15 s. This was repeated at temperatures ranging from 20 to 45 °C. FRET was observed as a decrease in Cy3 emission and an increase in Cy5 emission. As a control for photobleaching, a 15-mer poly(rA) oligonucleotide labeled with Cy3 was used in place of the 5′-vRNA using the same conditions. Emission traces shown are an average of three replicates. To determine the rate of RNA interaction, the Cy3 emission intensity over time was fitted to a single exponential equation and the rate of decrease in Cy3 emission was taken as the rate of interaction of the 5′-vRNA and 3′-vRNA. The rate was plotted versus temperature for all figures. This rate also corresponded to the rate of increase in the Cy5 acceptor though the magnitude of change was smaller. Three independent experiments were averaged to obtain rates. Only Cy3 emission was used for analysis.
UV Melting
The 5′-vRNA and 3′-vRNA sequences used for FRET were synthesized for further experiments without the fluorophore label (Integrated DNA Technologies Inc.). Melting experiments were carried out as described (17). Briefly, the 5′-vRNA and 3′-vRNA were mixed at equal concentrations based on their A280 before being concentrated by a centrifugal vacuum. Samples were then resuspended in the previously described buffer. Melting was also measured for each sequence individually as well as a looped promoter similar to that used for the NMR structure (8). Optical melting measurements were done at 280 nm with a heating rate of 1 °C/min from 0 to 90 °C on a Beckman DU 640 spectrophotometer and repeated using decreasing concentrations of RNA. Because of the use of magnesium in the buffer, which leads to hydrolysis at high temperatures, new samples were used for each melt. Data were analyzed, and thermodynamic parameters were derived using MeltWin 3.5 (17).
Secondary Structure Predictions
Secondary structures shown in Fig. 4 were predicted by free energy minimization using RNAstructure v5.2 (18). The structures predicted to be more stable at 37 °C are shown.
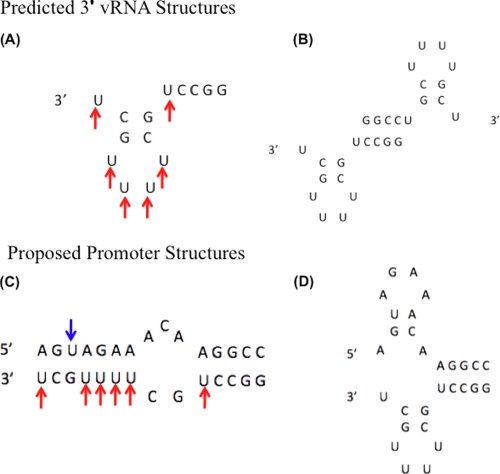
FIGURE 4.
Predicted and proposed RNA secondary structures. A and B, predicted structures formed by the 3′-vRNA sequence in the absence of the 5′-vRNA. C, panhandle structure predicted to form with 5′-vRNA and 3′-vRNA. D, functional corkscrew model of promoter when bound by polymerase complex (6). Arrows indicate uridines in 3′-vRNA (red) and 5′-vRNA (blue) sequences, which are potential sites for CMCT modification if unpaired.
Chemical Probing of Promoter RNA
1-Cyclohexyl-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate (CMCT) was purchased from Sigma Aldrich. 200 picomoles of each RNA were diluted in 18 μl of buffer. The RNA was annealed by heating at 65 °C for 2 min followed by slow cooling to 25 °C. 2 μl of 250 mm CMCT were added to the annealed RNA. Although CMCT preferentially reacts with unpaired uridines, it can also modify uridines at the end of helices as well as unpaired guanosines with higher concentrations and longer incubation times (19). To ensure modification of only unpaired uridines the RNA was incubated with 25 mm CMCT for 15 min. The reaction was then stopped with the addition of 10 μl of 5 m sodium acetate and 150 μl of 100% ethanol. The RNA was precipitated at −80 °C for at least 2 h then pelleted by centrifugation at 16,000 × g. This precipitation step was repeated three times to remove remaining salt from the RNA. The RNA was resuspended in 3 μl of RNase-free water before MALDI-TOF MS analysis.
