Abstract
Proteinase-activated receptor 1 (PAR1) induces activation of platelet and vascular cells after proteolytic cleavage of its extracellular N terminus by thrombin. In pathological situations, other proteinases may be generated in the circulation and might modify the responses of PAR1 by cleaving extracellular domains. In this study, epitope-tagged wild-type human PAR1 (hPAR1) and a panel of N-linked glycosylation-deficient mutant receptors were permanently expressed in epithelial cells (Kirsten murine sarcoma virus-transformed rat kidney cells and CHO cells). We have analyzed the role of N-linked glycosylation in regulating proteinase activation/disarming and cell global expression of hPAR1. We reported for the first time that glycosylation in the N terminus of hPAR1 downstream of the tethered ligand (especially Asn75) governs receptor disarming to trypsin, thermolysin, and the neutrophil proteinases elastase and proteinase 3 but not cathepsin G. In addition, hPAR1 is heavily N-linked glycosylated and sialylated in epithelial cell lines, and glycosylation occurs at all five consensus sites, namely, Asn35, Asn62, Asn75, Asn250, and Asn259. Removing these N-linked glycosylation sequons affected hPAR1 cell surface expression to varying degrees, and N-linked glycosylation at extracellular loop 2 (especially Asn250) of hPAR1 is essential for optimal receptor cell surface expression and receptor stability.
Keywords: Calcium Imaging, G Protein-coupled Receptors (GPCRs), Glycosylation, Post-translational Modification, Thrombin, Neutrophil Proteinase, Proteinase-activated Receptor 1, Thermolysin
Introduction
Proteinases can exert many biological effects on cells by virtue of their ability to cleave a plethora of protein substrates. Our understanding of proteinase signaling has advanced since the discovery of a small family of G protein-coupled receptors termed proteinase-activated receptors (PARs)2 (1–3). Classically, thrombin activates PAR1 by proteolytic cleavage of the PAR1 N-terminal domain between Arg41 and Ser42 to expose a new N terminus that acts as a tethered ligand and subsequently triggers receptor activation (2). Thrombin is a serine proteinase generated at the sites of vascular injury during the coagulation cascade and activates various types of cells in the vasculature like platelets and endothelial cells (2, 3). In addition to thrombin, other proteinases may be generated in the circulation under pathological situations; any proteinase that cleaves the correct peptide bond within the N terminus of PAR1 may be able to expose the tethered ligand and then initiate intracellular signaling to provoke a cellular response (1, 2, 4–8). Conversely, proteinases can also disarm PAR1 by proteolytic removal of the tethered ligand domain from the receptor (4, 9–14). Such disarmed receptors remain at the cell surface but are no longer available for activation by activating proteinases (e.g. thrombin); they can, however, be activated by the corresponding PAR1 activating peptides (1, 2, 15). Moreover, some of those proteinases (e.g. trypsin and neutrophil proteinases) have been reported to activate and/or disarm PAR1 in different cell systems (2, 4, 8, 13, 14, 16–18). Activation and desensitization of PAR1 must be well controlled in order to eliminate the potential pathological damage resulting from unchecked receptor signaling (14).
Our previous pharmacological studies have reported that activation of hPAR2 by mast cell tryptase can be regulated by receptor N-terminal glycosylation (19, 20). To our knowledge, hPAR1 contains a total of five predicted sites for N-linked glycosylation; three sequons are located on the receptor N terminus (Asn35-Ala36-Thr37, Asn62-Glu63-Ser64, and Asn75-Lys76-Ser77), and two are located within extracellular loop 2 (ECL2; Asn250-Ile251-Thr254, and Asn259-Glu260-Thr261) (Fig. 1) (21). PAR1, downstream of the tethered ligand, possesses two N-linked glycosylation sequons (Asn62 and Asn75), which are located along the N terminus in a domain where disarming proteinases may target. In addition, previous studies demonstrated that thermolysin and neutrophil proteinases target specific sites that are close proximally to these glycosylation sequons (Fig. 1) (12, 13, 16). Hence, we have analyzed the role of N-linked glycosylation in regulating cell surface expression and proteinase activation/disarming of hPAR1. We have permanently expressed epitope-tagged wild-type or glycosylation-deficient mutant hPAR1 in KNRK and CHO (Pro5 and Lec2) cell lines and sought to determine (i) the influence of N-linked glycosylation sequons on the receptor cell surface expression and signaling and (ii) the role of N-terminal glycosylation sequons, especially Asn62 and Asn75, in regulating receptor signaling by neutrophil proteinases and thermolysin.
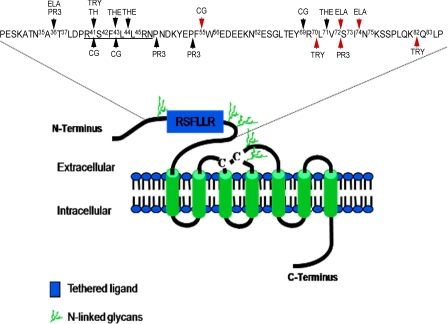
FIGURE 1.
Representative model of hPAR1 displaying the five potential N-linked glycosylation sequons and potential proteinase cleavage sites at the extracellular N terminus. Three sequons are located on the receptor N terminus (Asn35-Ala36-Thr37, Asn62-Glu63-Ser64, and Asn75-Lys76-Ser77) and two are located within extracellular loop 2 (ECL2; Asn250-Ile251-Thr254 and Asn259-Glu260-Thr261). Of the sequons at the N terminus, one (Asn35-Ala36-Thr37) is located N-terminal of the cleavage/activation site (R41↓S42FLLR), and the remaining two (Asn62-Glu63-Ser64 and Asn75-Lys76-Ser77) are located downstream of the cleavage/activation site. The disulfide bridge is shown by the two cysteines (C-C). The red arrows indicate the preferential cleavage site for each proteinase. TH, thrombin; TRY, trypsin; THE, thermolysin; PR3, proteinase 3; CG, cathepsin G; ELA, elastase. This figure was adapted from Refs. 4, 12, 13, and 29.
EXPERIMENTAL PROCEDURES
Materials
DMEM, sodium pyruvate, penicillin, streptomycin, amphotericin B, heat-inactivated FCS, non-enzymic cell dissociation solution, Geneticin (G418), Opti-MEM medium, Lipofectamine 2000, dNTPs, oligo(dT)12–18 primer, AccuPrime DNA polymerase, and Fluo-3 acetoxymethyl ester were all purchased from Invitrogen. Primers were designed “in house” and purchased from MWG-Biotech (Ebersberg, Germany). PCR purification kits, gel extraction kits, and plasmid isolation kits were all bought from Qiagen (Crawley, UK). XL1-Blue supercompetent cells and the QuikChange® site-directed mutagenesis kit were purchased from Stratagene Europe (Amsterdam, The Netherlands). The rapid DNA ligation kit was bought from Roche Applied Science. All restriction enzymes were purchased from New England Biolabs (Hitchin, UK). Anti-PAR1 ATAP-2 monoclonal antibody was purchased from Zymed Laboratories Inc. (San Francisco, CA). PAR1 activating peptide TFLLR-NH2 was purchased from Peptides International. Enhanced yellow fluorescent protein (eYFP) was obtained from BD Biosciences Clontech. Proteinase 3 was obtained from Athens Research and Technology, Inc. (Athens, Greece). Human leukocyte elastase and cathepsin-G were purchased from Elastin Products Co. Inc. M-Per mammalian protein extraction reagent and the ProFound HA.11 immunoprecipitation kit were purchased from Pierce. All other chemicals and reagents were purchased from Sigma-Aldrich unless otherwise stated.
Generation of cDNAs Encoding WT hPAR1E
Using an overlapping PCR approach, eYFP was fused to the C terminus of WT hPAR1 (pro-opiomelanocortin-M1-hPAR1-HA.11 in pcDNA3.1; C-terminally tagged with an HA.11 epitope and N-terminally tagged with an M1 epitope and a pro-opiomelanocortin) to generate WT hPAR1E (pro-opiomelanocortin-M1-hPAR1-HA.11-eYFP). The WT hPAR1E was subsequently amplified and purified and then sequenced by MWG-Biotech.
Generation of Glycosylation-deficient Mutant Cell Lines
All glycosylation-deficient mutants were generated using the QuikChange® site-directed mutagenesis kit and confirmed by MWG-Biotech sequencing. The oligonucleotides were designed to replace the asparagine residues at positions Asn35, Asn62, Asn75, Asn250, and Asn259 with glutamine residues. Single-site mutants (N35Q, N62Q, N75Q, N250Q, and N259Q) were first constructed. In order to elucidate the cumulative effects of a lack of glycosylation at multiple sites, four receptors with multiple glycosylation mutations (N62Q/N75Q, N35Q/N62Q/N75Q, N250Q/N259Q, and N35–259Q) were constructed. In addition, the alanine mutations were created at amino acid positions Phe55-Trp56 or Val72-Ser73 of N62Q/N75Q to generate cathepsin G mutant (F55A/W56A) or proteinase 3 mutant (V72A/S73A). KNRK and CHO cells permanently expressing wild-type or mutant hPAR1 cell lines were generated and cultured as described previously (Table 1) (19, 20).
