Abstract
Dyskeratosis congenita (DC) is a progressive and heterogeneous congenital disorder that affects multiple systems and is characterized by bone marrow failure and a triad of abnormal skin pigmentation, nail dystrophy, and oral leukoplakia. One common feature for all DC patients is abnormally short telomeres and defects in telomere biology. Most of the known DC mutations have been found to affect core components of the telomerase holoenzyme. Recently, multiple mutations in the gene encoding the telomeric protein TIN2 have been identified in DC patients with intact telomerase genes, but the molecular mechanisms underlying TIN2 mutation-mediated DC remain unknown. Here, we demonstrate that ectopic expression of TIN2 with DC missense mutations in human cells led to accelerated telomere shortening, similar to the telomere phenotypes found in DC patients. However, this telomere shortening was not accompanied by changes in total telomerase activity, localization of TIN2, or telomere end protection status. Interestingly, we found TIN2 to participate in the TPP1-dependent recruitment of telomerase activity. Furthermore, DC mutations in TIN2 led to its decreased ability to associate with TERC and telomerase activity. Taken together, our data suggest that TIN2 mutations in DC may compromise the telomere recruitment of telomerase, leading to telomere shortening and the associated pathogenesis.
Keywords: Cellular Regulation, Chromatin, DNA Damage, Signal Transduction, Telomere
Introduction
Dyskeratosis congenita (DC)3 patients exhibit a plethora of disease features in addition to the cutaneous symptoms, including mental retardation, pulmonary fibrosis, liver cirrhosis, premature aging phenotypes, and predisposition to cancer (1, 2, 8). Abnormally short telomere length is a hallmark of DC (3, 4). With progressive telomere attrition, the disease becomes more severe, with an earlier onset in successive generations of affected families (9, 10). The telomere shortening phenotype indicates defects in telomere maintenance. Here, considerable genetic heterogeneity exists, where mutations have been identified in genes encoding various subunits of the telomerase holoenzyme, including TERC, TERT, DKC1, NOP10, and NHP2 (5, 9, 11–14). The core telomerase comprises the TERC RNA and TERT reverse transcriptase and extends telomeres by adding G-rich repeats to the ends of chromosomes (15). Telomerase activity is observed in stem cells and in certain proliferative tissues such as the hematopoietic and dermatological systems (16, 17), all of which are affected in DC patients. The mutations found in telomerase subunits of DC patients have been shown to reduce TERC levels and telomerase activity (9, 18, 19), leading to shortened telomeres and ultimately defects in proliferative tissues.
Recently, novel heterozygous mutations have been identified in the TINF2 gene of certain DC patients (6, 7, 20). These patients have significantly shorter telomeres compared with control populations and no additional mutations in TERC, TERT, or DKC1 (7). These patients had normal TERC levels (6), although the status of telomerase activity was not studied. The prevalence of TINF2 mutations was estimated to be 11% in all DC patients (6). In addition to DC, mutations of TINF2 were also identified in patients with other diseases associated with bone marrow failure (e.g. ataxia-pancytopenia and aplastic anemia) (6, 21, 22).
TIN2 is the protein product of TINF2 and a subunit of the six-protein complex shelterin/telosome (23, 24). This complex protects telomere ends (25) and cooperates with the telomerase to maintain the telomeres (26–29). TIN2 plays a central role in the assembly and function of the six-protein complex, connecting the double-stranded DNA-binding proteins TRF1 (telomere repeat binding factor) and TRF2 to the single-stranded DNA-binding unit TPP1/POT1. However, the relationship between TIN2 dysfunction and DC and the consequences of TIN2 mutations remain to be elucidated.
Here, we report our investigation of the mechanism through which TIN2 mutations may contribute to telomere shortening. Using telomerase-positive human cells that ectopically expressed TIN2 mutants, we recapitulated the telomere shortening observed in DC patients and provided the potential links between such defects and the recruitment of telomerase.
EXPERIMENTAL PROCEDURES
Vectors and Cell Lines
cDNAs encoding human wild-type and mutant TIN2 (K280E, R282H, and R282S) and dyskerin were cloned into a pCL-based retroviral vector (c-terminal FLAG or 2×FLAG) for transient expression in 293T cells or stable expression in HTC75 cells. cDNAs encoding human TRF1, TRF2, and TPP1 were cloned into the vector pDEST-27 (GST tag; Invitrogen) for expression in 293T cells. Doxycycline-inducible cell lines were established using HT1080 cells. Various TIN2 (wild-type, K280E, R282H, R282S, and Δ90) and TPP1 (wild-type, ΔOB, ΔC22, and ΔOBΔC22) constructs were cloned into a pHAGE-based SFB tag lentiviral vector (C-terminal S-, FLAG-, and streptavidin-binding tag; doxycycline-inducible expression) (30). For protein expression induction, 8.5–200 ng/ml doxycycline was used. TPP1-ΔOB contains residues 244–544. TPP1-ΔC22 includes residues 1–522. TPP1-ΔOBΔC22 spans residues 244–522. The knockdown shRNA sequences of TIN2 (5′-GGAGCACATTCTTTGCCTG-3′), TPP1 (5′-GTGGTACCAGCATCAGCCTT-3′), and GFP (5′-CACAAGCTGGAGTACAACT-3′) were cloned into a retroviral vector as described previously (31).