MALDI-TOF Mass Spectometry
1 μl of a matrix solution containing 20 mm 3-hydroxypicolinic acid (MS grade), 50 mm diammonium citrate (ultra pure MS grade), and 25% acetonitrile (HPLC grade) was mixed with 1 μl of RNA on a steel target plate (Bruker) and allowed to dry completely. MS measurements were done with a Bruker Autoflex III Smartbeam. Singly charged RNA oligonucleotides were detected with the negative ion polarity mode. Three RNA standards (a 5-mer, 12-mer, and 20-mer) were used for calibration. Analysis was carried out using FlexAnalysis software (Bruker).
RESULTS
Time-resolved FRET Analysis for Detecting Formation of IAV Promoter
First, we tested if the conserved terminal ends of the viral RNA genome can interact independently in solution without IAV RNA polymerase or other factors using a trFRET based system. As shown in Fig. 1A, a 15-mer RNA encoding the 5′-end IAV sequence (5′-vRNA) was conjugated at its 3′-end to Cy3, which serves as a donor fluorophore, and a 14-mer RNA encoding the 3′-end viral sequence (3′-vRNA) was conjugated to the Cy5 acceptor fluorophore at its 5′-end. This trFRET system allowed for detection of the initial association of the two independent RNAs by monitoring the emission of both Cy3 and Cy5 on a millisecond time scale. The two labeled RNAs were mixed at temperatures ranging from 20° to 45 °C. RNA duplex formation was monitored by the decrease in emission intensity of the Cy3 donor and the increase in emission intensity of the Cy5 acceptor over time. As shown by Cy3 emission in Fig. 1B, FRET was detected up to 42 °C, indicating that the 5′-vRNA and 3′-vRNA physically interact in solution. Above 42 °C, no significant change in donor fluorescence over time was observed, indicating that the RNAs are no longer interacting at these higher temperatures. As a control for photobleaching and the effect of temperature on emission, a Cy3-labeled 15-mer poly(rA) was mixed with the Cy5-labeled 3′-vRNA. As shown in Fig. 1C, this control experiment showed no decrease in donor emission upon mixing and similar decreases in donor emission at time 0 due to increased temperature. This control experiment confirms that the decrease in Cy3 emission shown in Fig. 1B is due to the molecular interaction between the two IAV promoter RNAs, and more importantly, that the interaction can occur at physiological temperatures without other viral and cellular factors.
Effect of a 3′-vRNA Mutation Known to Impair Viral Promoter Function on Formation of an RNA Duplex in Solution
Next, we investigated the effects of two previously characterized promoter mutations on duplex formation, 3′-G3U and 3′-G3U/C8A (Fig. 2A). It was previously shown that the 3′-G3U mutation inactivated IAV promoter function whereas the combined 3′-G3U/C8A mutations, which restored a proposed base pair in the corkscrew model, showed wild-type levels of reporter gene expression (12). Our trFRET analysis revealed that both 3′-G3U and 3′-G3U/C8A mutant RNAs were able to form a duplex with the 5′-vRNA at temperatures up to 42 °C, as observed for the wild-type sequences (data not shown). To examine whether these 3′-vRNA mutations affect the kinetics of promoter duplex formation, the rates of change in donor emission were analyzed for both wild type and mutant promoters. As shown in Fig. 2B, the two mutated 3′-vRNA sequences formed a duplex slightly faster than the wild-type sequence. Overall, this demonstrates that the 3′-G3U sequence efficiently forms a duplex structure and that the lack of promoter activity is not due to the failure of the RNAs to associate.
