Abstract
We previously demonstrated that the receptor for the complement component C1q (gC1qR) is a lipid raft protein that is indispensable for adipogenesis and insulin signaling. Here, we provide the first report that gC1qR is an essential component of lamellipodia in human lung carcinoma A549 cells. Cell-surface gC1qR was concentrated in the lamellipodia along with CD44, monosialoganglioside, actin, and phosphorylated focal adhesion kinase in cells stimulated with insulin, IGF-1, EGF, or serum. The growth factor-induced lamellipodia formation and cell migration were significantly decreased in gC1qR-depleted cells, with a concomitant blunt activation of the focal adhesion kinase and the respective receptor tyrosine kinases. Moreover, the gC1qR-depleted cells exhibited a reduced proliferation rate in culture as well as diminished tumorigenic and metastatic activities in grafted mice. We therefore conclude that cell-surface gC1qR regulates lamellipodia formation and metastasis via receptor tyrosine kinase activation.
Keywords: Cell Migration, Lipid Raft, Receptor Tyrosine Kinase, Signal Transduction, Tumor Metastases, CD44, gC1qR
Introduction
Cell migration is essential for various functions such as tissue and organ development, wound healing, inflammation, blood vessel formation, and cancer cell metastasis. Directed cell migration is initiated by the formation of veil-like lamellipodia, which are F-actin projections on the leading edges of moving cells that provide temporary focal adhesion sites for cells to move themselves toward a chemical signal. Lamellipodia formation is regulated by different molecules, including chemokine receptors, CD44, ezrin/radixin/moesin, and Rho family GTPases (1–3).
Lipid rafts are cholesterol- and glycosphingolipid-rich plasma membrane domains that are involved in various cellular events, such as growth factor signaling, cell migration, and cancer metastasis (4–6). Lipid rafts play important roles in multiple stages of the migration process, such as cellular adhesion, polarization, and lamellipodia formation (7–9). For example, various lamellipodia components, including chemokine receptors, NADPH oxidase, Rac, Cdc42, CD44, and ezrin/radixin/moesin, are regulated in lipid rafts (9–11). Lipid rafts, as detected by the localization of cholera toxin B (a GM1-binding protein) and laurdan (a lipid raft-specific two-photon dye), are condensed in the lamellipodia during cell migration (12, 13), indicating that lipid raft structure is critical for lamellipodia formation.
Using comparative two-dimensional electrophoresis of lipid rafts isolated from 3T3-L1 preadipocytes and adipocytes, we previously identified a lipid raft protein that has been termed gC1qR2 (receptor for complement component C1q), HABP-1 (hyaluronic acid binding protein-1), or p32 (14). In addition to its role as a receptor for C1q, gC1qR associates with various extracellular matrix components and viral proteins (15, 16). Several recent reports have implicated the surface expression of gC1qR in tumor progression; indeed, gC1qR is up-regulated in adenocarcinoma cells and various tumor tissues (17, 18). In addition, cell-surface gC1qR serves as a receptor for the tumor homing peptide Lyp-1, which suggests involvement in tumor malignancy (18–20). However, the precise mechanism of gC1qR action in tumorigenesis remains to be elucidated. Thus, we investigated the role of cell-surface gC1qR in A549 lung adenocarcinoma cells. The transient and stable knockdown of gC1qR revealed that gC1qR serves as a critical regulator for lamellipodia formation, cell adhesion, and cell migration through its association with the lipid rafts. We further confirmed that gC1qR positively regulates the ligand-dependent activation of receptor tyrosine kinases (RTKs). Finally, we demonstrated that cell-surface gC1qR was indispensable for tumorigenesis and metastasis in nude mice.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The antibodies used in this study include the following: anti-IGFR and anti-phosphotyrosine protein (Transduction Laboratories); anti-IR, anti-gC1qR, protein A- and protein G-conjugated agarose beads (Upstate Biotechnology); anti-Akt, anti-phospho-Akt (Ser-473), anti-FAK, anti-phospho-FAK (Tyr-397 or Tyr-925), and anti-integrin β1 (Cell Signaling Biotechnology); anti-CD44, anti-cytochrome c, anti-Erk, anti-phospho-Erk, anti-EGFR, anti-flotillin, anti-pan-Ras, and anti-β-actin (Santa Cruz Biotechnology); and Alexa 488-conjugated mouse and Alexa 555-conjugated rabbit secondary antibodies, Alexa 555-conjugated CTB, and Alexa 555-conjugated phalloidin (Invitrogen). Protease and phosphatase inhibitor mixtures were purchased from Roche Applied Science. Human recombinant IGF and EGF proteins were obtained from R&D Systems. Synthetic siRNA against gC1qR (si-gC1qR) and scrambled sequence RNA (si-con) were from Ambion; biotin, collagen, poly-d-lysine, gelatin, and insulin were acquired from Sigma.
Cell Culture and Electroporation
A549 lung adenocarcinoma cells were purchased from ATCC and cultured in RPMI 1640 medium supplemented with 1% penicillin/streptomycin and 10% fetal calf serum (Jeil Biosciences). The stable cell line sh-gC1qR was established from A549 cells transfected with plasmids psh-gC1qR or pSilencer control (Ambion) and cultured in media containing 1 μg/ml puromycin for 3 weeks. The plasmids psh-gC1qR were constructed to express shRNA with target sequences of 5′-GCATC CCACC AACAT TTGAT T and 5′-CCGAC GGAGA CAAAG CTTTC T, which were derived from complementary sequences located at exon 3 and the junction between exons 1 and 2 of gC1qR (GenBankTM accession number NM_001212), respectively. For siRNA-induced silencing, A549 cells (3 × 105 cells) were electroporated with 120 pmol of either si-gC1qR or si-con using the MicroPorator MP-100 system (Digital Bio Technology), according to the manufacturer's instructions.