TABLE 1.
KNRK and CHO fibroblast (Pro5 and Lec2) permanently expressing wild type (top) or N-linked glycosylation mutant (bottom) hPAR1 cell clones generated for this study
In order to obtain permanent receptor-expressing cell lines, cells expressing high levels of hPAR1 were isolated by FACS using the ATAP-2 anti-PAR1 antibody. The glycosylation mutants were named with an N followed by a number related to the relative position of the potential glycosylation site and a Q. The alanine mutations were created at amino acid position Phe55-Trp56 or Val72-Ser73 of N62Q/N75Q to generate cathepsin G mutant (F55A/W56A) or proteinase 3 mutant (V72A/S73A).
| Name of clone | Receptor cell surface expression level | Cell type |
|---|---|---|
| WT | Maximum level of WT hPAR1 cell surface expression | KNRK and CHO |
| WT hPAR1E | Maximum level of WT hPAR1E cell surface expression | KNRK and CHO |
| WT-H | Higher level of WT hPAR1 cell surface expression | KNRK |
| WT-M | Medium level of WT hPAR1 cell surface expression | KNRK |
| WT-L | Low level WT hPAR1 cell surface expression | KNRK |
| Name of clone | Consensus sequence(s) disrupted | Cell type |
|---|---|---|
| N35Q | Asn35 | KNRK |
| N62Q | Asn62 | KNRK |
| N75Q | Asn75 | KNRK |
| N250Q | Asn250 | KNRK |
| N259Q | Asn259 | KNRK |
| N62Q/N75Q | Asn62 and Asn75 | KNRK |
| N35Q/N62Q/N75Q | Asn35, Asn62, and Asn75 | KNRK |
| N250Q/N259Q | Asn250 and Asn259 | KNRK |
| N35–259Q | Asn35, Asn62, Asn75, Asn250, and Asn259 | KNRK |
| N35QhPAR1E | Asn35 | KNRK |
| N62Q/N75QhPAR1E | Asn62 and Asn75 | KNRK |
| N250Q/N259QhPAR1E | Asn250 and Asn259 | KNRK |
| N35–259QhPAR1E | Asn35, Asn62, Asn75, Asn250, and Asn259 | KNRK |
| Cathepsin G mutant | Asn62, Asn75, Phe55, and Trp56 | KNRK |
| Proteinase 3 mutant | Asn62, Asn75, Val72, and Ser73 | KNRK |
Flow Cytometry Analysis (FACS)
WT and mutant cell lines (∼90% confluence) with matched receptor expression were selected and used for further studies by FACS (BD Biosciences) as we described previously (19). The anti-PAR1 ATAP-2 monoclonal antibody (1 μg/ml) and anti-mouse FITC-conjugated antibody (1:1000 dilution) were applied to assess the levels of receptor cell surface expression. The data were expressed as the median fluorescence of positive minus the median fluorescence of the EV (pcDNA3.1 transfected KNRK) cells.
Confocal Microscopy Analysis
For the WT hPAR1eYFP and mutant cell lines, the light emitted by the eYFP (530 nm) fused to the receptor was used to assess receptor expression. Cells on coverslips were fixed by incubation in formaldehyde (3%) in PBS for 15 min at room temperature and then permeabilized in PBS containing 0.2% Triton X-100 for 10 min at room temperature. The coverslips were then mounted with anti-fade and fixed to slides. The cells were visualized by a Nikon Eclipse (TE2000-E) microscope with a Bio-Rad Radiance 2100 scanning system and lasers.
Western Blot Analysis
HA.11-tagged wild-type and glycosylation-deficient mutants of hPAR1E were immunoprecipitated using a ProFound HA.11 immunoprecipitation kit as per the manufacturer's protocol. Protein samples (20 μg) were separated on a 10% SDS-polyacrylamide gel before transfer to Hybond C PVDF membrane. The membrane was subsequently blocked with PBS containing 5% nonfat milk before incubation overnight at 4 °C with the mouse monoclonal HA.11 antibody (1:1000 dilution in PBS/Tween 20 (0.1%) containing 2% nonfat milk). Blots were washed with PBS/Tween 20 (0.1%) for 1 h, replacing the buffer every 15 min, before a final incubation with the peroxidase-conjugated goat anti-mouse IgG (1:4000 in PBS/Tween 20 (0.1%) containing 2% nonfat milk) for 1 h. Following a further wash in PBS/Tween 20 (0.1%) for 1 h, bands were visualized using ECL (Amersham Biosciences).
Calcium Signaling Assay
The calcium signaling assay was performed as we described previously (19). Functional receptor activity was assessed by challenging cells with the PAR1 activating peptide TFLLR-NH2 (100 μm) and thrombin (5 nm). To assess whether a test proteinase was amputating the tethered ligand from the receptor (disarming), cells were first challenged by the addition of the test proteinase for 2 min. Successful amputation of the tethered ligand was then monitored by a subsequent application of thrombin (5 nm). The calcium signal in response to thrombin will be ablated if the tethered ligand has been removed from the receptor by the test proteinase. To demonstrate that the disarmed PAR1 is still functional and present at the cell surface, 100 μm TFLLR-NH2 was applied after thrombin challenge. A calcium signal triggered by the TFLLR-NH2 is indicative of a functional PAR1 at the cell surface, which is unresponsive to thrombin.
Calculations and Statistical Analysis
Data were analyzed using GraphPad Prism software. Statistical tests were carried out depending on the specifics of the data sets to be compared. When two cell lines subjected to the same treatment were compared, a paired Student's t test was adopted. The comparison of a group of cell lines subjected to the same treatment was performed by one-way analysis of variance followed by Tukey's multiple comparison test. In order to assess the changes over a period of time using a specific treatment or to assess a change over a concentration range between two different cell lines, two-way analysis of variance tables were adopted.
RESULTS
N-Linked Glycosylation at Specific Sites Regulates hPAR1 Cell Surface Expression
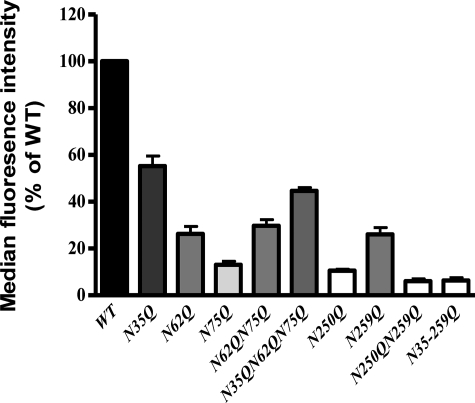
FACS demonstrated that removing any of the glycosylation sequons resulted in a significant decrease (p < 0.001) in hPAR1 cell surface expression in KNRK cells (Fig. 2). N35Q displayed the greatest cell surface expression; N250Q displayed the lowest expression when compared with the other single mutants, N35Q, N62Q, N75Q, N250Q, and N259Q (percentage relative to WT ± S.E.: 55 ± 4, 26 ± 3, 13 ± 1.5, 10.5 ± 0.7, and 26 ± 3% (n = 3) respectively). Removal of both glycosylation sequons on ECL2 (N250Q/N259Q) resulted in nearly total loss of cell surface expression (percentage relative to WT ± S.E.: 6 ± 1% (n = 3)). In contrast, removing all glycosylation sequons on the N terminus (N35Q/N62Q/N75Q) still resulted in nearly 50% cell surface expression compared with WT expression (percentage relative to WT ± S.E.: 44.7 ± 1% (n = 3)). Removing all glycosylation sequons resulted in a receptor (N35–259Q) that was expressed at a level similar to that of the N250Q/N259Q mutant (percentage relative to WT ± S.E.: 6 ± 1% (n = 3)).
FIGURE 2.
Cell surface expression of WT hPAR1 (WT) and glycosylation-deficient mutant hPAR1 receptor in KNRK cells assessed by FACS. Cells at ∼90% confluence were harvested and incubated with the ATAP-2 antibody before incubation with an anti-mouse FITC-conjugated antibody. Results are expressed as a percentage of the median fluorescence obtained with WT hPAR1 cells. The bars represent mean ± S.E. (error bars) of measurements for three independent experiments. The overall comparison showed that there were significant differences between 10 groups (p < 0.0001). Tukey's multiple comparison test indicated that the cell surface expression of all of the mutant receptors was significantly different from WT hPAR1 (p < 0.001).