Effect of the 3′-G3U Mutation on the Stability of the Promoter RNA Duplex
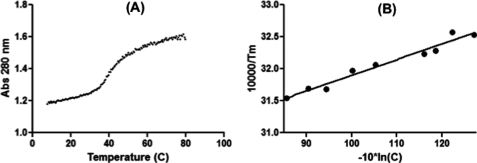
One possible reason for the lack of promoter activity associated with the 3′-G3U promoter is that it forms a thermodynamically less stable duplex than the wild type sequence. To determine the thermodynamic parameters of the promoter RNA duplex, optical melting measurements were performed. This technique measures the UV absorbance of RNA as a function of temperature to calculate the free energy change (ΔG°), entropy (ΔS°), and enthalpy (ΔH°), as well as the melting temperature (Tm) for structure formation (20). First, the stability of the conserved wild-type 5′-vRNA and 3′-vRNA duplex was determined. A representative melting curve of a single RNA concentration is shown in Fig. 3A. Nine concentrations were used to derive a van't Hoff plot of the concentration dependence of melting temperature (Fig. 3B). By averaging the results of the curve fits and fitting the van't Hoff plot, two sets of thermodynamic parameters can be derived. The entropy (ΔS°) and enthalpy (ΔH°) values are used to calculate the free energy change (ΔG°). Importantly, as shown in Table 1, these methods yielded ΔH° values for the 5′-vRNA and 3′-vRNA, which agree within 15% indicating that there is likely a transition from a single structured state to an unstructured state. This confirms that the viral promoter RNAs interact at biologically relevant temperatures with a Tm of 42.2 °C (at 1 × 10−4 m total oligonucleotide concentration). We also confirmed the relevance of our system by using a looped RNA harboring both 5′- and 3′-promoter sequences, which is similar to that used in the published NMR structure (8). Indeed, the addition of a tetraloop to our promoter sequences resulted in the apparent Tm increasing to 49.4 °C. This added stabilization is expected and further confirms that the panhandle structure is stable at physiological temperatures. However, further thermodynamic characterization of this hairpin could not be performed, as this structure does not melt cooperatively. The stem region close to the loop likely melts at a higher temperatures than the less stable terminal stem. This leads to more complex melting curves that cannot be fit by the currently available software (20).
FIGURE 3.
UV melting of promoter duplex. A, 5′-vRNA and 3′-vRNA sequences were combined and the absorbance at 280 nm was monitored over increasing temperatures. Shown is a representative melt curve. B, van't Hoff plot of UV melting data. Melt curves were recorded for nine concentrations of RNA and then the dependence of 1/Tm on concentration (C) was plotted.
TABLE 1.
Summary of UV melting data
| Sequence | TM−1 vs log(CT/a)a |
Average of curve fits |
||||||
|---|---|---|---|---|---|---|---|---|
| −ΔG°37 | −ΔH°37 | −ΔS° | TMb | −ΔG°37 | −ΔH°37 | −ΔS° | TMb | |
| kcal/mol | kcal/mol | eu | °C | kcal/mol | kcal/mol | eu | °C | |
| 5′-vRNA/3′-vRNA | 7.88 ± 0.11 | 81. ± 5.8 | 235.9 ± 18.7 | 42.2 | 7.77 ± 0.24 | 70.8 ± 10.1 | 203.1 ± 32.0 | 42.5 |
| 5′-vRNA/3′-G3U | 8.75 ± 0.13 | 64.8 ± 4.2 | 180.5 ± 13.1 | 48.0 | 8.65 ± 0.34 | 63.8 ± 10.3 | 177.8 ± 32.2 | 47.7 |
| 5′-vRNA/3′-G3U,C8A | 8.70 ± 0.10 | 78.3 ± 4.6 | 224.5 ± 14.6 | 45.9 | 8.72 ± 0.26 | 85.2 ± 11.4 | 246.6 ± 36.0 | 45.2 |
| 5′-vRNA | NDc | NDc | ||||||
| 3′-vRNA | 8.05 ± 0.13 | 88.12 ± 5.39 | 258.3 ± 17.02 | 46.6 | 7.34 ± 0.37 | 51.3 ± 7.92 | 41.7 ± 25.60 | 47.4 |
a a is 1 for self-complementary and 4 for non-self-complementary.
b Calculated for an RNA concentration of 1.0 × 10−4 m.
c No secondary structures were detected.
This thermodynamic analysis was also used to compare the stability of duplexes formed by the mutated promoter sequences as well as to determine whether the 5′-vRNA or 3′-vRNA sequences form any self-complementary structure. The thermodynamic parameters obtained are summarized in Table 1. Both 3′-G3U and 3′-G3U/C8A promoters yielded structures with a slightly higher Tm and more favorable ΔG° than the wild-type promoter, supporting that these mutant promoters do not form less stable duplexes than the wild-type promoter. This is consistent with the trFRET data indicating that both mutant 3′-vRNAs form a duplex with the 5′-vRNA more efficiently than the wild-type RNA (Fig. 2). It is not clear at this point if this increased stability is significant enough to have an impact on polymerase binding or initiation of transcription.