Isolation of Detergent-resistant Lipid Rafts
Lipid rafts were isolated as described previously (21). Briefly, A549 cells, grown to 70–80% confluence in seven 150-mm dishes, were lysed with 1 ml of lysis buffer (1% Triton X-100, 25 mm HEPES, pH 6.5, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF and protease inhibitor mixture) and subjected to discontinuous sucrose gradient ultracentrifugation (40, 30, and 5%) using an SW41 Ti rotor (39,000 rpm) for 18 h at 4 °C. After centrifugation, the sucrose gradients were fractionated into 13 fractions, including the pellet fraction. An opaque buoyant band corresponding to the lipid rafts was collected at the interface between the 30 and 5% sucrose gradients.
Immunoblotting, Immunoprecipitation, Immunofluorescence, and Cell-surface Biotinylation
Cells were lysed with SDS lysis buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 1 mm PMSF, and protease mixture) for 30 min at 4 °C. After microcentrifugation (14,000 rpm) for 10 min at 4 °C, the whole cell lysates (supernatants) were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were blocked for 1 h at 37 °C with 5% w/v dry milk in TBS buffer. Subsequent incubations with primary and secondary antibodies were conducted for 1 h at room temperature. The signals were visualized with an automatic image analysis system (LAS 3000, Fuji Life Science). For immunoprecipitation, 500 μg of the whole cell lysate was incubated with 2 μg of the designated antibody for 18 h at 4 °C and then incubated with 30 μl of protein A-agarose (50%, w/v) for 1 h at 4 °C. The immunoprecipitates were analyzed by immunoblotting with an anti-tyrosine phosphoprotein antibody. For immunostaining, cells were fixed with 3.7% paraformaldehyde for 20 min, permeabilized with 1% Triton X-100 for 5 min or left un-permeabilized, and then blocked with blocking buffer (5% BSA in PBS) for 1 h. Subsequent incubation with primary and fluorescein-conjugated secondary antibodies was conducted for 1 h at room temperature. Immunofluorescence images were captured by confocal microscopy (Zeiss LSM 510 META). Cell-surface biotinylation was performed according to the method of Bae et al. (22). After surface biotin labeling, cells were lysed with the SDS lysis buffer described above.
Cell Attachment Assay
The wells of 98-well culture plates were left uncoated or coated with collagen (5 μg/ml), gelatin (1 μg/ml), or poly-d-lysine (50 μg/ml) for 60 min. The wells were subsequently blocked for 30 min at room temperature with 100 μl of heat-denatured BSA (10 mg/ml) to avoid nonspecific cell adhesion. Approximately 2.5 × 104 cells were plated in each well and incubated with an incubation buffer (HBS: 150 mm NaCl, 25 mm HEPES, pH 7.4, and 2 mm EDTA) at 37 °C for 30 min. Wells were then washed twice with HEPES-buffered saline (25 mm HEPES, pH 7.4, and 150 mm NaCl). Unbound and loosely bound cells were removed by shaking, and the remaining cells were then fixed with 3.7% formaldehyde. The wells were washed three times with 500 μl of distilled water, and the attached cells were stained with 100 μl of 0.1% (w/v) Crystal Violet in 200 mm MES, pH 6.0, for 1 h at room temperature. Excess dye was removed by washing three times with 500 μl of distilled water, and the bound dye was solubilized with 100 μl of 10% (v/v) acetic acid. The absorbance was measured at 570 nm.
Wound Healing and Transwell Migration Assay
For the wound healing assay, cells were seeded at a high density on 12-well culture plates. The next day, the cells were serum-starved for an additional 18 h. After scraping the cell monolayer with a sterile micropipette tip, the cells were treated with growth factors, including insulin (100 nm), IGF (20 ng/ml), EGF (50 ng/ml), and serum (10% v/v). After 30 h, images were captured to determine the migratory activity of the cells, and the healed area was measured. The transwell migration assay was conducted using the method described by the manufacturer (Costar) with slight modifications. A549 cells stably transfected with sh-con and sh-gC1qR were trypsinized and resuspended in RPMI 1640 media supplemented with 0.5% BSA and 0.1% serum. In total, 5 × 104 cells were plated in 0.3 ml of media in the upper chamber (8 μm pore size) of each well; the lower side of the chamber was coated with collagen. A total of 0.3 ml of serum-free RPMI 1640 media, containing the above-mentioned growth factors, was added to the lower chamber to induce migration. After incubation for 18 h at 37 °C, the cells that remained on the upper surface were removed with a cotton swab, and the cells that had migrated through the filter were stained with hematoxylin (Sigma). Images of the stained cells were captured, and the number of cells in three different fields was counted per filter for quantification.
MTT and Soft Agar Assays
For the MTT assay, sh-con and sh-gC1qR A549 cells were seeded at 1 × 104 cells/well on a 96-well plate, grown for 48 h, and subsequently serum-starved for 18 h. The cells were treated with FCS (10%), insulin (100 nm), IGF (20 ng/ml), or EGF (50 ng/ml). After treatment for 3 days, the cells were treated with 5 μg/ml MTT for 4 h at 37 °C. MTT-formazan crystals were dissolved in DMSO and determined by reading absorbance at 570 nm using a spectrophotometer. For the soft agar assay, 0.6% agarose (2 ml/well) was added to a 60-mm dish and left to solidify at room temperature. Approximately 5 × 104 A549 cells stably transfected with sh-con or sh-gC1qR were resuspended in 2 ml of top agar (0.4%) and plated on top of the soft agar. The plate was incubated at room temperature for an additional 15 min, and the cells were subsequently maintained in culture for 3 weeks. Images of the colonies were obtained under light microscopy.