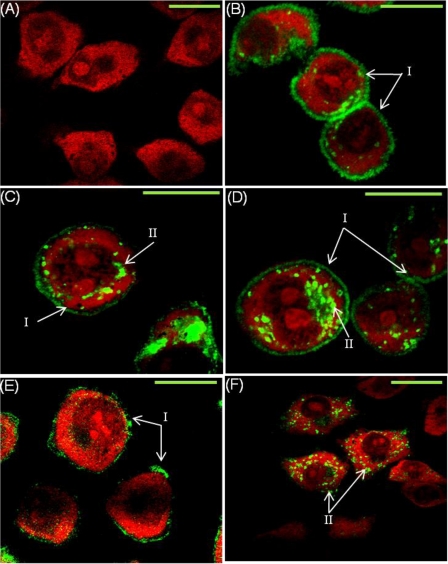
Analysis of eYFP-tagged hPAR1 localization in permeabilized cells by confocal microscopy demonstrated that all hPAR1E types (WT and glycosylation-deficient mutant hPAR1) with the exception of N35–259QhPAR1E were expressed on the cell surface; however, different levels of membrane localization and cytosolic retention were observed (Fig. 3). There was no detectable fluorescence (eYFP signaling) observed in EV cells (Fig. 3A). WT hPAR1E displayed strong cell membrane expression with little receptor observed in the cytoplasm (Fig. 3B). Removal of sequon Asn35 in the receptor N terminus resulted in the appearance of receptor within the cytosol (Fig. 3C). Removal of both sequons (Asn62 and Asn75) after the tethered ligand of hPAR1 resulted in a large amount of receptor being found in the cytosol (Fig. 3D). The relative intensity of the receptor membrane localization and global cellular distribution appeared to be reduced significantly after removal of ECL2 glycosylation sequons (Asn250 and Asn259) (Fig. 3E). Removal of all hPAR1 glycosylation sequons (Asn35, Asn62, Asn75, Asn250, and Asn259) resulted in a dramatic loss of cell surface expression with the highest level of receptor cytosolic retention (Fig. 3F).
FIGURE 3.
Confocal microscopy for WT hPAR1E and glycosylation-deficient mutants in KNRK cells. eYFP-tagged PAR1-expressing KNRK cells were grown on coverslips before fixing and permeabilizing. Cells were stained with propidium iodide prior to visualizing. eYFP is visualized here in green and propidium iodide in red. I, cell surface expression; II, internal receptor expression. Green bar on the right of each panel, 10 μm. A, EV; B, WT hPAR1E; C, N35QhPAR1E; D, N62Q/N75QhPAR1E; E, N250Q/N259QhPAR1E; F, N35–259QhPAR1E. The images are representative of four independent experiments.
hPAR1 Is a Heavily Glycosylated and Sialylated Receptor
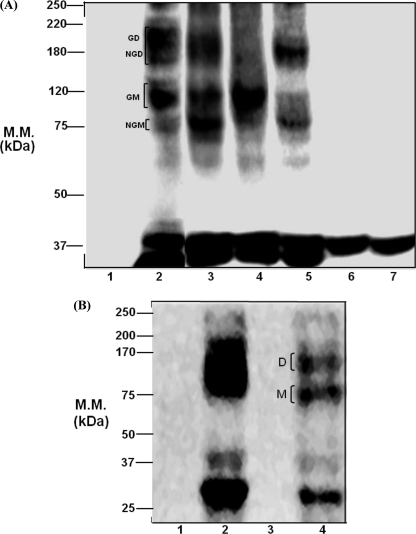
All of the glycosylation-deficient mutant receptors displayed different degrees of loss in molecular mass; no bands were detected in EV cells (Fig. 4A). The molecular mass of WT hPAR1E migrated as a broad band ranging from ∼37 to ∼250 kDa, with the majority of the receptor being observed from ∼75 to ∼220 kDa. This is a pattern similar to that of N62QhPAR1E. N62Q/N75QhPAR1E migrated from ∼37 to ∼120 kDa. N35Q/N62Q/N75QhPAR1E migrated with a molecular mass from ∼37 to ∼200 kDa, with the majority of the receptor being observed from ∼75 to ∼120 kDa. The predicted molecular mass of the fused hPAR1eYFP is approximately ∼70 to ∼80 kDa (hPAR1 is ∼47 kDa, and eYFP protein is ∼31 kDa). We proposed that the band at ∼75 kDa may represent an unglycosylated monomer of PAR1; the bands around ∼80 to ∼130 kDa may represent a glycosylated monomer of PAR1; and the bands around ∼150 to ∼220 kDa may represent glycosylated and unglycosylated dimers of PAR1 (Fig. 4). When removing all three glycosylation sites (Asn35, Asn62, and Asn75) in the N terminus of the receptor, the banding pattern changed, with only glycosylated dimer and unglycosylated monomer present. Removal of glycosylation at sites Asn62 and Asn75 produced a single band at ∼120 kDa representing a glycosylated monomer of PAR1. Only one band migrated at ∼37 kDa from both N250Q/N259QhPAR1E and N35–259QhPAR1E. A number of minor bands that may represent either eYFP protein or proteolytic degradation products of hPAR1 were routinely observed around ∼37 kDa in all lanes.
FIGURE 4.
Western blot analysis for wild-type and glycosylation-deficient mutant hPAR1. A, immunoprecipitates from KNRK cell lines expressing HA.11-tagged hPAR1 with the potential N-linked glycosylation sequons removed were analyzed by SDS-PAGE (10% gel) and immunoblotted using HA.11 monoclonal antibody. Lane 1, EV; lane 2, WT hPAR1E; lane 3, N62QhPAR1E; lane 4, N62Q/N75QhPAR1E; lane 5, N35Q/N62Q/N75QhPAR1E; lane 6, N250Q/N259QhPAR1E; lane 7, N35–259QhPAR1E. B, protein samples of hPAR1E-transfected CHO (Pro5 and Lec2) cell lines were immunoprecipitated by HA.11 immunoprecipitation and were analyzed by SDS-PAGE (10% gel) and immunoblotted using a monoclonal HA.11 antibody. Lane 1, Pro5; lane 2, Pro5-PAR1E; lane 3, Lec2; lane 4, Lec2-PAR1E. GD, glycosylated dimer; NGD, nonglycosylated dimer; GM, glycosylated monomer; NGM, nonglycosylated or minimally glycosylated monomer; M, nonglycosylated or minimally glycosylated monomer; D, PAR1 dimer; M.M., molecular mass. Results are representative of three separate experiments.
To determine the contribution of sialic acid to the molecular mass of hPAR1, we performed Western blot analysis for WT hPAR1E-transfected CHO cells (Pro5 and Lec2) (Fig. 4B). No bands were detected in the non-transfected Lec2 and Pro5. Pro5-PAR1E migrated as a broad high molecular weight band ranging from ∼25 to ∼200 kDa; the molecular mass of Lec2-PAR1E ranged from ∼25 to ∼150 kDa. Two bands were routinely observed in the ∼25 to ∼37 kDa range in both Pro5-PAR1E and Lec2-PAR1E.
The N-Linked Glycosylation Located after the Tethered Ligand of hPAR1 Regulates hPAR1 Signaling
To investigate whether N-linked glycosylation at individual residues had a role in regulating receptor function, we constructed calcium signaling concentration-effect curves for thrombin, trypsin, and TFLLR-NH2 in WT hPAR1 and hPAR1 glycosylation mutant KNRK cell lines. The EC50 values of WT hPAR1 and mutant receptors for the calcium signaling assay are displayed in Table 2.
TABLE 2.