Immunoprecipitation, Western Blotting, and Antibodies
Co-immunoprecipitation studies were performed as described previously (30), where cells transiently expressing various proteins were lysed in 1× buffer containing 1 m Tris-HCl (pH 8.0), 1 mm EDTA, 100 mm NaCl, 0.5% Nonidet P-40, 1 mm DTT, and proteinase inhibitor mixture. GST fusion proteins were pulled down by glutathione-agarose beads (Molecular Probes). The antibodies used were HRP-conjugated anti-GST polyclonal antibody (GE Healthcare), HRP-conjugated anti-FLAG antibody M2 (Sigma), rabbit anti-FLAG polyclonal antibody (Sigma), goat anti-actin polyclonal antibody (Santa Cruz Biotechnology), rabbit anti-tubulin monoclonal antibody (Epitomics, Inc.), mouse anti-TRF2 monoclonal antibody (Calbiochem), rabbit anti-RAP1 polyclonal antibody (Bethyl Laboratories), rabbit anti-TPP1 and anti-TIN2 polyclonal antibodies (32), and goat anti-TRF1 antibody (31).
Telomeric Repeat Amplification Protocol (TRAP)
To measure total cellular telomerase activity, TRAP assays were performed using the gel-based TRAPeze telomerase detection kit (Millipore) as described previously (27). Briefly, cells were lysed in CHAPS buffer and centrifuged. The supernatant was collected and diluted before the two-step TRAP assay. The amplified DNA was resolved on a 12% Tris/borate/EDTA polyacrylamide gel and visualized with SYBR Green I (Invitrogen).
Real-time Quantitative PCR-based TRAP (Q-TRAP) Assay
5–10 × 106 cells were lysed in high salt buffer (20 mm HEPES (pH 7.9), 0.42 m KCl, 25% glycerol, 0.2% Nonidet P-40, 0.1 mm EDTA, 1 mm DTT, and protease inhibitors) on ice for 30 min. The lysates were then diluted with 5 volumes of low salt buffer (20 mm HEPES (pH 7.9), 100 mm KCl, 25% glycerol, 0.1 mm EDTA, 1 mm DTT, and protease inhibitors) and centrifuged at 14,000 rpm for 10 min at 4 °C. The supernatant was collected for immunoprecipitations with the appropriate antibodies. FLAG- or SFB-tagged proteins were immunoprecipitated by anti-FLAG antibody M2-agarose beads (Sigma) and eluted in 180 μl of elution buffer (25 mm Tris-HCl (pH 7.4), 136 mm NaCl, 2.6 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10% glycerol, 1 mm DTT, and protease and RNase inhibitor) with FLAG or 3×FLAG peptides (Sigma), respectively. The eluants were then diluted 3–5-fold before being used for Q-TRAP. For cell lines in which TIN2 expression was induced to endogenous levels, no dilution of the FLAG eluant was needed prior to Q-TRAP.
Q-TRAP was carried out essentially as described (33). Each 25-μl Q-TRAP reaction contained 2 μl of FLAG-eluted immunoprecipitates, 100 ng of TS primer (5′-AATCCGTCGAGCAGAGTT-3′), 100 ng of ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′), and 1 mm EGTA in SYBR Green PCR Master Mix (Applied Biosystems). The reaction mixtures were incubated at 30 °C for 30 min and then PCR-amplified in 40 cycles at 95 °C for 15 s and 60 °C for 60 s using an ABI StepOnePlus real-time PCR system (Applied Biosystems).
Real-time Quantitative PCR (qPCR)
Real-time qPCR was performed as described previously (34). Total RNA was isolated with the RNeasy mini kit (Qiagen). For qPCR of TERC following immunoprecipitation, the eluted immunoprecipitates were incubated at 85 °C for 7 min and then directly used for reverse transcription using the iScript cDNA synthesis kit (Bio-Rad). Real-time qPCR amplification was carried out using SYBR Green Master Mix and the ABI StepOnePlus real-time PCR system. For detection of TERC, the primers used were 5′-TCTAACCCTAACTGAGAAGGGCGTAG-3′ (forward) and 5′-GTTTGCTCTAGAATGAACGGTGGAAG-3′ (reverse) (35). The 18 S primers used were 5′-CGAACGTCTGCCCTATCAACTT-3′ (forward) and 5′-ACCCGTGGTCACCATGGTA-3′ (reverse).
TRF Assay
HTC75 cells stably expressing control and FLAG-tagged wild-type and mutant TIN2 proteins were generated and passaged for TRF analysis as described previously (32). The results were analyzed using TeloRun (36).
Immunofluorescence
Cells were grown on glass coverslips, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and then blocked with 3% goat serum. To detect telomere dysfunction-induced foci, cells were co-stained with anti-TRF2 monoclonal antibody and anti-53BP1 polyclonal antibody (27). DAPI was used to visualize the nuclei.
Proliferation Assays
HTC75 cells expressing wild-type or mutant TIN2 were plated onto 6-well plates at 5 × 104 cells/well and maintained for 1 week. At the indicated time points, cells were trypsinized and counted.
Cell Cycle Analysis
Cells were fixed with cold 70% ethanol and stained with 50 μg/ml propidium iodide in PBS containing 0.2 mg/ml DNase-free RNase A. Cells were then analyzed with an LSR II flow cytometer (BD Biosciences) and FlowJo software (Tree Star, Inc.).