Tumorigenesis and Metastasis in Nude Mice
Six-week-old female BALB/c athymic mice were purchased from Orient Co. and maintained at 22 ± 2 °C and 50 ± 10% humidity under a 12-h light/12-h dark regimen. The Institutional Animal Care and Use Committee of the Korea Institute of Radiological and Medical Science approved the studies, which were performed under the guidelines for the use and care of laboratory animals. For the tumorigenesis experiment, sh-con or sh-gC1qR A549 cells (4 × 106 cells in 0.1 ml of PBS) were subcutaneously injected into the right hind leg of the mice (n = 4). Tumor volume was measured every other day for 1–7 weeks post-injection using a digital caliper and was calculated according to the following equation: V = 0.5 × (width2 × length). For the metastasis experiment, sh-con or sh-gC1qR A549 cells (5 × 105 cells in 0.1 ml of PBS) were injected into the tail vein of mice (n = 5). Mice were sacrificed 28 days post-injection, and the number of liver metastatic foci was macroscopically counted.
RESULTS
Localization of gC1qR in Lamellipodia
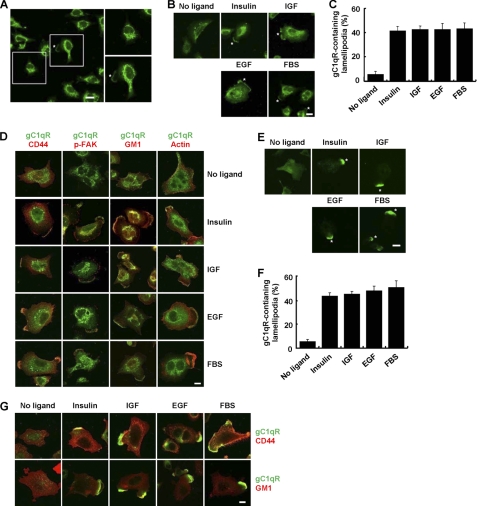
To evaluate the cellular function of cell-surface gC1qR, we first examined the localization of gC1qR in nonconfluent A549 human lung adenocarcinoma cells by immunofluorescence. Although gC1qR was mainly found in intracellular compartments, a small portion was detected in a patched area of the plasma membrane that might represent a lamellipodium (Fig. 1A). Because lamellipodia are formed after growth factor stimulation, we next investigated whether cellular exposure to growth factors could induce the translocation of gC1qR into lamellipodia. Nonconfluent A549 cells were serum-starved and treated with serum or growth factors such as insulin, insulin-like growth factor (IGF), and epidermal growth factor (EGF). After treatment, the cellular localization of gC1qR was monitored in permeabilized A549 cells by immunofluorescence. As shown in Fig. 1B, gC1qR was condensed in the protruding lamellipodia after the treatment with serum or growth factors. Expression of gC1qR in the lamellipodia was observed in less than 5% of serum-starved cells, whereas gC1qR expression was observed in the lamellipodia of more than 40% of the growth factor-stimulated A549 cells (Fig. 1C). Interestingly, gC1qR colocalized with raft/lamellipodial components, such as CD44, GM1, phospho-FAK (pFAK), and actin, upon cellular exposure to growth factors (Fig. 1D, and supplemental Fig. S1A). These results indicate that the translocation of gC1qR corresponded to the condensation of lipid rafts at the leading edges of cells upon growth factor stimulation.
FIGURE 1.
Cell-surface gC1qR is recruited to the lamellipodia of motile A549 cells. A, rapidly proliferating nonconfluent A549 cells were permeabilized with 0.1% Triton X-100 and analyzed by immunofluorescence with an anti-gC1qR antibody. Boxed regions are enlarged to show the differential distribution patterns of the gC1qR. An asterisk indicates the localization of gC1qR in lamellipodia. Scale bar, 20 μm. B and C, nonconfluent A549 cells were serum-starved for 18 h, followed by stimulation with insulin (100 nm), IGF (20 ng/ml), EGF (50 ng/ml), and serum (10%) for 20 min. The permeabilized cells were analyzed by immunofluorescence using an anti-gC1qR antibody (B). Asterisks indicate gC1qR-containing lamellipodia. Scale bar, 20 μm. The ratio of the number of cells with gC1qR-containing lamellipodia to the total number of cells is shown (C). D, colocalization of gC1qR with lamellipodia markers such as CD44, p-FAK (Tyr-397), GM1, and actin in lamellipodia. Nonconfluent A549 cells were serum-starved and stimulated with serum or various growth factors as indicated in B. The cellular localization of gC1qR, CD44, and p-FAK was determined after permeabilization by immunofluorescence using anti-gC1qR, anti-CD44, and anti-p-FAK antibodies. Rhodamine-conjugated CTB was used to detect GM1, whereas rhodamine-conjugated phalloidin was used to detect actin. Scale bar, 20 μm. The supplemental Fig. S1A shows the fluorescence imaging for each molecule. E and F, nonconfluent A549 cells were serum-starved and stimulated with serum or growth factors as indicated in B. The cellular localization of gC1qR was determined in nonpermeabilized cells by immunofluorescence with an anti-gC1qR antibody (E). Asterisks indicate gC1qR-containing lamellipodia. Scale bar, 20 μm. The ratio of the number of cells with gC1qR-containing lamellipodia to the total number of cells is shown (F). G, surface expression of gC1qR with CD44 in lamellipodia. Nonconfluent A549 cells were serum-starved and stimulated with serum or various growth factors as indicated in B. The surface expression of gC1qR and CD44 was determined in nonpermeabilized cells by immunofluorescence. Scale bar, 20 μm. The supplemental Fig. S1B shows the fluorescence imaging for each molecule.
Next, we monitored the surface expression of gC1qR in nonpermeabilized A549 cells by immunofluorescence. Cell-surface gC1qR appeared in lamellipodia after cellular stimulation with serum or growth factors (Fig. 1E). Fig. 1F shows that more than 40% of the cells expressed gC1qR in their lamellipodia after the treatment, whereas less than 5% of the serum-starved cells expressed gC1qR. The cell-surface gC1qR colocalized with the cell-surface CD44 and GM1 after cellular exposure to serum or growth factors (Fig. 1G and supplemental Fig. S1B).