Interestingly, the 5′-vRNA formed no detectable self-complementary structure while the 3′-vRNA formed a structure with a Tm of 45.6 °C and a ΔG° of −8.05 kcal/mol at 37 °C, though the lack of agreement between the average of the curve fits and the van't Hoff plot suggests that more than one structure is forming. However, given the good agreement of the thermodynamic data for the combination of the 3′-vRNA and 5′-vRNA suggests that any alternative 3′-vRNA structures are likely only a small proportion of the structured RNA when both the sequences are present. We employed secondary structure prediction software to model the self-structure formation of the 3′-vRNA sequence. This analysis yielded two possible structures. The first is a hairpin loop with base pairing between nucleotides 2 and 9 as well as nucleotides 3 and 8 (Fig. 4A). This hairpin loop could also form a dimer with the palindromic CCGG stretch (Fig. 4B). These alternative structures may explain the slower rate of association for the wild-type RNA at low temperatures observed in Fig. 2B. These 3′-vRNA self-structures may also be destabilized by the G3U and G3U/C8A 3′ sequences, which could explain the slightly higher Tm observed for these sequences when compared with the wild-type sequence (Table 1). In summary, the data presented in Fig. 3 demonstrate that the 3′-G3U and G3U/C8A mutations do not decrease, and possibly increase, the stability of the promoter duplex.
MALDI-TOF MS Analysis with CMCT Chemical Probing to Examine the Structure of the Promoter Duplex in Solution
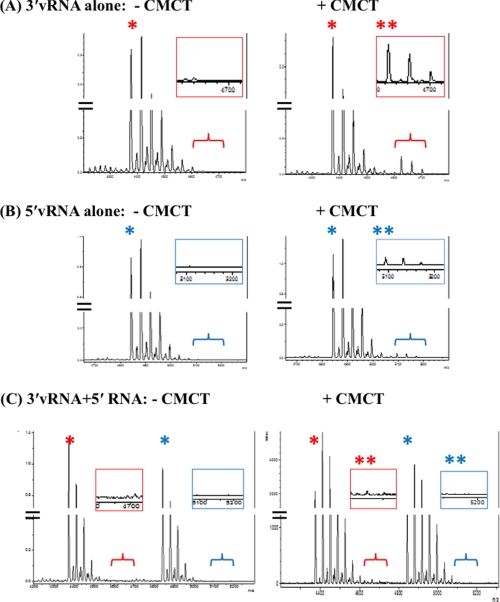
We next investigated if the predicted structures of the IAV promoter discussed above form in solution under our experimental conditions. For this we employed a chemical probing method combined with MALDI-TOF MS. In this analysis, CMCT chemically modifies unpaired uridines resulting in modified RNAs with a higher mass than the unmodified RNAs. These molecular weight (M.W.) shifts can be detected by MS analysis (21). As expected from the sequence (Fig. 1A), CMCT should be able to modify the 5′-vRNA sequence in solution, as UV melting experiments (Table 1) indicated it is not structured. Although UV melting revealed structure in the 3′-vRNA sequence, both predicted secondary structures (Fig. 4, A and B) have several unpaired uridines that would be accessible to modification. This was confirmed using 25 mm CMCT incubated with either the 5′-vRNA or 3′-vRNA at 37 °C for 15 min. CMCT modifications were then detected with MALDI-TOF MS. As shown in Fig. 5A, the spectrum of unmodified 3′-vRNA showed a peak at M.W. 4,375 (see “*”), with several slightly higher molecular weight peaks due to salt binding. When incubated with CMCT, a peak appeared at M.W. 4,627 (“**”). This corresponds to a single CMCT modification leading to a 252 Da increase in mass, again together with several higher M.W. peaks due to salt binding. A similar labeling pattern was observed for the 5′-vRNA alone (Fig. 5B) with a peak at M.W. 4,843 and several salt-bound peaks. When incubated with CMCT peaks were observed corresponding to an increase in mass due to modification (see “**” in inset). This confirms that under these conditions unpaired uridines in both the 3′-vRNA and 5′-vRNA are modified by CMCT.
FIGURE 5.