RESULTS
DC Mutations in TIN2 Lead to Telomere Shortening
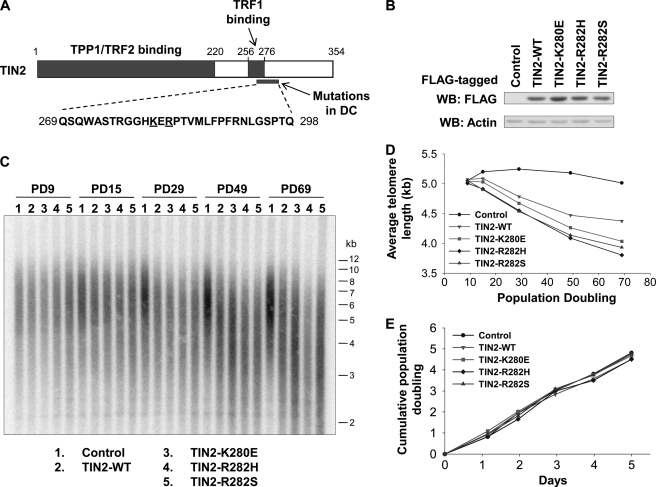
To date, a total of 18 TIN2 mutations in 43 DC patients have been reported (supplemental Fig. S1A) (6, 7, 20). These mutations are tightly clustered in a small region (amino acids 269–298) that is highly conserved across mammalian species (Fig. 1A and supplemental Fig. S1B). Of these, Lys-280 and Arg-282 appear to be mutation hot spots, with K280E, R282H, and R282S mutations found in ∼50% of the patients. We first studied these three missense mutations in telomerase-positive human cells to determine whether telomere length maintenance was affected. Human fibrosarcoma HTC75 cells stably expressing wild-type or mutant TIN2 proteins that were FLAG-tagged at the C terminus were generated. These exogenous TIN2 proteins were expressed at a level comparable with each other (Fig. 1B). As shown in Fig. 1 (C and D), although telomere length remained fairly constant in control cells, expression of wild-type TIN2 led to gradual and progressive shortening of telomeres, consistent with the role of TIN2 as a negative regulator of telomere length (37, 38). In comparison, expression of the TIN2 mutants led to accelerated telomere shortening. In fact, this phenotype was apparent as early as population doubling (PD) 15. It is possible that this difference in telomere length resulted from differences in cell proliferation. However, the growth rates and cell cycle progression of wild-type and mutant TIN2 cells were indistinguishable from those of control cells (Fig. 1E and supplemental Fig. S2A). Taken together, our results indicate that ectopic expression of TIN2 mutants in HTC75 cells recapitulated the telomere shortening defect in DC, providing a useful system to study the underlying mechanism of TIN2 dysfunction and DC.
FIGURE 1.
Ectopic expression of TIN2 DC mutants results in telomere shortening. A, schematic representation of TIN2 domain organization. The region frequently mutated in DC is shown. The amino acid positions studied here are underlined. B, Western blot (WB) analysis of HTC75 cells expressing FLAG-tagged wild-type or mutant TIN2. Control, empty vector. Anti-actin antibodies were used for loading controls. C, telomere restriction fragment analysis of cells from B. D, quantification of C to determine average telomere length. E, growth curve plot of cells from B.
DC Mutations in TIN2 Do Not Grossly Affect Telomerase Activity, Telomere End Protection, or Other Key Telomeric Proteins
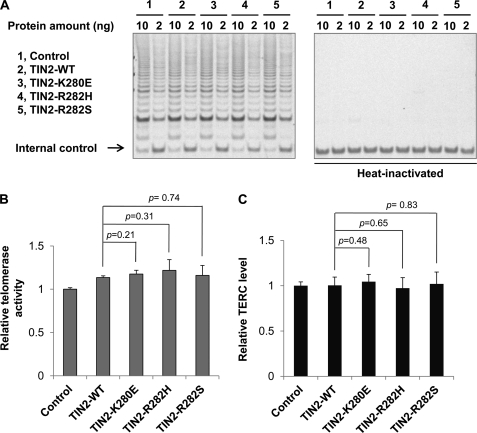
DC patients with mutations in TERC, TERT, DKC1, NOP10, and NHP2 exhibit reduced telomerase activity and decreased TERC levels (9, 13, 14, 18, 19, 39–41). We therefore tested whether TIN2 mutants could also affect TERC levels and telomerase activity. As shown in Fig. 2 (A and B), expression of TIN2 missense mutants did not appear to affect total telomerase activities as measured by TRAP assays. In addition, real-time PCR analysis revealed no significant difference in TERC levels among cells expressing wild-type or mutant TIN2 (Fig. 2C). Although TRAP assays are limited in terms of precise quantification, these data are nonetheless consistent with findings in DC patients with TIN2 mutations (6) and suggest that the molecular mechanism responsible for TIN2-related DC may differ from that for classic DC.
FIGURE 2.
DC mutations in TIN2 do not affect the overall telomerase activity. A, TRAP assays were carried out with extracts from HTC75 cells expressing vector alone (Control) or FLAG-tagged wild-type and mutant TIN2. Heat-inactivated samples served as negative controls. B, the telomerase activities of cells from A were quantified. Error bars represent S.E. from three independent experiments. p values were calculated using Student's t test. C, the levels of TERC transcription in cells from A were determined by qPCR. The results were normalized to the transcriptional level of 18 S RNA. Error bars represent S.E. from triplicates of two independent experiments. p values were calculated using Student's t test.