Localization of gC1qR in Lipid Rafts
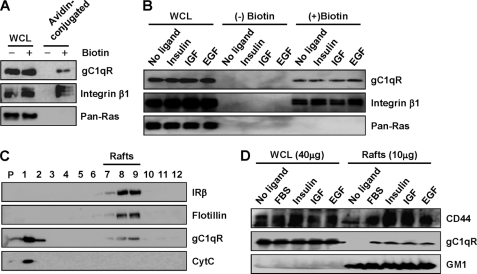
We further confirmed the surface expression of gC1qR in rapidly proliferating nonconfluent A549 cells by surface biotin labeling of gC1qR. After cells were labeled with membrane-impermeable biotin, the whole cell lysates were precipitated with avidin-conjugated agarose beads, and the precipitates were analyzed by immunoblotting. As shown in Fig. 2A, gC1qR and integrin β1 were surface biotin-labeled, whereas the intracellular protein Ras was not. Because surface gC1qR was concentrated in the lamellipodia after treatment with serum or growth factors (Fig. 1, E–G), it is tempting to speculate that intracellular gC1qR might be translocated to the cell surface upon exposure to serum or growth factors. To directly address this possibility, we monitored the change in the surface expression of gC1qR after serum or growth factor stimulation. As shown in Fig. 2B, the amount of surface biotin-labeled gC1qR and integrin β1 did not change even after treatment with serum or growth factors. This result indicates that the gC1qR condensed in the lamellipodia after serum or growth factor stimulation originated from the cell surface and not from intracellular compartments.
FIGURE 2.
Cell-surface gC1qR is recruited to detergent-resistant lipid rafts after growth factor stimulation. A, rapidly proliferating nonconfluent A549 cells were surface-biotinylated with membrane-impermeable biotin for 2 h. Surface-biotinylated proteins were analyzed by immunoblotting with anti-gC1qR, integrin β1, and Pan-Ras antibodies. B, nonconfluent cells were serum-starved, subsequently treated with serum or growth factors as indicated in Fig. 1B, and surface-biotinylated. The surface-biotinylated proteins were analyzed by immunoblotting. WCL, whole cell lysate. C, detergent-resistant lipid rafts were isolated from rapidly proliferating nonconfluent cells. After sucrose gradient ultracentrifugation, the fractionated samples (from bottom to top) were analyzed by immunoblotting with anti-gC1qR, anti-insulin receptor β subunit (IRβ), anti-flotillin, and anti-cytochrome c antibodies. P indicates pellet. D, nonconfluent cells were serum-starved for 18 h and stimulated with serum or growth factors as indicated in Fig. 1B. Detergent-resistant lipid rafts were isolated from the cells and analyzed by immunoblotting with anti-CD44 and anti-gC1qR antibodies. GM1 was analyzed by blotting with biotin-labeled CTB and consecutive avidin-conjugated HRP.
Because gC1qR colocalized with CD44 and GM1 (Fig. 1), which are concentrated in lipid rafts (11, 23), gC1qR might be translocated to the rafts during lamellipodia formation upon cellular exposure to serum or growth factors. Thus, we examined whether gC1qR is associated with detergent-resistant lipid rafts. Detergent-resistant lipid rafts were prepared from rapidly proliferating nonconfluent A549 cells, based on detergent resistance and low density on sucrose gradient ultracentrifugation, and analyzed by immunoblotting. The gC1qR protein was recovered with raft marker proteins, such as flotillin and IRβ in the raft fractions, whereas the nonraft protein cytochrome c was not recovered (Fig. 2C). Next, we monitored the amount of gC1qR in lipid rafts after treatment with serum or growth factors. The amount of gC1qR was dramatically increased along with CD44 in the raft fractions after treatment (Fig. 2D), indicating that treatment with serum or growth factors induces the condensation of gC1qR in lamellipodial lipid rafts.
Knockdown of gC1qR Prevents Ligand-induced Lamellipodia Formation
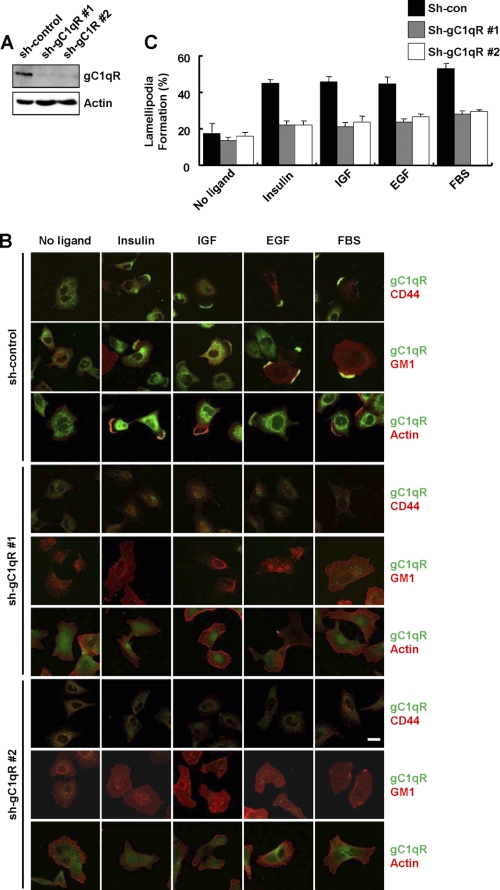
Because cell-surface gC1qR was condensed in the lamellipodia after treatment with serum or growth factors, gC1qR might regulate growth factor-induced lamellipodia formation. To evaluate this possibility, we monitored serum- or growth factor-induced lamellipodia formation after stable knockdown of gC1qR. The knockdown of gC1qR was achieved in A549 cells by small hairpin RNA (shRNA). As shown in Fig. 3A, the expression of gC1qR was significantly reduced in two stable cell clones transfected with sh-gC1qR. Lamellipodia formation was monitored by the cellular localization of CD44, actin, and GM1 in A549 cells that were serum-starved and then stimulated by serum or growth factors. After cellular exposure to serum or growth factors, the patched aggregation of CD44, actin, and GM1 in lamellipodia appeared in the control cells but not in gC1qR-depleted cells (Fig. 3B). Indeed, lamellipodia appeared in more than 44% of the control cells (sh-con) but in less than 24% of the gC1qR knockdown cells (sh-gC1qR) after ligand activation (Fig. 3C). These data indicate that gC1qR is essential for ligand-induced lamellipodia formation.