Chemical probing of promoter sequences with CMCT to modify unpaired uridines. X-axis is mass to charge ratio. Y-axis is arbitrary unit of intensity. A, 3′-vRNA: Unmodified (left). Incubated with CMCT (right). B, 5′-vRNA: Unmodified (left). Incubated with CMCT (right) (C) 5′-vRNA and 3′-vRNA annealed: Unmodified (left). Incubated with CMCT (right). * indicates peak corresponding to mass of unlabeled RNA. ** indicates where peaks corresponding to a CMCT-modified RNA would be observed.
The predicted panhandle duplex (Fig. 4C) has no unpaired uridines and thus we predicted that the CMCT modification observed with the individual RNAs seen in Fig. 5, A and B should not be detected if this duplex is forming in the solution. This would also confirm that the corkscrew conformation (Fig. 4D) is not present in solution. When 5′-vRNA and 3′-vRNA were annealed (Fig. 5C, left panel) both 5′-vRNA specific and 3′-vRNA specific MS peaks, identical to those observed for the unmodified individual RNAs (left panels of Fig. 5, A and B), were observed. However, when the 5′-vRNA and 3′-vRNA duplex was incubated with CMCT, the distinct higher M.W. peaks corresponding to a modified uridine (see “**” in right panels of Fig. 5, A and B), were no longer detected (Fig. 5C, right panel). Thus, this chemical probing analysis suggests that the IAV promoter RNAs form a panhandle-like duplex in solution which protects the uridines in both strands from CMCT modification.
DISCUSSION
While morbidity and mortality due to influenza continue to have a large impact on public health, the molecular mechanisms underlying many steps of influenza A virus replication are not well understood. Although only the 3′-end of the viral genome segment is necessary as a template for RNA synthesis, the polymerase complex is not active without being bound to both the 5′- and 3′-terminal ends of the vRNA. Indeed, these terminal sequences are highly conserved among IAV strains and essential for viral replication (4).
However, it remained unclear if the promoter RNAs are able to form a duplex without stabilization by cellular or viral factors at physiologic temperatures. Indeed, data from the trFRET analysis (Fig. 1) and UV melting (Fig. 3) clearly suggest that the 5′-vRNA and 3′-vRNA can form a duplex in solution at temperatures at which viral replication occurs. We also confirmed that the looped RNA used in the NMR study is structured at physiological temperatures. These data, along with the chemical probing data (Fig. 5), suggest that the panhandle duplex structure, which was observed at 4 °C by the NMR analysis (8), is likely present at higher, more biologically relevant temperatures, and thus may play a role in initiation of RNA synthesis and/or polymerase binding during viral assembly. This information is valuable for any further studies using these sequences including evaluating the impact of the 3′ mutations on polymerase binding.
The sequence and base pairing requirements for a functional promoter have been studied using mutagenesis (12–13). Importantly, it is not clear which step(s) of IAV RNA synthesis the characterized mutations impede. A single base change in the 3′ sequence (G3U) drastically reduced promoter activity in a reporter gene assay, but this does not appear to be due to the inability of this sequence to form an RNA duplex in this study. Indeed, both the nonfunctional G3U and functional G3U/C8A 3′ sequences interacted with the 5′-vRNA sequence with similar rates (Fig. 2B) and did not show decreased stability (Table 1). One possible explanation is that there is a difference in the way the nonfunctional sequence is bound by the polymerase complex.
The previous studies using a reporter gene assay found that there was not a strict sequence requirement for the promoter, but rather that base pairing was necessary to form the corkscrew structure. This fits with a sequential model of the RNA interacting with the polymerase complex (12). However, given this lack of sequence requirement, it is surprising that this region is highly conserved. Possibly, this region may need to form two stable structures during the course of viral replication. A panhandle-like duplex may form between the terminal ends of newly synthesized genome segments. This structure may then be bound by the polymerase complex, facilitating the transition to the corkscrew formation of the RNA. This transition also leaves the 3′-end unpaired, allowing for the re-initiation of the next round of RNA synthesis. It has also been shown that the functional activities of the viral polymerase are differentially activated when it binds to the promoter sequences sequentially versus when it can bind the promoter sequence simultaneously (14). The formation of an RNA duplex in solution could therefore be important for regulating the different activities of the polymerase (such as capped mRNA binding and endonuclease activity), but this may be missed when using a reporter gene assay.