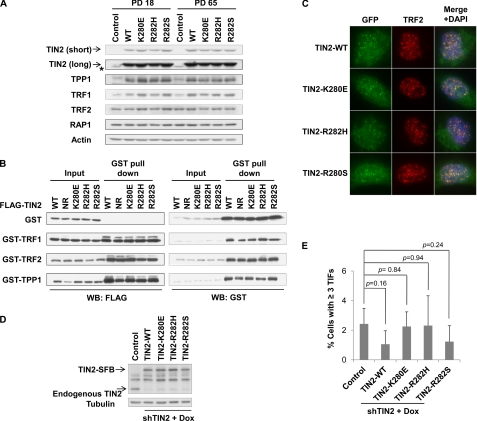
Changes in any of the six core telomeric proteins (TIN2, TRF1, TRF2, RAP1, TPP1, and POT1) are known to affect each other. For example, depletion of TIN2 by siRNA results in a reduction of the TRF1 protein level (37), and the stability of TPP1 is dependent on TIN2 levels and vice versa (28). Furthermore, the abundance of individual core telomeric proteins has a great influence on telomere length homeostasis. Inhibition of TRF1, TRF2, RAP1, TIN2, TPP1, or POT1 leads to elongated telomeres (37, 42–45), whereas overexpression of TRF1, TRF2, and TIN2 leads to telomere shortening (38, 46–48). We therefore postulated that DC mutations of TIN2 might affect the expression of TIN2 and other telomeric proteins. To this end, we analyzed the expression of the six core telomeric proteins by immunoblotting using PD18 and PD65 of the cells from Fig. 1. The expression of wild-type and mutant TIN2 proteins appeared similar and did not change with long-term passage (Fig. 3A). As expected (28, 37), ectopic expression of TIN2 (either wild-type or mutant) led to an increased total amount of endogenous TRF1 and TPP1, but not TRF2 and RAP1 (Fig. 3A). Importantly, even at PD65, when telomere length differed significantly between the cells (Fig. 1D), little variation in the protein levels of TRF1, TRF2, RAP1, and TPP1 could be observed in wild-type versus mutant TIN2 cells (Fig. 3A and supplemental Fig. S2B). These data indicate that TIN2 mutations have little effect on the expression or stability of the other core telomeric proteins.
FIGURE 3.
DC mutations in TIN2 do not affect the expression of key telomeric proteins, their interaction with TIN2, or telomere end protection. A, the expression levels of the indicated endogenous telomeric proteins in control (empty vector) cells and cells expressing wild-type or mutant TIN2 were analyzed by Western blotting. Cells from early (PD18) and late (PD65) passages were examined. Arrowheads indicate FLAG-tagged wild-type and mutant TIN2. The asterisk indicates endogenous TIN2. short, short exposure; long, long exposure. B, TIN2 mutants maintained their ability to interact with TRF1, TRF2, and TPP1. FLAG-tagged wild-type TIN2 or mutants were transiently coexpressed with GST alone or with GST-tagged TRF1, TRF2, and TPP1 in 293T cells. GST fusion proteins were pulled down with glutathione beads, and the precipitated proteins were detected by Western blotting (WB). NR, nonrelevant sample. C, TIN2 mutants were targeted to the telomeres. Cells expressing GFP-tagged wild-type TIN2 or mutants were immunostained with anti-TRF2 antibodies (red). D, shown is the establishment of HT1080 cells with inducible TIN2 expression and stable TIN2 shRNA (shTIN2) knockdown. Expression of SFB-tagged wild-type or mutant TIN2 was induced with doxycycline (Dox). Control, empty vector. Anti-tubulin antibodies were used as a loading control. E, shown is the quantification of telomere dysfunction-induced focus assays performed with cells from D. Only cells with three or more 53BP1 foci that also co-localized with TRF2 foci were scored. Error bars represent S.E. (n = 3). p values were calculated using Student's t test. TIFs, telomere dysfunction-induced foci.
TIN2 interacts with TRF2 and TPP1 through its N terminus and with TRF1 through a domain located close to the DC mutation hot spot (Fig. 1A) (23, 31, 32, 37, 38, 45, 48–50). It has been postulated that TRF1-TIN2 interaction might be altered by TIN2 mutations (7). To test this idea, we carried out pulldown experiments in HEK293 cells transiently coexpressing TIN2 and other telomeric proteins. FLAG-tagged wild-type TIN2 or mutants were coexpressed with GST-tagged TRF1, TRF2, or TPP1. Consistent with our data on endogenous proteins (Fig. 3A), TIN2 mutants had no impact on the expression of GST-tagged TRF1, TRF2, or TPP1 (Fig. 3B). Furthermore, these mutants retained their ability to interact with other telomeric proteins (Fig. 3B). Co-immunoprecipitation experiments using differently tagged TIN2 and telomeric proteins showed similar results (supplemental Fig. S3). Together, these results support the notion that the interaction with core telomeric proteins remains intact for TIN2 with DC mutations.
An additional level of regulation of telomeric proteins involves subcellular localization. Although the six core telomeric proteins are all targeted to the telomeres, some of them can be found in other cellular compartments as well (51–54). For instance, TIN2, TPP1, and POT1 can localize to both the cytoplasm and nucleus (54). A single amino acid mutation in the nuclear export signal of TPP1 can disrupt its nuclear export as well as telomere maintenance (54). To address whether a similar phenomenon can be observed with TIN2 DC mutations, we introduced GFP-tagged wild-type and mutant TIN2 into HTC75 cells. In all cases, GFP signals could be found in both the nucleus and cytoplasm (Fig. 3C), as reported previously for TIN2 (54). Furthermore, the nuclear signals of either wild-type or mutant TIN2 exhibited punctate patterns that overlapped with TRF2 signals, indicating that the DC mutations did not alter the cellular localization of TIN2.