EC50 values for N-terminal glycosylation-deficient mutant hPAR1 and their respective matching cell surface expression of WT hPAR1 agonist concentration effect curves
Cell clones with matched receptor expression assessed by FACS analysis were selected for functional studies. Where it was not possible to match receptor expression between the WT hPAR1 cell line and a cell line expressing a mutant glycosylation receptor, the WT, WT-H, WT-M, and WT-L with different cell confluence in order to reduce receptor expression were used and thus match the cell surface expression of the mutant cell line. Reduced receptor cell surface expression of the WT-H, WT-M, and WT-L cell lines were confirmed by using FACS analysis.
| Cell lines with matched receptor expression | EC50 |
||
|---|---|---|---|
| TFLLR | Thrombin | Trypsin | |
| μm | nm | nm | |
| WT-H | 17 | 0.14 | 33 |
| N35Q | 16 | 0.2 | 43 |
| WT-L | 27 | 0.2 | 39 |
| N62Q | 29 | 0.4 | 43 |
| N75Q | 31 | 0.2 | 41 |
| WT-M | 22 | 0.12 | 26 |
| N62Q/N75Q | 30 | 0.3 | 42 |
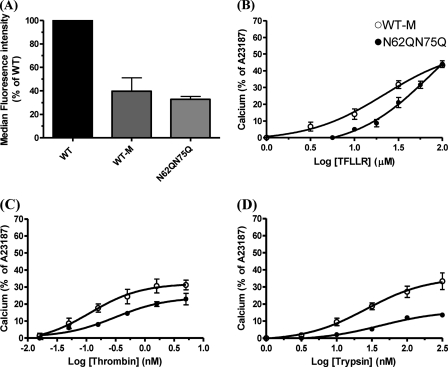
N62Q/N75Q concentration effect curves were compared with WT-M because they displayed similar cell surface expression (p > 0.05) (Fig. 5A). TFLLR-NH2 displayed a slight rightward shift in activating N62Q/N75Q compared with WT-M, stimulating a calcium signal at 3–100 μm for WT-M and 10–100 μm for N62Q/N75Q, respectively, but achieved the same maximum response of 42 ± 1.5% of A23187 at 100 μm in both cell lines (Fig. 5B). For N62Q/N75Q and WT-M, thrombin induced a calcium signal from 0.05 to 5 nm (Fig. 5C). However, the magnitude of the responses to thrombin in N62Q/N75Q was smaller than those obtained for WT-M (24 ± 2.6% of A23187 and 31 ± 1.8% of A23187, respectively) (p > 0.05). The EC50 value for thrombin in N62Q/N75Q was 0.3 nm, and the EC50 value in WT-M was 0.12 nm. Interestingly, the N62Q/N75Q displayed reduced sensitivity toward trypsin compared with WT-M, and the maximal response achieved was only 14% of A23187 compared with 32 ± 3% of A23187 for the WT-M (p < 0.001) (Fig. 5D). In addition, the EC50 value for trypsin in N62Q/N75Q was significantly greater than WT-M (42 and 26 nm, respectively). However, further analysis of N62Q and N75Q cell lines revealed no observable changes in receptor function for TFLLR-NH2, thrombin, or trypsin when compared with their matched WT hPAR1 cell line (data not shown).
FIGURE 5.
Cell surface expression and calcium signaling analysis of WT-M and N62Q/N75Q. A, comparison of WT-M and glycosylation-deficient mutant N62Q/N75Q KNRK cell surface expression. Cells at ∼90% confluence were harvested and incubated with the ATAP-2 antibody before incubation with an anti-mouse FITC-conjugated antibody. Cell surface expression was assessed by FACS analysis. Results are expressed as a percentage of the median fluorescence obtained with WT cells. B–D, calcium signaling in the WT-M and glycosylation mutant N62Q/N75Q in response to TFLLR-NH2, thrombin, and trypsin. Results are expressed as the means ± S.E. of at least four independent experiments.
Removal of N-Linked Glycosylation Asn62 and/or Asn75 from hPAR1 Enhances the Ability of Trypsin to Disarm the Receptor
Having found that trypsin shows reduced activity toward N62Q/N75Q by a calcium signaling assay, we then explored the ability of trypsin to disarm N62Q/N75Q, N62Q, and N75Q.
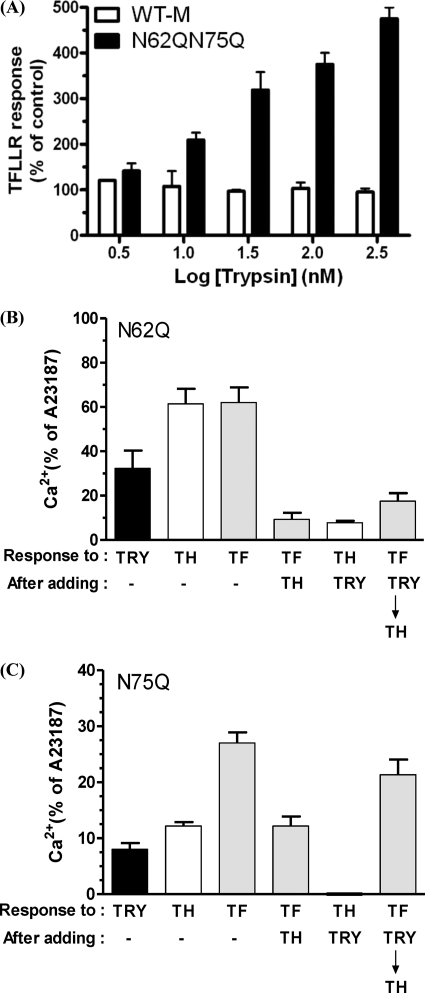
Fig. 6A shows the TFLLR-NH2 (100 μm) responses following trypsin (3, 10, 30, 100, and 300 nm) and then thrombin (5 nm) challenge in WT-M and N62Q/N75Q cells (i.e. trypsin and then thrombin and then TFLLR-NH2). For WT-M, the TFLLR-NH2 responses remained the same after challenging the cells with trypsin from 3 to 300 nm. Challenging the N62Q/N75Q cells with 3 nm trypsin followed by 5 nm thrombin and then 100 μm TFLLR-NH2 triggered a calcium signal that was similar to that seen for WT-M cells. However, a robust increase in the magnitude of TFLLR-NH2-triggered calcium signal was observed after challenging the N62Q/N75Q cells with 300 nm trypsin, which was over 4-fold greater than that observed for WT-M cells (p < 0.05).
FIGURE 6.
A, analysis of trypsin disarming in WT-M and N62Q/N75Q. Calcium imaging of Fluo-3-loaded cells was used to assess 100 μm TFLLR-NH2 responses following trypsin and then 5 nm thrombin challenge in WT hPAR1- and hPAR1(N62Q/N75Q)-transfected KNRK cells. The cellular response to TFLLR-NH2 after the addition of thrombin but without pretreatment with trypsin is used as a percentage control. B, analysis of trypsin disarming in N62Q. Shown is hPAR1(N62Q)-transfected KNRK cell calcium signaling response following the addition of 100 nm trypsin, 5 nm thrombin, and 100 μm TFLLR-NH2 and then 2 μm A23187. C, analysis of trypsin disarming in N75Q. Shown is hPAR1(N75Q)-transfected KNRK cell calcium signaling response following the addition of 100 nm trypsin, 5 nm thrombin, 100 μm TFLLR-NH2, and then 2 μm A23187. TRY, trypsin; TH, thrombin; TF, TFLLR-NH2. The data sets are expressed as mean ± S.E. (error bars) from three separate experiments.
We next assessed whether individual glycosylation at Asn62 or Asn75 is responsible for regulating trypsin disarming of hPAR1 (Fig. 6, B and C). Challenging N62Q cells with 5 nm thrombin or 100 μm TFLLR-NH2 resulted in a calcium response of 61.4 ± 6.7% of A23187 and 62 ± 6.9% of A23187, respectively (Fig. 6B). Challenging N62Q cells with 100 nm trypsin resulted in an observable calcium signal (32 ± 8% of A23187). The addition of thrombin following trypsin challenge resulted in a marked reduction (p < 0.001) in the calcium response to thrombin compared with the response obtained with thrombin added alone (7.8 ± 1% of A23187 versus 61.4 ± 6.7% of A23187). The addition of TFLLR-NH2 following trypsin and thrombin resulted in a response that was slightly increased (p > 0.05) in magnitude compared with the response obtained to TFLLR-NH2 following just thrombin addition (17.5 ± 3.6% of A23187 versus 9 ± 3% of A23187) but was significantly reduced (p < 0.001) compared with the response of TFLLR-NH2 added alone (17.5 ± 3.6% of A23187 versus 62 ± 6.9% of A23187).
Challenging N75Q cells with trypsin only resulted in a response of 8 ± 1% of A23187 (Fig. 6C). Challenging N75Q cells with TFLLR-NH2 or thrombin alone resulted in a calcium response of 27 ± 2% of A23187 and 12 ± 0.7% of A23187, respectively. The addition of TFLLR-NH2 following thrombin addition resulted in a calcium response of 12 ± 1.7% of A23187. The addition of thrombin following the addition of trypsin resulted in no calcium response. Finally, challenging the cells with TFLLR-NH2 following trypsin and thrombin addition triggered a calcium response of 21 ± 2.7% of A23187, which was of a magnitude comparable (p > 0.05) with the response obtained with TFLLR-NH2 alone.