Previous work used crosslinking of the promoter RNA to the polymerase to estimate binding affinity (22). Given the possibility of a conformational change between two RNA structures, it seems likely that a mutation such as the 3′-G3U, which still forms a duplex, may not affect the initial binding of the polymerase complex. It could, however, affect the stability of the corkscrew conformation of the promoter, possibly leading to a failure to initiate RNA synthesis. In addition, the promoter sequences of IAV gene segments are only partially complementary and a fully double-stranded promoter is not functional and will not be bound by the polymerase (23). This supports the idea that a less stable duplex may facilitate the transition between these two RNA promoter structures (Fig. 4, C and D) during the initiation of RNA synthesis. Finally, while the biophysical data presented in this study support the formation of a panhandle-like structure in solution at physiological temperatures, much work is needed to mechanistically understand what factors influence the regulation of viral mRNA production versus genome replication, as well as assembly of the viral polymerase and RNA segments for packaging.
This work was supported, in whole or in part, by a National Institutes of Health (NIH) contract granted to the New York Influenza Center of Excellence (NIH/NIAID HHSN266200700008C), the Oral Cellular and Molecular Biology Training Grant (T32 DE007202), GM22939 (to D. H. T.), and R01HG004002 (to D. H. M.).
- IAV
- influenza A virus
- FRET
- Förster resonance energy transfer
- CMCT
- 1-cyclohexyl-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate.
REFERENCES
- 1. Ishihama A. (1996) Biochimie 78, 1097–1102 [DOI] [PubMed] [Google Scholar]
- 2. Herz C., Stavnezer E., Krug R., Gurney T., Jr. (1981) Cell 26, 391–400 [DOI] [PubMed] [Google Scholar]
- 3. Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. (1981) Cell 23, 847–858 [DOI] [PubMed] [Google Scholar]
- 4. Fodor E., Pritlove D. C., Brownlee G. G. (1994) J. Virol. 68, 4092–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kobasa D., Kawaoka Y. (2005) Curr. Mol. Med. 5, 791–803 [DOI] [PubMed] [Google Scholar]
- 6. Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8140–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee K. H., Seong B. L. (1998) J. Gen. Virol. 79, 1923–1934 [DOI] [PubMed] [Google Scholar]
- 8. Bae S. H., Cheong H. K., Lee J. H., Cheong C., Kainosho M., Choi B. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim H. J., Fodor E., Brownlee G. G., Seong B. L. (1997) J. Gen. Virol. 78, 353–357 [DOI] [PubMed] [Google Scholar]
- 10. Leahy M. B., Dobbyn H. C., Brownlee G. G. (2001) J. Virol. 75, 7042–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leahy M. B., Pritlove D. C., Poon L. L., Brownlee G. G. (2001) J. Virol. 75, 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flick R., Hobom G. (1999) J. Gen. Virol. 80, 2565–2572 [DOI] [PubMed] [Google Scholar]
- 13. Flick R., Neumann G., Hoffmann E., Neumeier E., Hobom G. (1996) RNA. 2, 1046–1057 [PMC free article] [PubMed] [Google Scholar]
- 14. Cianci C., Tiley L., Krystal M. (1995) J. Virol. 69, 3995–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klumpp K., Ruigrok R. W., Baudin F. (1997) EMBO. J. 16, 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baudin F., Bach C., Cusack S., Ruigrok R. W. (1994) EMBO. J. 13, 3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McDowell J. A., Turner D. H. (1996) Biochemistry. 35, 14077–14089 [DOI] [PubMed] [Google Scholar]
- 18. Reuter J. S., Mathews D. H. (2010) BMC. Bioinformatics. 11, 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. (1987) Nucleic. Acids Res. 15, 9109–9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schroeder S. J., Turner D. H. (2009) Methods Enzymol. 468, 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turner K. B., Yi-Brunozzi H. Y., Brinson R. G., Marino J. P., Fabris D., Le Grice S. F. (2009) RNA. 15, 1605–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. González S., Ortin J. (1999) J. Virol. 73, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiley L. S., Hagen M., Matthews J. T., Krystal M. (1994) J. Virol. 68, 5108–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]