The six core telomeric proteins help protect chromosome ends from being recognized as damaged DNA (25), and TIN2 plays an integral part in this process. It has been shown that either N- or C-terminal truncation of TIN2 can lead to exposed telomere ends and activation of DNA damage response, as evidenced by the detection of telomere dysfunction-induced foci (50, 55). This is probably due to the disruption of TRF1-TIN2 or TRF2-TIN2 interactions (50, 55). To rule out the compensatory effects of endogenous TIN2, we generated HT1080 cells in which endogenous TIN2 was stably knocked down by shRNA expression. Wild-type and mutant TIN2 proteins under the control of a tetracycline-inducible promoter were then introduced into these cells. As shown in Fig. 3D, these cells expressed wild-type and mutant TIN2 at levels similar to endogenous TIN2. We found no significant increase in cells containing telomere dysfunction-induced foci when mutant TIN2 expression was induced (Fig. 3E), indicating that DC mutations have minimal impact on TIN2-mediated telomere protection and therefore are unlikely to disrupt TIN2 interaction with other core telomeric proteins.
TIN2 with DC Mutations Is Compromised in Its Ability to Associate with the Telomerase
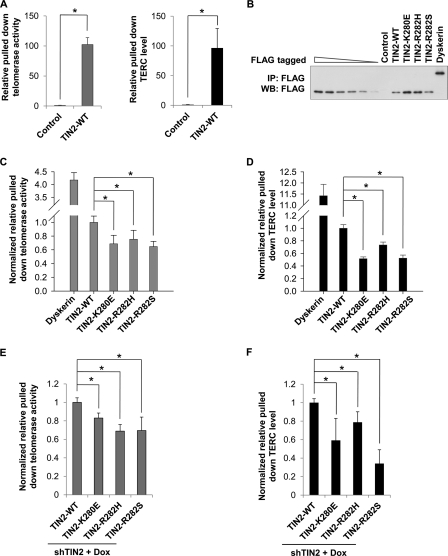
Mutant TIN2 cells exhibit accelerated shortening of telomeres, which is not accompanied by detectable changes in telomere end protection or total telomerase activity. We therefore postulated that DC mutations in TIN2 might affect telomerase recruitment rather than its activity per se. The TIN2-interacting protein TPP1 can interact directly with the telomerase and mediate telomerase recruitment to the telomeres (27–29). Given the physical and functional links between TIN2 and TPP1, it is possible that TIN2 might also associate with the telomerase and participate in telomerase recruitment. To test this possibility, we assayed for telomerase activity in FLAG-TIN2 immunoprecipitates from HTC75 cells by real-time qPCR. As shown in Fig. 4A, anti-FLAG-TIN2 immunoprecipitation was able to bring down a significant amount of telomerase activity as well as TERC compared with controls.
FIGURE 4.
DC mutations in TIN2 compromise its ability to associate with the telomerase. A, wild-type TIN2 could bring down telomerase activity. FLAG-tagged wild-type TIN2 was immunoprecipitated from HTC75 cells with anti-FLAG antibodies, eluted with FLAG peptides, and used for Q-TRAP and TERC reverse transcription-PCR. B, anti-FLAG immunoprecipitates (IP) from HTC75 cells expressing wild-type or mutant TIN2 proteins were eluted with FLAG peptides for quantitative immunoblotting. TIN2 protein mixtures were serially diluted to obtain a calibration curve to normalize the protein amount in each immunoprecipitate. Dyskerin was included as a positive control for subsequent analysis. WB, Western blot. C, shown are the results from Q-TRAP analysis of the FLAG immunoprecipitates from B. The results were normalized based on the protein amount calculated in B. D, shown are the results from real-time qPCR analysis of the level of TERC in the FLAG immunoprecipitates from B. The results were normalized to the protein amount in the immunoprecipitates. E and F, examination of TIN2-associated telomerase activity and TERC levels in TIN2-inducible cells from supplemental Fig. S4. Cells stably expressing TIN2 shRNA (shTIN2) and doxycycline (Dox)-inducible SFB-tagged TIN2 proteins (supplemental Fig. S4) were treated with doxycycline, followed by immunoprecipitation with anti-FLAG antibodies. The immunoprecipitates were eluted with 3×FLAG peptides and used in Q-TRAP for telomerase activity (E) and qPCR for TERC levels (F) as described above. In A and C–F, error bars represent S.E. from triplicates of two independent experiments. p values were calculated using Student's t test. *, p < 0.05.
We next examined whether DC mutations would affect the amount of TERC or telomerase activity that associated with TIN2. The qPCR results from these experiments were normalized to the amount of proteins that were immunoprecipitated from each cell line (Fig. 4B). Compared with wild-type TIN2, the TERC-binding telomerase subunit dyskerin was able to bring down considerably more telomerase activity and TERC (Fig. 4C), consistent with a previous report (56). On the other hand, DC mutations led to an ∼40% decrease in both the level of telomerase activity (Fig. 4C) and the amount of TERC (Fig. 4D) that could be brought down by TIN2. To exclude the possibility that overexpression of TIN2 may alter telomerase regulation, we used TIN2 shRNA knockdown cells (Fig. 3D) to express SFB-tagged wild-type and mutant TIN2 at levels comparable with endogenous TIN2 (supplemental Fig. S4A). We immunoprecipitated TIN2 complexes from these cells and found that DC mutations in TIN2 did not affect the level of co-precipitated endogenous TPP1 (supplemental Fig. S4B). In contrast, a significant reduction in the level of telomerase activity (Fig. 4E) and amount of TERC (Fig. 4F) was observed in the immunoprecipitates from mutant TIN2 cells. This finding is consistent with our observation when wild-type or mutant TIN2 was transiently coexpressed with TERT and TPP1 (supplemental Fig. S5). In these co-precipitation experiments, all three TIN2 DC mutants appeared to associate with less TERT. Collectively, these results indicate a defect in the ability of TIN2 DC mutants to recruit the telomerase.