Elastase, Proteinase 3, and Thermolysin but Not Cathepsin G Displayed Enhanced Ability to Disarm hPAR1(N62Q/N75Q)
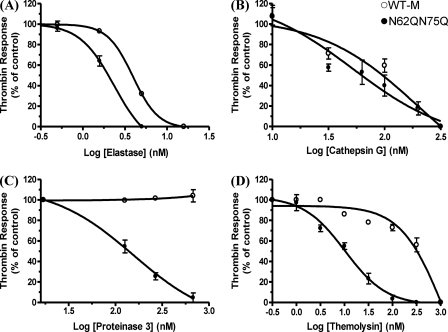
Effective proteinase receptor disarming was assessed by thrombin activation of the cells following proteinase addition. For N62Q/N75Q and WT-M, significant differences (p < 0.0001) in responses to 5 nm thrombin following the addition of elastase at different concentrations were observed (Fig. 7A). For WT-M, no changes were observed in thrombin-triggered calcium responses after the addition of 1 nm elastase, and the thrombin-triggered calcium response was totally ablated after the addition of 15 nm elastase. The thrombin response curve for N62Q/N75Q shifted left half a log when compared with WT-M. For N62Q/N75Q, the calcium signaling response to thrombin started to decrease after challenging the cells with 1 nm elastase (∼80% of control thrombin response). The thrombin-triggered calcium response was totally ablated after challenging cells with 5 nm elastase.
FIGURE 7.
Analysis of WT-M and hPAR1(N62Q/N75Q) calcium signaling following proteinase stimuli. WT-M and N62Q/N75Q in KNRK cells were challenged by the addition of elastase (A), the addition of cathepsin G (B), the addition of proteinase 3 (C), or the addition of thermolysin (D) followed by the addition of 5 nm thrombin and finally the addition of 2 μm A23187. Results are expressed as mean ± S.E. from at least three independent experiments.
For N62Q/N75Q and WT-M, no significant differences (p > 0.05) in responses to 5 nm thrombin following the addition of cathepsin G from 10 to 300 nm were observed (Fig. 7B). The thrombin-triggered calcium response was totally ablated after the addition of 300 nm cathepsin G for both N62Q/N75Q and WT-M.
For N62Q/N75Q and WT-M, significant differences (p < 0.0001) in responses to 5 nm thrombin following the addition of proteinase 3 were observed (Fig. 7C). For WT-M, no changes were observed in the thrombin-triggered calcium response following the addition of 1000 nm proteinase 3. In contrast, significant decreases in the response to thrombin were observed in N62Q/N75Q following the addition of proteinase 3 from 1 to 1000 nm. The thrombin-triggered calcium signal was totally abolished after the addition of 1000 nm proteinase 3.
For N62Q/N75Q and WT-M, significant differences (p < 0.0001) in responses to 5 nm thrombin following the addition of thermolysin at different concentrations were observed (Fig. 7D). The thrombin response curve after adding thermolysin at increasing concentration for N62Q/N75Q shifted 30-fold to the left when compared with WT-M. For N62Q/N75Q, the calcium signaling response to thrombin started to decrease after application of thermolysin at 3 nm (∼80% of control thrombin response). The thrombin-triggered calcium response was totally ablated after challenging N62Q/N75Q with 300 nm thermolysin. However, for WT-M, the thrombin-triggered calcium responses remained elevated at thermolysin concentrations up to 30 nm. The calcium signaling response to thrombin started to decrease following the 100 nm thermolysin addition (∼80% of thrombin control). The thrombin-triggered calcium response in WT-M was totally ablated after the addition of 1000 nm thermolysin.
Molecular Evidence That Cathepsin G and Proteinase 3 Are Disarming hPAR1(N62Q/N75Q)
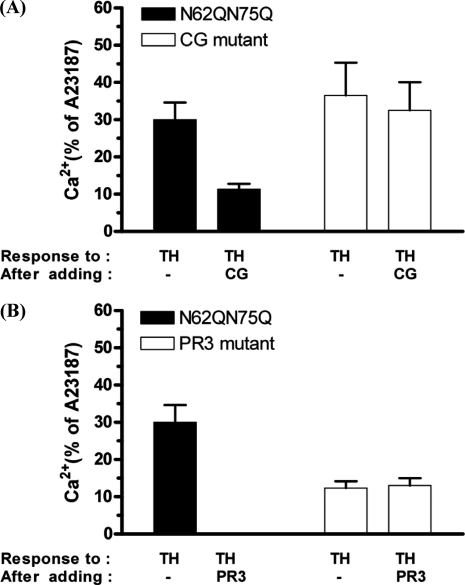
The addition of 100 nm cathepsin G to N62Q/N75Q cells before the addition of 5 nm thrombin markedly reduced (p < 0.001) the response to thrombin compared with the response obtained with thrombin added alone (11 ± 2% of A23187 and 30 ± 5% of A23187, respectively) (Fig. 8A). In contrast, the prior addition of cathepsin G failed to reduce the thrombin calcium signal in cathepsin G mutant cells when compared with the control (32.5 ± 8% of A23187 and 36.5 ± 8% of A23187, respectively) (p > 0.05).
FIGURE 8.
Molecular evidence that cathepsin G and proteinase 3 disarm hPAR1(N62Q/N75Q). CG mutant (A) and proteinase 3 mutant (B) cells loaded with Fluo-3 were stimulated by the addition of either 100 nm cathepsin G or 300 nm proteinase 3 for 2 min and then challenged with 5 nm thrombin, followed by 100 μm TFLLR-NH2 and then 2 μm calcium ionophore (A23187). TH, thrombin; CG, cathepsin G; PR3, proteinase 3. Results are expressed as the mean ± S.E. (error bars) of at least two independent experiments.
No observable calcium response to thrombin was observed when 300 nm proteinase 3 had been added to the N62Q/N75Q cells prior to thrombin challenge (Fig. 8B). In contrast, the prior addition of proteinase 3 failed to reduce the thrombin calcium signal in the proteinase 3 mutant cells, when compared with the response obtained with thrombin added alone (13 ± 2% of A23187 and 12.33 ± 2.5% of A23187, respectively) (p > 0.05).
Elastase, Cathepsin G, Proteinase 3, and Thermolysin Mainly Disarm hPAR1(N75Q)
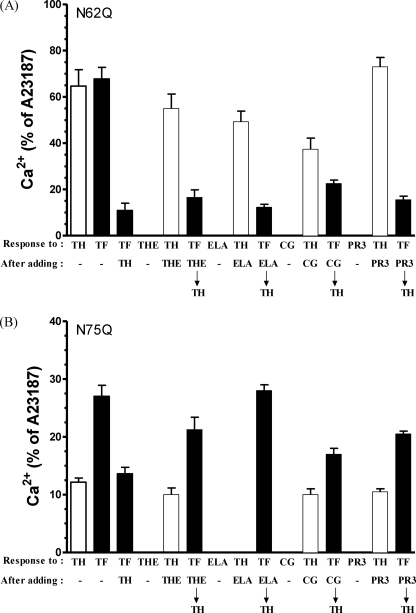
The ability of thermolysin and neutrophil serine proteinases to activate/disarm N62Q and N75Q was investigated (Fig. 9).
FIGURE 9.
Analysis of hPAR1(N62Q) and hPAR1(N75Q) calcium signaling following proteinase stimuli. N62Q (A) and N75Q (B) cells loaded with Fluo-3 were stimulated by the addition of either 50 nm thermolysin, 1.5 nm elastase, 100 nm cathepsin G, or 300 nm proteinase 3 for 2 min and then challenged with 5 nm thrombin, followed by 100 μm TFLLR-NH2 and then 2 μm A23187. TRY, trypsin; TH, thrombin; TF, TFLLR-NH2; THE, thermolysin; ELA, elastase; CG, cathepsin G; PR3, proteinase 3. Results are expressed as the mean ± S.E. (error bars) of at least three independent experiments.
Challenging N62Q cells with 5 nm thrombin or 100 μm TFLLR-NH2 resulted in a calcium response of 64.7 ± 7% of A23187 and 67.8 ± 5% of A23187, respectively (Fig. 9A). In addition, challenge with TFLLR-NH2 following the thrombin addition resulted in a calcium response of 9 ± 3% of A23187. Thermolysin (50 nm) had no observable effect on calcium signaling in N62Q. Challenge with thrombin following the thermolysin addition resulted in a calcium response at 53.5 ± 4.6% of A23187, which is comparable with that obtained with thrombin added alone (64.7 ± 7% of A23187). The addition of TFLLR-NH2 following thermolysin and thrombin resulted in a similar calcium response to the response obtained with TFLLR-NH2 following just thrombin challenge (13.6 ± 4% of A23187 and 9 ± 3% of A23187, respectively). The addition of 1.5 nm elastase had no observable effect on calcium signaling in N62Q cells. The addition of thrombin following elastase addition resulted in a calcium response of 49 ± 3.5% of A23187. The addition of TFLLR-NH2 following the disarming with elastase and thrombin resulted in a similar calcium response to the TFLLR-NH2 response obtained following just thrombin alone 12 ± 1% of A23187 and 9 ± 3% of A23187, respectively). The addition of 100 nm cathepsin G had no observable effect on calcium signaling in N62Q cells. After challenging N62Q cells with cathepsin G, the addition of thrombin resulted in a small decrease in the calcium response compared with that obtained with thrombin added alone (39 ± 3.8% of A23187 and 64.7 ± 7% of A23187, respectively). The addition of TFLLR-NH2 following the addition of thermolysin and thrombin triggered a calcium response at 16 ± 6% of A23187 that was slightly larger than that obtained with TFLLR-NH2 following thrombin alone (9 ± 3% of A23187). The addition of 300 nm proteinase 3 had no observable effect on calcium signaling in N62Q cells. Challenge with thrombin following proteinase 3 addition resulted in a calcium response of 67 ± 6.7% of A23187, which is comparable with that obtained when thrombin was added alone (64.7 ± 7% of A23187). The addition of TFLLR-NH2 following the addition of proteinase 3 and thrombin resulted in a calcium response similar to that obtained with TFLLR-NH2 following the thrombin addition (11.6 ± 4% of A23187 and 9 ± 3% of A23187, respectively).