FIGURE 3.
Knockdown of gC1qR prevents growth factor-induced lamellipodia formation. A, expression of gC1qR was determined by immunoblot analysis of whole cell lysates obtained from a A549 clone expressing a scrambled control shRNA (sh-con), and two A549 clones stably expressing shRNA for gC1qR (sh-gC1qR #1 and sh-gC1qR #2). Actin was used as a loading control. B and C, after 18 h of serum starvation, sh-con and sh-gC1qR cells were stimulated with serum or growth factors as indicated in Fig. 1B. After permeabilization, the cellular colocalization of gC1qR with CD44, actin, and GM1 was determined by immunofluorescence using anti-gC1qR, anti-CD44, rhodamine-conjugated phalloidin, and rhodamine-conjugated CTB, respectively (B). Scale bar, 20 μm. The supplemental Fig. S2 shows the fluorescence imaging of each molecule. The ratio of the number of cells with GM1-containing lamellipodia to the total number of cells is given (C).
Knockdown of gC1qR Inhibits Cell Adhesion and Migration
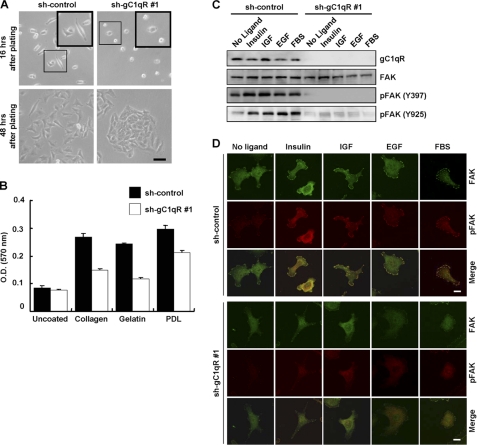
Given the accumulating evidence that gC1qR plays a role in C1q-mediated cell adhesion and spreading (24), we tested whether gC1qR participates in these processes. We first observed the cellular morphology of sh-con and sh-gC1qR A549 cells at 16 and 48 h after plating. Sixteen h after plating, the sh-gC1qR cells displayed significantly decreased adhesion when compared with the sh-con cells (upper panels in Fig. 4A). In addition, at 48 h after plating, the growth pattern of the sh-gC1qR cells was apparently different from that of the control cells. As shown in the lower panels of Fig. 4A, sh-gC1qR A549 cells had an aggregate growth pattern with tight cell-cell contacts, whereas sh-con cells had a normally dispersed growth pattern. The reduction of cell adhesion in the gC1qR-depleted cells was further quantified in cellular adhesion assays on culture plates coated with various extracellular matrix components such as collagen, gelatin, and poly-d-lysine. As shown in Fig. 4B, the sh-gC1qR cells apparently adhered less to wells coated with various extracellular matrix components than the sh-con cells.
FIGURE 4.
Knockdown of gC1qR disturbs cell adhesion. A, adhesion and growth pattern of A549 cells after gC1qR knockdown. After plating of sh-con and sh-gC1qR #1 A549 cells (3 × 105 cells, respectively) on a 6-well plate for 16 h (upper panels) and 48 h (lower panels), the cell adhesion and growth pattern was observed by microscopy. The boxed regions in the upper two panels are enlarged and presented in separate boxes. Scale bar, 100 μm. B, for the adhesion assay, sh-con and sh-gC1qR #1 A549 cells (3 × 105 cells) were seeded on 6-well plates that were left uncoated or coated with collagen (5 μg/ml), gelatin (1 μg/ml), or poly-d-lysine (PDL) (50 μg/ml). After 30 min, the plates were stained with crystal violet. Absorbance at 570 nm was determined using a spectrophotometer. C and D, after serum starvation for 18 h, sh-con and sh-gC1qR #1 A549 cells were stimulated with serum or growth factors as indicated in Fig. 1B. The expression of gC1qR, FAK, and p-FAK on Tyr-397 and Tyr-925 was determined from whole cell lysates by immunoblotting (C). The cellular localization of FAK and p-FAK (Tyr-397) was determined by immunofluorescence (D).
To dissect the underlying mechanism of gC1qR regulation in cell adhesion, we tested whether gC1qR knockdown could affect the activity of FAK, which is a critical regulator of cell adhesion. After treatment with serum or growth factors, phosphorylation of FAK at tyrosine 397 and 925 was significantly increased in sh-con cells but was unchanged in sh-gC1qR cells (Fig. 4C). In addition, immunofluorescence analysis with an anti-pFAK antibody showed that gC1qR knockdown abolished ligand-induced FAK activation (Fig. 4D).
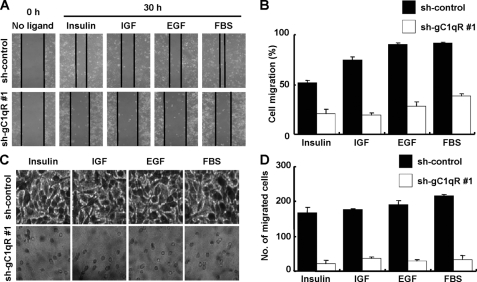
Based on the finding that gC1qR is critically involved in cell adhesion, we further investigated the effect of gC1qR knockdown on cell migration. Wound healing and transwell migration assays showed that ligand-induced cell migration was drastically affected by the stable depletion of gC1qR (Fig. 5, A–D). These data indicate that gC1qR mediates cell migration via lamellipodia formation.
FIGURE 5.