The TIN2/TPP1 Heterodimer Recruits the Telomerase
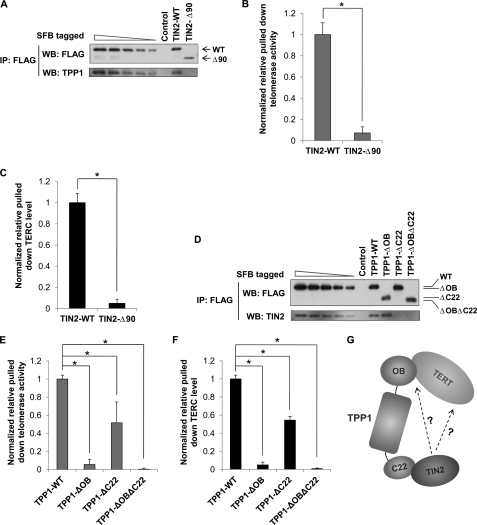
We have shown previously that TPP1 can directly interact with the telomerase through its OB (oligonucleotide- and oligosaccharide-binding) fold and thereby recruit the telomerase to the telomeres (27). Furthermore, Abreu et al. (28) have demonstrated that knockdown of TIN2 leads to attenuated telomere recruitment of the telomerase. Coupled with the fact that TIN2 and TPP1 can interact with each other, our findings here raise the possibility that TIN2-telomerase association may require TIN2-TPP1 interaction. To test this idea, we investigated whether TIN2 mutants defective in TPP1 binding could still associate with the telomerase. The N-terminal region of TIN2 mediates TIN2-TPP1 binding (Fig. 1A) (48). Deletion of this domain in TIN2 (TIN2-Δ90) abrogated its ability to associate with TPP1 (Fig. 5A). Importantly, TIN2-Δ90 could no longer bring down telomerase activity or TERC (Fig. 5, B and C). Furthermore, knockdown of TPP1 led to diminished association of TIN2 with the telomerase (supplemental Fig. S6), consistent with previous work demonstrating the importance of TPP1 in telomerase recruitment (28). Taken together, our data suggest that TIN2-TPP1 interaction is important for TIN2 association with the telomerase.
FIGURE 5.
TIN2/TPP1 heterodimer recruits telomerase. A, anti-FLAG immunoprecipitates (IP) were prepared from HT1080 cells expressing inducible SFB-tagged wild-type TIN2 and TIN2-Δ90 and then eluted with 3×FLAG peptides for quantitative immunoblotting. The wild-type TIN2 immunoprecipitates were serially diluted to obtain a calibration curve to estimate the protein amount in each immunoprecipitate. WB, Western blot. B, Q-TRAP was carried out using the immunoprecipitates in A. The results were normalized to the protein amount. C, real-time qPCR was performed using the immunoprecipitates in A to determine TERC levels. The results were normalized to protein amount in the immunoprecipitates. D, anti-FLAG immunoprecipitates from HT1080 cells expressing inducible SFB-tagged wild-type or mutant TPP1 were eluted with 3×FLAG peptides for quantitative immunoblotting. The wild-type TPP1 immunoprecipitates were serially diluted to obtain a calibration curve to estimate the protein amount in each immunoprecipitate. E, Q-TRAP was carried out using the immunoprecipitates in D to determine telomerase activity. The results were normalized to the protein amount. F, real-time qPCR was performed using the immunoprecipitates in D to determine TERC levels. The results were normalized to 18 S RNA expression and protein amount in the immunoprecipitates. G, shown is a schematic model for TIN2-mediated telomerase recruitment and regulation. In B, C, E, and F, error bars represent S.E. from triplicates of three independent experiments. p values were calculated using Student's t test. *, p < 0.05.
To further investigate the contribution of TIN2 in telomerase recruitment, we quantified and compared the telomerase activity and the amount of TERC that associated with TPP1 and its deletion mutants. Consistent with previous studies, truncation of the TPP1 N-terminal OB fold (TPP1-ΔOB) that is important for telomerase recruitment had no effect on TIN2-TPP1 binding (Fig. 5D), and it drastically reduced TPP1-associated telomerase activity and TERC (∼20-fold) (Fig. 5, E and F). In comparison, deletion of the C-terminal TIN2-interacting domain of TPP1 (TPP1-ΔC22) abolished its ability to interact with endogenous TIN2 (Fig. 5D). Notably, this mutant (TPP1-ΔC22) was also compromised in its ability to bring down telomerase activity and TERC (2-fold) (Fig. 5, E and F). This level of change is comparable with that observed in TIN2 DC mutants (Fig. 4, C and D). Collectively, these observations suggest that TIN2 may facilitate telomerase recruitment or modulate telomerase activity through its interaction with TPP1, and this function may be impaired in TIN2 DC mutants, leading to defective telomerase regulation and telomere length maintenance.