Challenging N75Q cells with thrombin or TFLLR-NH2 resulted in a calcium response of 12 ± 0.7% of A23187 and 27 ± 2% of A23187, respectively. The addition of TFLLR-NH2 following thrombin challenge resulted in a calcium response of 13.6 ± 1% of A23187 (Fig. 9B). No observable calcium response was detected after challenging N75Q cells with thermolysin. After challenging N75Q cells with thermolysin, the addition of thrombin resulted in a calcium response that was comparable with that obtained to thrombin alone (10 ± 1% of A23187 and 12 ± 0.7% of A23187, respectively), and finally challenging the cells with TFLLR-NH2 triggered a calcium response of 21 ± 2% of A23187. No observable calcium response was detected after challenging N75Q cells with elastase or with thrombin following the addition of elastase. In addition, the response to TFLLR-NH2 following elastase and thrombin challenge was the same as that obtained with TFLLR-NH2 alone (28 ± 1% of A23187 versus 27 ± 2% of A23187).
The addition of cathepsin G had no observable effect on calcium signaling in N75Q cells. Challenging cells with thrombin following the addition of cathepsin G resulted in a calcium response of 10 ± 1% of A23187, which was comparable with that obtained with thrombin alone (12 ± 0.7% of A23187). The addition of TFLLR-NH2 following the addition of cathepsin G and thrombin triggered a calcium response, which was of a magnitude similar to that obtained with TFLLR-NH2 following thrombin alone (17 ± 1 and 13.6 ± 1% of A23187, respectively). The addition of proteinase 3 had no observable effect on calcium signaling in N75Q cells. The addition of thrombin after challenging N75Q cells with proteinase 3 resulted in a very small decrease in calcium response compared with that obtained with thrombin alone (10 ± 0.5% of A23187 and 12 ± 0.7% of A23187, respectively). Finally, challenging the cells with TFLLR-NH2 following the proteinase 3 and thrombin addition triggered a calcium response of 20.5 ± 0.5% of A23187.
DISCUSSION
In the present study, we have demonstrated for the first time that N-linked glycans (Asn62 and Asn75) after the tethered ligand on the N terminus of hPAR1 regulate receptor signaling to trypsin, thermolysin, and the neutrophil proteinases elastase and proteinase 3 but not cathepsin G. In addition, we reported that hPAR1 is heavily N-linked glycosylated and sialylated in epithelial cell lines (KNRK and Pro5 cells), and glycosylation occurs at all five consensus sites, namely Asn35, Asn62, Asn75, Asn250, and Asn259. Removing N-linked glycosylation sequons at these positions affected hPAR1 cell surface expression to varying degrees, and N-linked glycosylation at ECL2 (especially Asn250) of hPAR1 might be essential for optimal receptor cell surface expression and receptor stability.
N-Linked glycosylation, which adds oligosaccharides to the nitrogen in the side-chain amide of asparagine residues, requires the asparagines in a consensus sequence of Asn-X-Ser/Thr (where X is any amino acid except proline) (22, 23). However, not all consensus sites are actually glycosylated because the oligosaccharide might be trimmed and elaborated during transit through the endoplasmic reticulum and the Golgi apparatus (23). Disrupting the glycosylation sites by mutagenesis, individually and/or in combination, allowed us to investigate the status and even the function of each specific putative N-linked glycosylation site.
We first examined the role of N-linked glycosylation in regulating the receptor cell surface expression and the receptor global distribution in KNRK cells. Interestingly, unlike in previous studies in HeLa (24), there is still a very high level of cell surface expression of hPAR1 when removing all glycosylation sites on the N terminus. The ECL2 single glycosylation mutant N250Q displayed the lowest level of cell surface expression of hPAR1 among all of the other single glycosylation mutants; furthermore, removing all glycosylation sequons on ECL2 resulted in similar low levels of receptor cell surface expression to the non-glycosylation mutant, where all glycosylation sequons had been deleted. N-Linked glycosylation of hPAR1 on ECL2 (especially Asn250) versus glycosylation in the N terminus might therefore be more important for receptor optimal cell surface expression. Glycosylation on ECL2 of PAR2 is also important for enhancing receptor expression but not regulating receptor signaling; more importantly, the ECL2 region is extremely sensitive to mutations, and mutating residues in and around the glycosylation sequon, even neutral mutations such as alanine (Ala) in the ECL2 glycosylation sequon, alters receptor structure and function (19, 24). The PAR1 glycosylation study on HeLa cells (24) reported that N-terminal N-linked glycosylation of PAR1 versus ECL2 N-linked glycosylation is critical for the regulation of PAR1 signaling and trafficking. In this study, mutants were generated by substituting Ala for Glu. It is therefore hard to determine whether the altered hPAR1 cell surface expression and signaling in HeLa cells are a result of the loss of N-linked glycosylation or due to the Ala substitution altering receptor structure and function. We have generated our N-linked glycosylation mutants by substituting Asn for Glu at all five potential N-linked glycosylation sites; Glu is chemically and structurally more similar to Asn. For this reason, we propose that this substitution itself will not lead to structural and functional differences between the mutants and wild-type hPAR1, and any differences detected should be due to the loss of glycosylation.
Using confocal microscopy, we demonstrated that the glycosylation-deficient mutants were abundant in the cytosol. Removal of a single N-terminal glycan resulted in observable receptor cytosolic retention, but removal of all glycosylation sites produced a receptor that could mainly be seen in the cytosol with minimal cell surface expression. Therefore, we conclude that N-linked glycosylation of hPAR1 at all sites might be required for the optimal receptor cell surface expression (24, 25).
Similar to the study of Soto and Trejo (24), we have demonstrated that in KNRK cells, hPAR1 is heavily glycosylated, and the glycosylation occurs at all five consensus sites. Removal of N-linked glycosylation from ECL2 or removal of all N-linked glycans from the receptor resulted in only a single Western blot band visualized at around 37 kDa compared with wild type or the N terminus mutants, which produced a second band at a lower molecular weight. It is well known that N-linked glycosylation can regulate receptor conformation, folding, or stability and even protect receptors from proteolytic degradation. The bands shown around ∼37 kDa may represent either eYFP protein or deglycosylation and/or proteolytic degradation products of hPAR1 (5, 14, 24, 26–28). We thus propose that N-linked glycosylation, especially on ECL2 (Asn250 and Asn259), of hPAR1 might be vital for receptor stability and even protect hPAR1 from endogenous proteolytic degradation. Furthermore, we expressed the hPAR1 in CHO cells (Pro5 and Lec2) in order to assess the sialylation status of hPAR1. The Pro5 cell line is the parent clone for the Lec2 cell line, which has a substantial loss in the ability to attach sialic acid to the terminal positions on oligosaccharides. Western blot analysis of hPAR1 in Lec2 cells revealed a protein that had a molecular mass that was up to ∼47 kDa lower than that observed for hPAR1 expressed in the Pro5 cells. Presumably, this significant loss of ∼47 kDa is due to the loss of receptor-associated sialic acid, and we can therefore conclude that hPAR1 in Pro5 cells, at least, is a heavily sialylated receptor.
In order to establish the role of N-linked glycosylation on the N terminus of hPAR1 in regulating receptor function, agonist concentration effect curves were produced for the glycosylation-deficient hPAR1 cell lines and their respective WT hPAR1 cell line to evaluate the coupling of the mutant receptors to calcium. Receptor cell surface expression for each mutant cell line was compared and matched by FACS to WT hPAR1 cells in order to ensure that any functional differences detected were not due to differences in receptor cell surface expression (19). Except for the N62Q/N75Q mutant, no significant changes in receptor function were detected when compared with their respective matching WT hPAR1 for all mutant cell lines. This N62Q/N75Q mutant displayed a slight reduction in sensitivity toward lower concentrations of TFLLR-NH2 and thrombin, although statistical analysis showed no significant differences in EC50 value and receptor maximum calcium signaling response toward 100 μm TFLLR-NH2 or 5 nm thrombin. The biggest reduction in agonist sensitivity was observed with trypsin activation. PAR1 coupling to calcium signaling appears not to be affected by the lack of glycosylation on the receptor. Even the N35–259Q mutant cell line still displayed a detectable calcium signal toward TFLLR-NH2 and thrombin (data not shown), thus adding strength to this conclusion.