Knockdown of gC1qR disturbs cell migration. The wound healing assay (A and B) and invasion assay (C and D) for sh-con and sh-gC1qR #1 A549 cells were performed as described under “Experimental Procedures.” The microscopic image in each panel is representative of three independent experiments (A and C). The quantification of cellular migration in the wound healing and invasion assays is presented in B and D, respectively.
Knockdown of gC1qR Blunts Ligand-induced RTK Activation
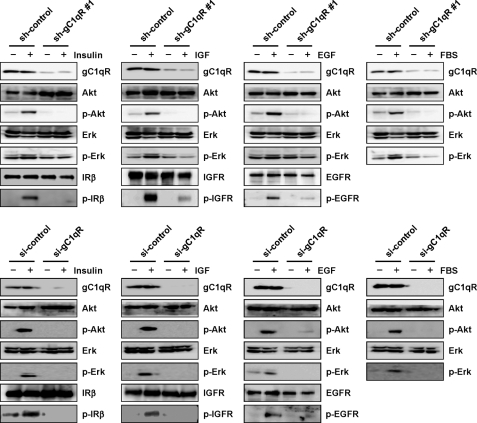
We previously demonstrated that gC1qR knockdown prevents the insulin-induced tyrosine phosphorylation of the IR in 3T3-L1 adipocytes (14). This finding led us to investigate whether gC1qR could also regulate growth factor-induced signal transduction because its knockdown severely decreased cell adhesion, migration, and lamellipodia formation in A549 cells upon growth factor stimulation (Figs. 3–5). Hence, we stimulated sh-con and sh-gC1qR cells with serum or growth factors after serum starvation. In contrast to sh-con cells, sh-gC1qR cells showed a lack of Akt and Erk phosphorylation after treatment (Fig. 6, upper panel). Moreover, ligand-induced tyrosine phosphorylation of IR, IGFR, and EGFR was abolished in sh-gC1qR cells. These results were reproduced in A549 cells with transient gC1qR knockdown (Fig. 6, lower panel). From these data, we can conclude that gC1qR plays an indispensable role in the activation of RTKs.
FIGURE 6.
Stable and transient knockdown of gC1qR decreases RTK activation. Control (sh-con and sh-con) and stably and transiently gC1qR-depleted (sh-gC1qR #1 and si-gC1qR) A549 cells were serum-starved for 18 h and then stimulated with insulin (100 nm), IGF (20 ng/ml), EGF (50 ng/ml), or serum (10%) for 15 min. The expression of gC1qR, Akt, phospho-Akt (p-Akt on Ser-473), Erk, and phospho-Erk (p-Erk) was determined by immunoblotting. Tyrosine phosphorylation of IRβ, IGFR, and EGFR was determined by immunoprecipitation following immunoblotting with an anti-phosphotyrosine antibody.
Knockdown of gC1qR Inhibits Tumor Formation and Metastatic Activities
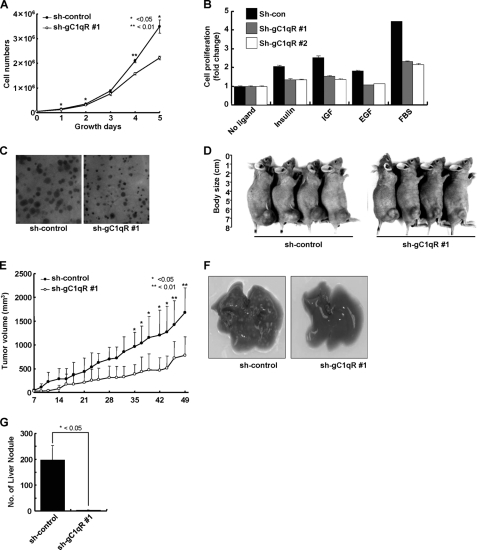
We measured the cell growth rate by counting the number of sh-con and sh-gC1qR A549 cells during culture in culture media containing 10% fetal bovine serum. As shown in Fig. 7A, sh-gC1qR cells exhibited a reduced growth rate when compared with sh-con cells; ∼40% fewer sh-gC1qR cells were observed after 5 days. The MTT assay also showed that the gC1qR knockdown decreased serum- or ligand-dependent cellular proliferation (Fig. 7B). Moreover, in the foci formation assay, sh-gC1qR cells had smaller foci than sh-con cells (Fig. 7C).
FIGURE 7.
Knockdown of gC1qR abolished tumorigenesis and metastasis of A549 cells in nude mice. A, proliferation of sh-con and sh-gC1qR #1 A549 cells. Both cell lines were seeded at 5 × 104 cells per 60-mm dish. The total cell number was counted on the indicated days. B, growth factor-dependent proliferation of sh-con and sh-gC1qR A549 cells. These cell lines were seeded at 5 × 103 cells per 96-well plate and cultured for 48 h. The cells were serum-starved for 18 h and then treated with serum or growth factors. After 72 h, the MTT assay was performed as described under “Experimental Procedures.” Cell proliferation was expressed as a fold change relative to nontreated cells. C, anchorage-independent growth of sh-con and sh-gC1qR #1 A549 cells on soft agar. Photomicrographs of the colonies were obtained 3 weeks after plating (×40). D and E, photographs (D) and measurement of tumor volume (E). sh-con or sh-gC1qR #1 A549 cells (4 × 106 cells) were subcutaneously injected into the right hind legs of BALB/c athymic mice (n = 4). Photographs were taken on day 49, and the tumor volume was measured every other day until day 49. (*, p < 0.05, n = 4 per group.) Filled circle, sh-con; open circle, sh-gC1qR #1. F and G, macroscopic observation (F) and quantification (G) of liver metastasis. sh-con or sh-gC1qR A549 cells (5 × 105 cells) were injected into the tail veins of BALB/c athymic mice (n = 5), and liver nodules were counted 28 days post-injection. (*, p < 0.05, n = 5 per group.)
