Abstract
The mouse M-lysozyme gene is exclusively expressed in myeloid cells of the blood system being progressively turned on upon cell differentiation. In this study the mechanism controlling this tissue- and differentiation stage-specific gene expression was analyzed at the level of chromatin structure. A complex pattern consisting of constitutive and differentiation dependent DNasel hypersensitive sites (HSs) was found in a set of various myeloid cell lines, representing different stages of maturity. The chromatin of a lymphoid cell line, which does not express the lysozyme gene, is completely insensitive to DNasel digestion. Chromatin analysis of two multipotent hematopoietic stem cell lines which can be differentiated in vitro to mature myeloid cells confirmed that these identified DNasel HSs are specific for distinct differentiation stages, rather than being a characteristic feature of the cell lines. Additionally, the stem cell studies revealed that the hypersensitivity of the chromatin domain is already established at the multipotent stage. DNA fragments spanning a cell type- and differentiation stage-specific cluster of HSs in the 3' region of the gene showed enhancer activity in all cell types tested. In the light of this lack of specificity, we suggest that cell type-specific modification of the chromatin structure in this region may play a role in determining the binding of a widespread transcription factor, and hence contribute to the time specificity of lysozyme M gene expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baeuerle P. A., Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988 Apr 22;53(2):211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- Choi O. R., Engel J. D. Developmental regulation of beta-globin gene switching. Cell. 1988 Oct 7;55(1):17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Dente L., Cortese R. Cell-specific expression of a transfected human alpha 1-antitrypsin gene. Cell. 1985 Jun;41(2):531–540. doi: 10.1016/s0092-8674(85)80026-x. [DOI] [PubMed] [Google Scholar]
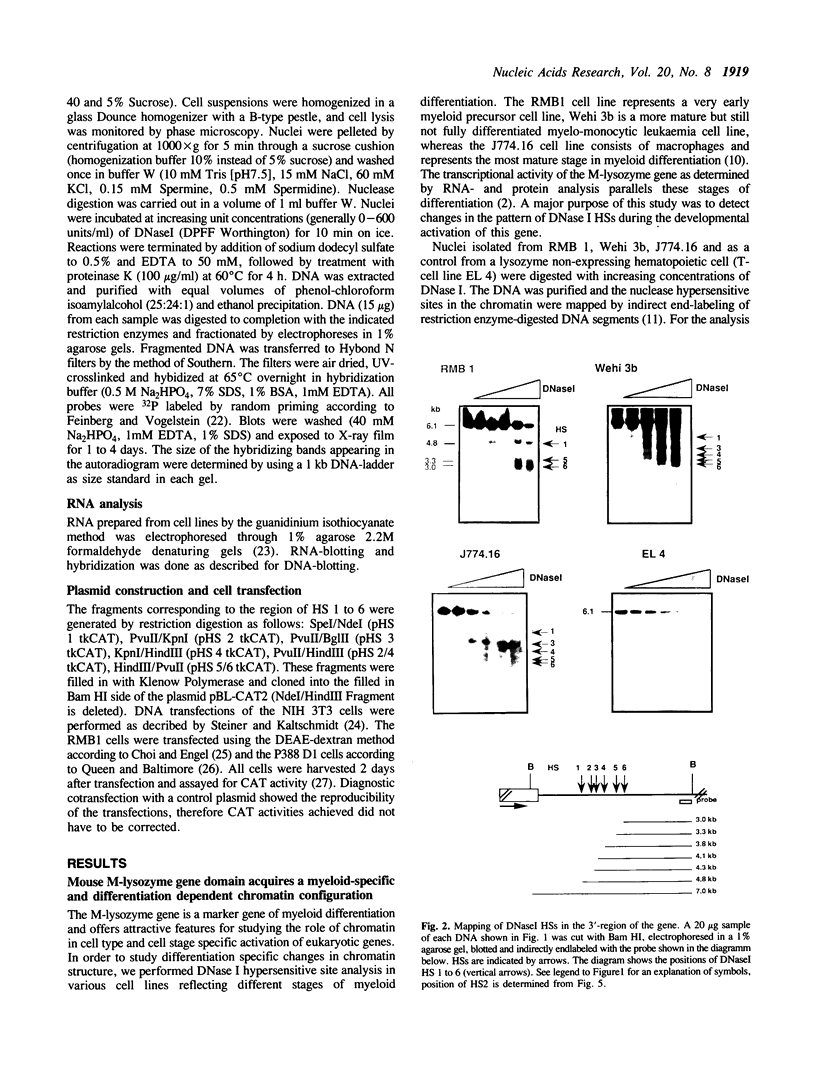
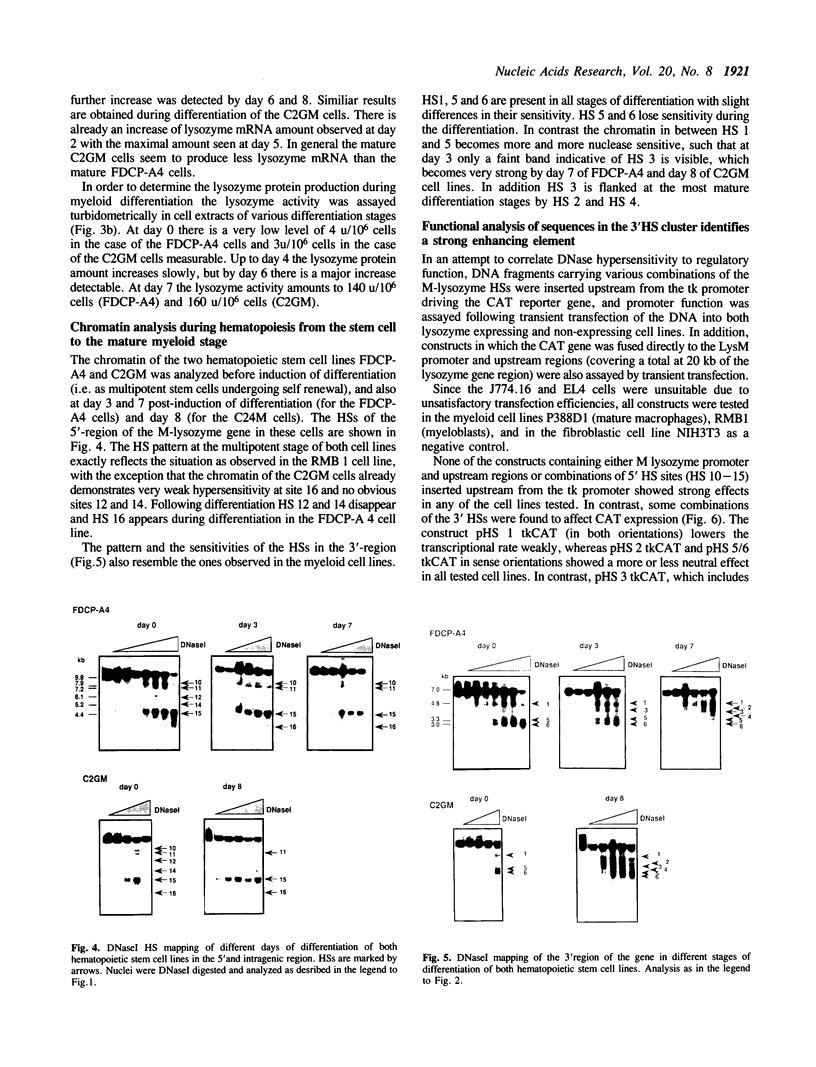