In our experiments, trypsin triggered calcium signaling in wild-type hPAR1 and its mutants to varying degrees. However, the TFLLR-NH2 calcium signaling response after the addition of trypsin and thrombin is much greater in N62Q/N75Q mutants compared with wild-type hPAR1. This just confirmed that trypsin disarms the mutant receptor, and the disarmed mutant receptors still remained on the cell surface but could not be activated by thrombin anymore because the tethered ligand had been removed; however, TFLLR will still activate those mutant receptors by binding to receptor ECL2. Trypsin can activate PAR1 by cleaving at the activating cleavage site Arg41-Ser42 to expose the tethered ligand (2, 4, 29). Moreover, trypsin has also been reported to cleave PAR1 at residues Arg70-Leu71 and Lys82-Gln83, which amputates the tethered ligand from the receptor (4) (Fig. 1). Furthermore, these residues are suggested to be cleaved by trypsin much faster than the residues Arg41-Ser42 (9). Given that these two potential trypsin cleavage sites are located close to Asn75, it is not surprising that in the N62Q/N75Q and N75Q mutants, trypsin can gain easy access to these cleavage sites and therefore disarm these mutant receptors. In contrast, when PAR1 is fully glycosylated at Asn75, these cleavage sites are subsequently shielded by the attached oligosaccharide, and thus trypsin disarms WT hPAR1 less efficiently. Interestingly, the previous study by Nakayama et al. (9) reported that 100 nm trypsin cleaved PAR1 but did not activate it in HUVEC cells, thus suggesting receptor disarming. We therefore assume that the glycosylation status of PAR1 in HUVEC cells must be different from our PAR1-KNRK system. Indeed, previous studies reported that PAR1 expressed in HUVECs and platelets migrated as a homogenous species with an apparent mass of 66 kDa (27). This molecular mass is considerably below that reported in other cell systems, including ours, and suggests that PAR1 is not so heavily glycosylated in these cell types. Therefore, we suggest that the Asn62 and Asn75 might not be fully glycosylated in HUVECs, and therefore these potential trypsin cleavage sites around Asn75 are more readily available for trypsin cleavage and subsequent disarms.
We have also shown that the neutrophil proteinases elastase, cathepsin G, and proteinase 3 do not activate PAR1 in our KNRK cell system but rather disarm the receptor. Like us, Renesto et al. (13) reported that elastase, cathepsin G, and proteinase 3 can cleave and inactivate PAR1 in platelets and human endothelial cells. In contrast, Suzuki et al. (8) reported that elastase can activate PAR1 in human lung epithelial cells and also mentioned that proteolytic cleavage of PAR1 by elastase might activate or inactivate the receptor depending on the site of cleavage. The authors also suggested that the extent of glycosylation of the receptor could influence the site and extent of cleavage of the receptor (8).
In our current study, we note that elastase disarms N62Q/N75Q better than WT hPAR1 at the same concentration, and 1.5 nm elastase disarms N75Q, but not N62Q. The mass spectrometry study by Renesto et al. (13) indicated that elastase cleaves the N terminus of hPAR1 at Val72-Ser73 and Ile74-Asn75, which are located after the tethered ligand. Furthermore, Loew et al. (4) reported that elastase cleaved the PAR1 exodomain at Ala36-Thr37, Val72-Ser73, and Ala86-Phe87 sites with preferential cleavage at Val72, which would disable the receptor. Therefore, after amputating the Asn75 glycosylation, the potential cleavage sites Val72-Ser73 might be more easily accessible for elastase cleavage.
Because cathepsin G has been reported to have both activating and disarming actions on PARs (12, 13, 30, 31), we tested the role of N-terminal glycosylation of hPAR1 in regulating this proteinase. We have demonstrated that cathepsin G could clearly disarm N62Q/N75Q and WT hPAR1 to the same efficiency. In addition, our data also revealed that cathepsin G at 100 nm disarms N62Q and N75Q. Like us, others have also reported that cathepsin G at 30 nm could disarm PAR1 (30). Molino et al. (12) reported that in addition to the Arg41-Ser42 site, cathepsin G cleaves PAR1 at Phe43-Leu44 and Phe55-Trp56, removing the tethered ligand and rendering the receptor unresponsive to thrombin. In 1997, the mass spectrometry study of Renesto et al. (13) indicated that cathepsin G cleaves PAR1 on platelets and endothelial cells downstream of the thrombin cleavage site at Phe55-Trp56 and Tyr69-Arg70. Thus, we then concluded that cathepsin G might have four potential cleavage sites in the N terminus of hPAR1 (Arg41-Ser42, Phe43-Leu44, Phe55-Trp56, and Tyr69-Arg70). Furthermore, the kinetic analyses of Loew et al. (4) showed that the preferential cathepsin G cleavage site for PAR1 is the Phe55-Trp56 site. Moreover, our finding with the cathepsin G mutant is in accord with a previous report that has demonstrated that Phe55-Trp56 is the main region cleaved within the receptor N terminus by cathepsin G (12). Also, our Western blot data revealed that Asn62 is not heavily glycosylated in our KNRK system. Therefore, we suggest that cathepsin G sensitivity toward hPAR1 is not regulated by N-linked glycosylation.
We have also assessed proteinase 3 inhibition of thrombin-induced Ca2+ mobilization in WT hPAR1, N62Q/N75Q, N62Q, and N75Q. Our data revealed that 300 nm proteinase 3 disarms N62Q/N75Q but not WT hPAR1, N62Q, or N75Q. It is well established that proteinase 3 has an elastase-like specificity for Ala, Ser, and Val at the P1 site (32). One previous study on human oral epithelial cells (33) also pointed out that proteinase 3 can rapidly cleave PAR2 between Arg and Ser and relatively inefficiently cleave between Lys and Val. Furthermore, proteinase 3 cleaved the peptide corresponding to the N terminus of PAR2 at Arg36-Ser37, indicating that the site of the PAR2 is structurally accessible by proteinase 3 (33). Interestingly, one previous study on synthetic peptides (13) reported that the cleavage site for proteinase 3 in the N terminus of PAR1 is Val72-Ser73. In addition, the kinetic analysis of Loew et al. (4) on the N terminus domain of PAR1 showed that proteinase 3 early cleavage sites for PAR1 are Ala36-Thr37, Pro48-Asn49, Val72-Ser73, and Ala92-Ser93, and the late cleavage site is Pro54-Phe55. Furthermore, Sokolova and Reiser (29) suggested in a recent review that proteinase 3 cleaves human PAR1 at Ala36-Thr37 and Val72-Ser73. Although there are differences, previous studies have agreed that the Val72-Ser73 might be the earliest cleavage site for proteinase 3. We therefore altered that site by creating a mutation, Ala72-Ala73, in our N62Q/N75Q (named the proteinase 3 mutant), and indeed the proteinase 3 almost immediately lost the ability to disarm the mutant receptor. As mentioned by Loew et al. (4), there exists a late cleavage site between Pro54 and Phe55; we therefore suggest that apart from the Val72-Ser73 cleavage site, there might exist relatively inefficient later cleavage events between Pro54 and Phe55 and possibly at Pro48-Asn49. Thus, there might be a possibility that if we expose PAR1 to proteinase 3 for a greater length of time, proteinase 3 might disarm WT hPAR1. Indeed, we did notice that this phenomenon happened when we preincubated PAR1 with proteinase 3 for more than 3 min (data not shown). We have shown that in WT hPAR1 cells, proteinase 3 does not disarm the receptor immediately (within 1 min); however, after the removal of the glycans in Asn62 and Asn75, proteinase 3 immediately disarmed the glycosylation mutant receptor and inhibited thrombin-induced calcium mobilization. We therefore suggest that glycosylation of Asn62 and Asn75 together regulate hPAR1 signaling to proteinase 3.