DISCUSSION
In this study, we investigated the mechanism by which DC missense mutants of TIN2 could trigger telomere shortening. We were able to recapitulate the telomere shortening phenotype found in DC patients by expressing TIN2 DC mutants in telomerase-positive human cells, thus providing support to the hypothesis that the mutations identified in TIN2 were indeed responsible for DC. We demonstrated here that TIN2 DC mutants did not affect total TERC levels, telomerase activity, or cell proliferation. Furthermore, despite the clustering of DC mutations in a region adjacent to the TRF1-binding motif in TIN2, no apparent effect was observed for TIN2 interaction with either TRF1 or other core telomeric proteins. These findings were further corroborated by the observation that TIN2 DC mutant-expressing cells did not exhibit telomere capping defects. Our work therefore underscores the mechanistic differences in telomere length dysregulation between classic DC and DC induced by TIN2 dysfunction.
We found that TIN2 could recruit the telomerase and that DC missense mutations significantly reduced its ability to associate with TERC and telomerase activity. Because TIN2 is crucial for anchoring the TPP1-POT1-telomerase complex to the telomeres (28, 54), DC mutations in TIN2 likely result in decreased telomerase activity that is recruited to the telomeres, which in turn leads to telomere shortening. Defective telomeric targeting of telomerase in patients with TIN2 missense mutations may in fact account for DC pathogenesis. This represents a new mechanism by which telomere dysfunction can trigger telomere attrition and possibly DC. Our findings also raise the possibility that mutations affecting telomerase trafficking may be identified in patients with DC as well. During the preparation of this manuscript, mutations of TCAB1 in DC patients were identified and found to cause telomerase mislocation (57).
How does TIN2 regulate the telomerase? Our data indicate that TIN2 recruits the telomerase in a TPP1-dependent manner. It has been shown previously that the OB domain of TPP1 is critical for telomerase recruitment and regulation. Here, we demonstrated that the TIN2-interacting domain on TPP1 also plays a role, and this regulation may be secondary to the activities mediated by the OB fold (Fig. 5G). It is possible that TIN2 may allosterically control telomerase recruitment and activity once it binds to TPP1 through the TIN2-interacting domain. However, a weak association between TIN2 and the telomerase cannot be ruled out. Alternatively, TIN2 may specifically regulate the telomerase that has been recruited to the telomeres rather than total telomerase activity. Moreover, additional analysis of the association between endogenous TIN2 mutants and telomerase may need to be carried out in cells from DC patients. These are areas that merit further investigation. The telomerase regulatory function of the TPP1 TIN2-interacting domain provides new insight into the molecular mechanisms underlining DC, adds another level of cross-talk between TPP1 and TIN2, and highlights the complexity of telomerase regulation in humans.
Supplementary Material
Acknowledgments
We thank Dr. Dan Liu for technical help and Dr. Liuh-Yow Chen and Yi Zhang for help with constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA133249 from NCI and Grant GM081627 from NIGMS. This work was also supported by Welch Foundation Grant Q-1673 and National Basic Research Program Grant 2010CB945400.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and additional references.
- DC
- dyskeratosis congenita
- TRAP
- telomeric repeat amplification protocol
- Q-TRAP
- real-time quantitative PCR-based TRAP
- qPCR
- quantitative PCR
- PD
- population doubling.
REFERENCES
- 1. Kirwan M., Dokal I. (2009) Biochim. Biophys. Acta 1792, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirwan M., Dokal I. (2008) Clin. Genet. 73, 103–112 [DOI] [PubMed] [Google Scholar]
- 3. Alter B. P., Baerlocher G. M., Savage S. A., Chanock S. J., Weksler B. B., Willner J. P., Peters J. A., Giri N., Lansdorp P. M. (2007) Blood 110, 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vulliamy T. J., Knight S. W., Mason P. J., Dokal I. (2001) Blood Cells Mol. Dis. 27, 353–357 [DOI] [PubMed] [Google Scholar]
- 5. Calado R. T., Young N. S. (2008) Blood 111, 4446–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walne A. J., Vulliamy T., Beswick R., Kirwan M., Dokal I. (2008) Blood 112, 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Savage S. A., Giri N., Baerlocher G. M., Orr N., Lansdorp P. M., Alter B. P. (2008) Am. J. Hum. Genet. 82, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Savage S. A., Alter B. P. (2008) Mech. Ageing Dev. 129, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Armanios M., Chen J. L., Chang Y. P., Brodsky R. A., Hawkins A., Griffin C. A., Eshleman J. R., Cohen A. R., Chakravarti A., Hamosh A., Greider C. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15960–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P. J., Dokal I. (2004) Nat. Genet. 36, 447–449 [DOI] [PubMed] [Google Scholar]
- 11. Vulliamy T., Marrone A., Goldman F., Dearlove A., Bessler M., Mason P. J., Dokal I. (2001) Nature 413, 432–435 [DOI] [PubMed] [Google Scholar]
- 12. Heiss N. S., Knight S. W., Vulliamy T. J., Klauck S. M., Wiemann S., Mason P. J., Poustka A., Dokal I. (1998) Nat. Genet. 19, 32–38 [DOI] [PubMed] [Google Scholar]