- Cross M., Mangelsdorf I., Wedel A., Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6232–6236. doi: 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M., Renkawitz R. Repetitive sequence involvement in the duplication and divergence of mouse lysozyme genes. EMBO J. 1990 Apr;9(4):1283–1288. doi: 10.1002/j.1460-2075.1990.tb08237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar V., Nandi A., Schildkraut C. L., Skoultchi A. I. Erythroid-specific nuclease-hypersensitive sites flanking the human beta-globin domain. Mol Cell Biol. 1990 Aug;10(8):4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990 Jun;2(3):437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ford A. M., Watt S. M., Furley A. J., Molgaard H. V., Greaves M. F. Cell lineage specificity of chromatin configuration around the immunoglobulin heavy chain enhancer. EMBO J. 1988 Aug;7(8):2393–2399. doi: 10.1002/j.1460-2075.1988.tb03084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Thompson C., Elder J. T., Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Heyworth C. M., Dexter T. M., Kan O., Whetton A. D. The role of hemopoietic growth factors in self-renewal and differentiation of IL-3-dependent multipotential stem cells. Growth Factors. 1990;2(2-3):197–211. doi: 10.3109/08977199009071506. [DOI] [PubMed] [Google Scholar]
- Leenen P. J., Jansen A. M., van Ewijk W. Murine macrophage cell lines can be ordered in a linear differentiation sequence. Differentiation. 1986;32(2):157–164. doi: 10.1111/j.1432-0436.1986.tb00568.x. [DOI] [PubMed] [Google Scholar]
- Maschek U., Pülm W., Hämmerling G. J. Altered regulation of MHC class I genes in different tumor cell lines is reflected by distinct sets of DNase I hypersensitive sites. EMBO J. 1989 Aug;8(8):2297–2304. doi: 10.1002/j.1460-2075.1989.tb08356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Ralph P., Moore M. A., Nilsson K. Lysozyme synthesis by established human and murine histiocytic lymphoma cell lines. J Exp Med. 1976 Jun 1;143(6):1528–1533. doi: 10.1084/jem.143.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooncer E., Heyworth C. M., Dunn A., Dexter T. M. Self-renewal and differentiation of interleukin-3-dependent multipotent stem cells are modulated by stromal cells and serum factors. Differentiation. 1986;31(2):111–118. doi: 10.1111/j.1432-0436.1986.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Stanley E. R., Heard P. M. Factors regulating macrophage production and growth. Purification and some properties of the colony stimulating factor from medium conditioned by mouse L cells. J Biol Chem. 1977 Jun 25;252(12):4305–4312. [PubMed] [Google Scholar]
- Steiner C., Kaltschmidt C. An automated method for calcium phosphate-mediated gene transfer. Trends Genet. 1989 May;5(5):138–138. doi: 10.1016/0168-9525(89)90053-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]