To examine the tumorigenic abilities of sh-con and sh-gC1qR cells, both cell lines were subcutaneously injected into BALB/c athymic mice, and tumor formation was monitored for 49 days. Differences in tumor volume were detected as early as 2 weeks after implantation. By 49 days, the size of sh-gC1qR cell-derived tumors was about ⅓ the size of the tumors derived from control cells (Fig. 7, D and E). To assess the metastatic potential of sh-gC1qR cells, the liver was examined 28 days after the intravenous injection of the cell lines into athymic mice. Although many nodules were observed in the livers of control animals, almost none were detected in of the livers of sh-gC1qR cell-injected animals (Fig. 7, F and G). These results demonstrate that gC1qR knockdown inhibits the tumor growth and metastatic potential of A549 cells in vivo.
DISCUSSION
It has been reported that gC1qR plays multiple roles in different subcellular locations. Accumulating evidence supports the expression of surface gC1qR in different mammalian cells. Cell-surface gC1qR is a receptor for diverse extracellular matrix proteins (hyaluronic acid, fibronectin, and vitronectin) (25–27), bacterial and viral pathogens (Plasmodium falciparum, Listeria monocytogenes, Staphylococcus aureus, human immunodeficiency virus, and Hantan virus) (28–32), and other ligands (complement component C1q, and tumor homing peptide Lyp-1) (18, 25). Consistent with these findings, gC1qR was dynamically condensed into lamellipodia along with lamellipodia components, such as GM1, CD44, pFAK, and F-actin, upon exposure to different growth factors in A549 cells (Fig. 1). Subsequent surface biotinylation and raft fractionation experiments confirm these results (Fig. 2). The prevention of lamellipodia formation and cell migration in gC1qR-depleted cells (Fig. 3) suggests a role for gC1qR on the cell surface.
It is known that many lamellipodial components such as CCR5, CD44, FAK, and cdc42 are concentrated in lipid rafts (11, 13, 33, 34). Disruption of rafts by methyl-β-cyclodextrin, a cholesterol-depleting agent, abolishes lamellipodia formation and impedes the migration of U-251 MG human glioblastoma cells and MCF7 breast adenocarcinoma cells (13, 35), substantiating the role of lipid rafts in the assembly of migration machinery. We previously demonstrated that gC1qR is a lipid raft protein in 3T3-L1 adipocytes; the protein disappeared from raft fractions upon methyl-β-cyclodextrin treatment (14). In A549 cells, the association of gC1qR with lipid rafts was dramatically increased in response to growth factor stimulation (Fig. 2). We also monitored raft dynamics in gC1qR-depleted cells expressing palmitoyl-GFP, a genuine raft marker. Growth factor-induced raft condensation at the leading edge is disrupted in these cells (data not shown). These results indicate that the regulatory role of gC1qR in lamellipodia formation could involve the rafts.
The molecular mechanism underlying the gC1qR regulation of lamellipodia formation and migration was identified by the disruption of RTK signaling in gC1qR-depleted cells (Fig. 6). Similar to our previous finding that gC1qR knockdown in adipocytes abrogates the proximal signaling of insulin (14), gC1qR knockdown also significantly prevented the activation of insulin receptor, IGFR, EGFR, and their downstream effectors in A549 cells. Because the reciprocal signaling cross-talk between the migration machinery and RTKs is critical for cellular migration, our results clearly indicate that gC1qR could regulate the migration machinery by coupling it with the signal transduction of various RTKs.
Recently, it has been shown that CD44, a major hyaluronan receptor, regulates various RTKs, including EGFR, FGF receptor, and c-Met, subsequent to ligand-induced cellular migration (36, 37). Interestingly, gC1qR and CD44 have been associated with several mutual cellular events, such as hyaluronic acid binding and cellular entry of IlnB in L. monocytogenes (29, 38), implying that there might be an interaction between these two molecules. It has also been suggested that CD44 could interact with gC1qR in U937 cells (39). In our results, both molecules were concentrated in the lamellipodia after ligand activation (Fig. 1). We also observed a molecular interaction between gC1qR and CD44 by immunoprecipitation in MCF-7 breast cancer cells (data not shown). Based on these findings, it could be hypothesized that the molecular association of gC1qR with CD44 is also required for ligand-induced RTK activation and cross-talk with the migration machinery. However, the precise mechanism underlying the role of a potential gC1qR-CD44 complex in RTK activation should be studied further.
Supplementary Material
This work was supported by the Basic Research Laboratory Program of the Korea Research Foundation Grant 0001197 (to Y.-G. K.), National R&D Program for Cancer Control, Ministry of Health, Welfare and Family Affairs Grant 09201800, and Nuclear Research and Development Program of Korea Research Foundation Grant 20100025943 (to J.-S. L.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- gC1qR
- receptor for complement component C1q
- GM1
- monosialoganglioside
- CTB
- cholera toxin subunit B
- RTK
- receptor tyrosine kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- EGFR
- EGF receptor
- IR
- insulin receptor
- IGFR
- IGF receptor.