- 13. Vulliamy T., Beswick R., Kirwan M., Marrone A., Digweed M., Walne A., Dokal I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8073–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walne A. J., Vulliamy T., Marrone A., Beswick R., Kirwan M., Masunari Y., Al-Qurashi F. H., Aljurf M., Dokal I. (2007) Hum. Mol. Genet. 16, 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Autexier C., Lue N. F. (2006) Annu. Rev. Biochem. 75, 493–517 [DOI] [PubMed] [Google Scholar]
- 16. Härle-Bachor C., Boukamp P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6476–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hiyama K., Hirai Y., Kyoizumi S., Akiyama M., Hiyama E., Piatyszek M. A., Shay J. W., Ishioka S., Yamakido M. (1995) J. Immunol. 155, 3711–3715 [PubMed] [Google Scholar]
- 18. Marrone A., Stevens D., Vulliamy T., Dokal I., Mason P. J. (2004) Blood 104, 3936–3942 [DOI] [PubMed] [Google Scholar]
- 19. Cerone M. A., Ward R. J., Londoño-Vallejo J. A., Autexier C. (2005) Cell Cycle 4, 585–589 [PubMed] [Google Scholar]
- 20. Sasa G., Ribes-Zamora A., Nelson N., Bertuch A. (2011) Clin. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du H. Y., Mason P. J., Bessler M., Wilson D. B. (2009) Pediatr. Blood Cancer 52, 687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsangaris E., Adams S. L., Yoon G., Chitayat D., Lansdorp P., Dokal I., Dror Y. (2008) Hum. Genet. 124, 507–513 [DOI] [PubMed] [Google Scholar]
- 23. Liu D., O'Connor M. S., Qin J., Songyang Z. (2004) J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- 24. de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 25. Palm W., de Lange T. (2008) Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 26. Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 27. Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007) Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 28. Abreu E., Aritonovska E., Reichenbach P., Cristofari G., Culp B., Terns R. M., Lingner J., Terns M. P. (2010) Mol. Cell. Biol. 30, 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tejera A. M., Stagno d'Alcontres M., Thanasoula M., Marion R. M., Martinez P., Liao C., Flores J. M., Tarsounas M., Blasco M. A. (2010) Dev. Cell 18, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim H., Lee O. H., Xin H., Chen L. Y., Qin J., Chae H. K., Lin S. Y., Safari A., Liu D., Songyang Z. (2009) Nat. Struct. Mol. Biol. 16, 372–379 [DOI] [PubMed] [Google Scholar]
- 31. O'Connor M. S., Safari A., Xin H., Liu D., Songyang Z. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11874–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004) Nat. Cell Biol. 6, 673–680 [DOI] [PubMed] [Google Scholar]
- 33. Herbert B. S., Hochreiter A. E., Wright W. E., Shay J. W. (2006) Nat. Protoc. 1, 1583–1590 [DOI] [PubMed] [Google Scholar]
- 34. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 35. Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., Harley C. B., Cech T. R. (1997) Science 277, 955–959 [DOI] [PubMed] [Google Scholar]
- 36. Ouellette M. M., Liao M., Herbert B. S., Johnson M., Holt S. E., Liss H. S., Shay J. W., Wright W. E. (2000) J. Biol. Chem. 275, 10072–10076 [DOI] [PubMed] [Google Scholar]
- 37. Ye J. Z., de Lange T. (2004) Nat. Genet. 36, 618–623 [DOI] [PubMed] [Google Scholar]
- 38. Kim S. H., Kaminker P., Campisi J. (1999) Nat. Genet. 23, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wong J. M., Collins K. (2006) Genes Dev. 20, 2848–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitchell J. R., Wood E., Collins K. (1999) Nature 402, 551–555 [DOI] [PubMed] [Google Scholar]
- 41. Wong J. M., Kyasa M. J., Hutchins L., Collins K. (2004) Hum. Genet. 115, 448–455 [DOI] [PubMed] [Google Scholar]
- 42. O'Connor M. S., Safari A., Liu D., Qin J., Songyang Z. (2004) J. Biol. Chem. 279, 28585–28591 [DOI] [PubMed] [Google Scholar]
- 43. Takai K. K., Hooper S., Blackwood S., Gandhi R., de Lange T. (2010) J. Biol. Chem. 285, 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Okamoto K., Iwano T., Tachibana M., Shinkai Y. (2008) J. Biol. Chem. 283, 23981–23988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004) Genes Dev. 18, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Steensel B., de Lange T. (1997) Nature 385, 740–743 [DOI] [PubMed] [Google Scholar]
- 47. Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M. R., Schnapp G., de Lange T. (2000) Mol. Cell. Biol. 20, 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Houghtaling B. R., Cuttonaro L., Chang W., Smith S. (2004) Curr. Biol. 14, 1621–1631 [DOI] [PubMed] [Google Scholar]
- 49. Ye J. Z., Donigian J. R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., de Lange T. (2004) J. Biol. Chem. 279, 47264–47271 [DOI] [PubMed] [Google Scholar]
- 50. Kim S. H., Beausejour C., Davalos A. R., Kaminker P., Heo S. J., Campisi J. (2004) J. Biol. Chem. 279, 43799–43804 [DOI] [PubMed] [Google Scholar]
- 51. Deng Z., Atanasiu C., Burg J. S., Broccoli D., Lieberman P. M. (2003) J. Virol. 77, 11992–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bradshaw P. S., Stavropoulos D. J., Meyn M. S. (2005) Nat. Genet. 37, 193–197 [DOI] [PubMed] [Google Scholar]
- 53. Kaminker P., Plachot C., Kim S. H., Chung P., Crippen D., Petersen O. W., Bissell M. J., Campisi J., Lelièvre S. A. (2005) J. Cell Sci. 118, 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen L. Y., Liu D., Songyang Z. (2007) Mol. Cell. Biol. 27, 5898–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim S. H., Davalos A. R., Heo S. J., Rodier F., Zou Y., Beausejour C., Kaminker P., Yannone S. M., Campisi J. (2008) J. Cell Biol. 181, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009) Science 323, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhong F., Savage S. A., Shkreli M., Giri N., Jessop L., Myers T., Chen R., Alter B. P., Artandi S. E. (2011) Genes Dev. 25, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.