Our data also revealed that thermolysin disarms N62Q/N75Q with 30-fold better efficiency than WT hPAR1. One previous study in insect SF9 cells (34) reported that thermolysin may represent an important mechanism of rapid receptor deactivation of the human thrombin receptor. Furthermore, like us, one previous study on PAR1 in astrocytes (35) found that treatment with thermolysin generated a thrombin-insensitive receptor, whereas the response to the activating peptide was not affected. In the literature, it is well established that the predicted thermolysin cleavage sites in PAR1 are within the tethered ligand domain (SFLLR) at Phe43-Leu44 and Leu44-Leu45, suggesting that thermolysin would be expected to remove the exodomain of PAR1 and destroy the tethered ligand and thus disarm the receptor (16, 36). Interestingly, in our study system, removing both of the glycosylation sequons (Asn62 and Asn75) resulted in a receptor that was more efficiently disarmed by thermolysin, whereas removal of glycosylation at the Asn62 sequon alone from the receptor had no observable effect on thermolysin disarming. However, the N75Q did display increased disarming compared with WT hPAR1, thus suggesting that this sequon regulates PAR1 disarming by thermolysin. Therefore, we predict that, apart from the well established cleavage sites, there might be some other thermolysin cleavage sites in the N terminus of PAR1, especially in the region of the glycosylation site Asn75. In theory, bacterial thermolysin cleaves peptide bonds N-terminally to the hydrophobic residues leucine and isoleucine, with some specificity for phenylalanine and valine (36). Therefore, we suggest that Leu71-Val72 in PAR1 may also be a potential thermolysin cleavage site, and it is very close to the Asn75 site. When amputating the Asn75 glycan, this potential thermolysin cleavage site (Leu71-Val72) will be more readily available for thermolysin cleavage.
In summary, hPAR1 is a heavily glycosylated receptor and is important for cell surface expression, receptor stability, and function. We show here for the first time that glycosylation in the N terminus of hPAR1 downstream of the tethered ligand (especially Asn75) governs receptor disarming to trypsin, thermolysin, and the neutrophil proteinases elastase and proteinase 3 but not cathepsin G. Thus, we concluded that N-linked glycosylation at the N terminus of hPAR1 plays a vital role in regulating the susceptibility to proteinase disarming of the receptor.
Footnotes
- PAR
- proteinase-activated receptor
- hPAR1
- human proteinase-activated receptor-1
- KNRK
- Kirsten murine sarcoma virus-transformed rat kidney
- eYFP
- enhanced yellow fluorescent protein
- hPAR1E
- human proteinase activated receptor-1 with eYFP tag
- ECL2
- extracellular loop 2
- HUVEC
- human umbilical vein endothelial cell
- N35–259Q
- N35Q/N62Q/N75Q/N250Q/N259Q mutant.
REFERENCES
- 1. Ramachandran R., Hollenberg M. D. (2008) Br. J. Pharmacol. 153, Suppl. 1, S263–S282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macfarlane S. R., Seatter M. J., Kanke T., Hunter G. D., Plevin R. (2001) Pharmacol. Rev. 53, 245–282 [PubMed] [Google Scholar]
- 3. Hirano K. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 27–36 [DOI] [PubMed] [Google Scholar]
- 4. Loew D., Perrault C., Morales M., Moog S., Ravanat C., Schuhler S., Arcone R., Pietropaolo C., Cazenave J. P., van Dorsselaer A., Lanza F. (2000) Biochemistry 39, 10812–10822 [DOI] [PubMed] [Google Scholar]
- 5. Vouret-Craviari V., Grall D., Chambard J. C., Rasmussen U. B., Pouysségur J., Van Obberghen-Schilling E. (1995) J. Biol. Chem. 270, 8367–8372 [DOI] [PubMed] [Google Scholar]
- 6. Suidan H. S., Bouvier J., Schaerer E., Stone S. R., Monard D., Tschopp J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8112–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molino M., Barnathan E. S., Numerof R., Clark J., Dreyer M., Cumashi A., Hoxie J. A., Schechter N., Woolkalis M., Brass L. F. (1997) J. Biol. Chem. 272, 4043–4049 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki T., Moraes T. J., Vachon E., Ginzberg H. H., Huang T. T., Matthay M. A., Hollenberg M. D., Marshall J., McCulloch C. A., Abreu M. T., Chow C. W., Downey G. P. (2005) Am. J. Respir. Cell Mol. Biol. 33, 231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakayama T., Hirano K., Shintani Y., Nishimura J., Nakatsuka A., Kuga H., Takahashi S., Kanaide H. (2003) Br. J. Pharmacol. 138, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schechter N. M., Brass L. F., Lavker R. M., Jensen P. J. (1998) J. Cell. Physiol. 176, 365–373 [DOI] [PubMed] [Google Scholar]
- 11. Ubl J. J., Grishina Z. V., Sukhomlin T. K., Welte T., Sedehizade F., Reiser G. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L1339–L1348 [DOI] [PubMed] [Google Scholar]
- 12. Molino M., Blanchard N., Belmonte E., Tarver A. P., Abrams C., Hoxie J. A., Cerletti C., Brass L. F. (1995) J. Biol. Chem. 270, 11168–11175 [DOI] [PubMed] [Google Scholar]
- 13. Renesto P., Si-Tahar M., Moniatte M., Balloy V., Van Dorsselaer A., Pidard D., Chignard M. (1997) Blood 89, 1944–1953 [PubMed] [Google Scholar]
- 14. Kuliopulos A., Covic L., Seeley S. K., Sheridan P. J., Helin J., Costello C. E. (1999) Biochemistry 38, 4572–4585 [DOI] [PubMed] [Google Scholar]
- 15. Norton K. J., Scarborough R. M., Kutok J. L., Escobedo M. A., Nannizzi L., Coller B. S. (1993) Blood 82, 2125–2136 [PubMed] [Google Scholar]
- 16. Déry O., Corvera C. U., Steinhoff M., Bunnett N. W. (1998) Am. J. Physiol. 274, C1429–C1452 [DOI] [PubMed] [Google Scholar]
- 17. Shpacovitch V., Feld M., Bunnett N. W., Steinhoff M. (2007) Trends Immunol. 28, 541–550 [DOI] [PubMed] [Google Scholar]
- 18. Hansen K. K., Saifeddine M., Hollenberg M. D. (2004) Immunology 112, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Compton S. J., Sandhu S., Wijesuriya S. J., Hollenberg M. D. (2002) Biochem. J. 368, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Compton S. J., Renaux B., Wijesuriya S. J., Hollenberg M. D. (2001) Br. J. Pharmacol. 134, 705–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Compton S. J. (2003) Drug Dev. Res. 59, 350–354 [Google Scholar]
- 22. Opdenakker G., Rudd P. M., Ponting C. P., Dwek R. A. (1993) FASEB J. 7, 1330–1337 [DOI] [PubMed] [Google Scholar]
- 23. Wheatley M., Hawtin S. R. (1999) Hum. Reprod. Update 5, 356–364 [DOI] [PubMed] [Google Scholar]
- 24. Soto A. G., Trejo J. (2010) J. Biol. Chem. 285, 18781–18793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tordai A., Brass L. F., Gelfand E. W. (1995) Biochem. Biophys. Res. Commun. 206, 857–862 [DOI] [PubMed] [Google Scholar]
- 26. Mize G. J., Harris J. E., Takayama T. K., Kulman J. D. (2008) Protein Expr. Purif. 57, 280–289 [DOI] [PubMed] [Google Scholar]
- 27. Brass L. F., Vassallo R. R., Jr., Belmonte E., Ahuja M., Cichowski K., Hoxie J. A. (1992) J. Biol. Chem. 267, 13795–13798 [PubMed] [Google Scholar]
- 28. Brass L. F., Pizarro S., Ahuja M., Belmonte E., Blanchard N., Stadel J. M., Hoxie J. A. (1994) J. Biol. Chem. 269, 2943–2952 [PubMed] [Google Scholar]
- 29. Sokolova E., Reiser G. (2007) Pharmacol. Ther. 115, 70–83 [DOI] [PubMed] [Google Scholar]
- 30. Ramachandran R., Sadofsky L. R., Xiao Y., Botham A., Cowen M., Morice A. H., Compton S. J. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L788–L798 [DOI] [PubMed] [Google Scholar]
- 31. Sambrano G. R., Huang W., Faruqi T., Mahrus S., Craik C., Coughlin S. R. (2000) J. Biol. Chem. 275, 6819–6823 [DOI] [PubMed] [Google Scholar]
- 32. Rao N. V., Wehner N. G., Marshall B. C., Gray W. R., Gray B. H., Hoidal J. R. (1991) J. Biol. Chem. 266, 9540–9548 [PubMed] [Google Scholar]
- 33. Uehara A., Sugawara S., Muramoto K., Takada H. (2002) J. Immunol. 169, 4594–4603 [DOI] [PubMed] [Google Scholar]
- 34. Chen X., Earley K., Luo W., Lin S. H., Schilling W. P. (1996) Biochem. J. 314, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ubl J. J., Sergeeva M., Reiser G. (2000) J. Physiol. 525, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamilton J. R., Chow J. M., Cocks T. M. (1999) Br. J. Pharmacol. 127, 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]