REFERENCES
- 1. Baumgartner M., Sillman A. L., Blackwood E. M., Srivastava J., Madson N., Schilling J. W., Wright J. H., Barber D. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13391–13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oliferenko S., Kaverina I., Small J. V., Huber L. A. (2000) J. Cell Biol. 148, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Haastert P. J., Devreotes P. N. (2004) Nat. Rev. Mol. Cell Biol. 5, 626–634 [DOI] [PubMed] [Google Scholar]
- 4. Kim B. W., Lee C. S., Yi J. S., Lee J. H., Lee J. W., Choo H. J., Jung S. Y., Kim M. S., Lee S. W., Lee M. S., Yoon G., Ko Y. G. (2010) Expert Rev. Proteomics 7, 849–866 [DOI] [PubMed] [Google Scholar]
- 5. Lingwood D., Simons K. (2010) Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 6. Simons K., Gerl M. J. (2010) Nat. Rev. Mol. Cell Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- 7. Giepmans B. N., van Ijzendoorn S. C. (2009) Biochim. Biophys. Acta 1788, 820–831 [DOI] [PubMed] [Google Scholar]
- 8. Saito T., Yokosuka T. (2006) Curr. Opin. Immunol. 18, 305–313 [DOI] [PubMed] [Google Scholar]
- 9. Ushio-Fukai M. (2006) Sci. STKE 2006, re8. [DOI] [PubMed] [Google Scholar]
- 10. Itoh K., Sakakibara M., Yamasaki S., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T. (2002) J. Immunol. 168, 541–544 [DOI] [PubMed] [Google Scholar]
- 11. Oliferenko S., Paiha K., Harder T., Gerke V., Schwärzler C., Schwarz H., Beug H., Günthert U., Huber L. A. (1999) J. Cell Biol. 146, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kindzelskii A. L., Sitrin R. G., Petty H. R. (2004) J. Immunol. 172, 4681–4685 [DOI] [PubMed] [Google Scholar]
- 13. Mañes S., Mira E., Gómez-Moutón C., Lacalle R. A., Keller P., Labrador J. P., Martínez-A C. (1999) EMBO J. 18, 6211–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim K. B., Kim B. W., Choo H. J., Kwon Y. C., Ahn B. Y., Choi J. S., Lee J. S., Ko Y. G. (2009) Proteomics 9, 2373–2382 [DOI] [PubMed] [Google Scholar]
- 15. Chowdhury A. R., Kamal A., Ghosh I., Datta K. (2009) in Hyaluronan Binding Protein 1 (HABP1/p32/gC1qR): A New Perspective in Tumor Development (Stern R. ed) pp. 51–68, Academic Press, Oxford [Google Scholar]
- 16. Ghebrehiwet B., Lim B. L., Kumar R., Feng X., Peerschke E. I. (2001) Immunol. Rev. 180, 65–77 [DOI] [PubMed] [Google Scholar]
- 17. Chen Y. B., Jiang C. T., Zhang G. Q., Wang J. S., Pang D. (2009) J. Surg. Oncol. 100, 382–386 [DOI] [PubMed] [Google Scholar]
- 18. Fogal V., Zhang L., Krajewski S., Ruoslahti E. (2008) Cancer Res. 68, 7210–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park J. H., von Maltzahn G., Xu M. J., Fogal V., Kotamraju V. R., Ruoslahti E., Bhatia S. N., Sailor M. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sánchez-Martín D., Cuesta A. M., Fogal V., Ruoslahti E., Alvarez-Vallina L. (2011) J. Biol. Chem. 286, 5197–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim K. B., Lee J. W., Lee C. S., Kim B. W., Choo H. J., Jung S. Y., Chi S. G., Yoon Y. S., Yoon G., Ko Y. G. (2006) Proteomics 6, 2444–2453 [DOI] [PubMed] [Google Scholar]
- 22. Bae T. J., Kim M. S., Kim J. W., Kim B. W., Choo H. J., Lee J. W., Kim K. B., Lee C. S., Kim J. H., Chang S. Y., Kang C. Y., Lee S. W., Lee S. W., Ko Y. G. (2004) Proteomics 4, 3536–3548 [DOI] [PubMed] [Google Scholar]
- 23. Kim S. I., Yi J. S., Ko Y. G. (2006) J. Cell. Biochem. 99, 878–889 [DOI] [PubMed] [Google Scholar]
- 24. Feng X., Tonnesen M. G., Peerschke E. I., Ghebrehiwet B. (2002) J. Immunol. 168, 2441–2448 [DOI] [PubMed] [Google Scholar]
- 25. Ghebrehiwet B., Lim B. L., Peerschke E. I., Willis A. C., Reid K. B. (1994) J. Exp. Med. 179, 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim B. L., Reid K. B., Ghebrehiwet B., Peerschke E. I., Leigh L. A., Preissner K. T. (1996) J. Biol. Chem. 271, 26739–26744 [DOI] [PubMed] [Google Scholar]
- 27. Gupta S., Batchu R. B., Datta K. (1991) Eur. J. Cell Biol. 56, 58–67 [PubMed] [Google Scholar]
- 28. Biswas A. K., Hafiz A., Banerjee B., Kim K. S., Datta K., Chitnis C. E. (2007) PLoS Pathog. 3, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun L., Ghebrehiwet B., Cossart P. (2000) EMBO J. 19, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nguyen T., Ghebrehiwet B., Peerschke E. I. (2000) Infect. Immun. 68, 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fausther-Bovendo H., Vieillard V., Sagan S., Bismuth G., Debré P. (2010) PLoS Pathog. 6, e1000975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi Y., Kwon Y. C., Kim S. I., Park J. M., Lee K. H., Ahn B. Y. (2008) Virology 381, 178–183 [DOI] [PubMed] [Google Scholar]
- 33. Baillat G., Siret C., Delamarre E., Luis J. (2008) Biochim. Biophys. Acta 1783, 2323–2331 [DOI] [PubMed] [Google Scholar]
- 34. Gingras D., Gauthier F., Lamy S., Desrosiers R. R., Béliveau R. (1998) Biochem. Biophys. Res. Commun. 247, 888–893 [DOI] [PubMed] [Google Scholar]
- 35. Murai T., Maruyama Y., Mio K., Nishiyama H., Suga M., Sato C. (2011) J. Biol. Chem. 286, 1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ponta H., Sherman L., Herrlich P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 37. Orian-Rousseau V. (2010) Eur. J. Cancer 46, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 38. Jung C., Matzke A., Niemann H. H., Schwerk C., Tenenbaum T., Orian-Rousseau V. (2009) Mol. Microbiol. 72, 1196–1207 [DOI] [PubMed] [Google Scholar]
- 39. Menzies C. A., David-Habiel, Glassberg P. F., Phuong-Nguyen, Jolyon-Jesty, Peerschke E. I. B., Berhane-Ghebrehiwet (2008) FASEB J. 22, 673 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.